Identification and functional analysis of genetic variants of ISL1 gene promoter in human atrial septal defects
Funding information: This work was supported by the National Natural Science Foundation of China (81870288 and 82170353); the Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2020-PT310-007); Tianjin Municipal Heath Commission and Tianjin Binhai New Area Health Commission (KJ20071 and 2019BWKY010); Tianjin Science and Technology Commission (18PTZWHZ00060); TEDA International Cardiovascular Hospital Internal Grant (2021-ZX-002); Academic Support Project for Top-notch Talents in Disciplines (majors) of Universities in Anhui Province (gxbjZD2022043); and The Major Project of Natural Science Foundation of the Department of Education of Anhui Province (KJ2019ZD32).
Funding information: National Natural Science Foundation of China, Grant/Award Numbers: 81870288, 82170353; Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences, Grant/Award Number: 2020-PT310-007; Tianjin Municipal Heath Commission and Tianjin Binhai New Area Health Commission, Grant/Award Numbers: KJ20071, 2019BWKY010; Tianjin Science and Technology Commission, Grant/Award Number: 18PTZWHZ00060; TEDA International Cardiovascular Hospital Internal Grant, Grant/Award Number: 2021-ZX-002; Academic Support Project for Top-notch Talents in Disciplines (majors) of Universities in Anhui Province, Grant/Award Number: gxbjZD2022043; The Major Project of Natural Science Foundation of the Department of Education of Anhui Province, Grant/Award Number: KJ2019ZD32
Abstract
Background
Atrial septal defect (ASD) is a common type of congenital heart disease. A gene promoter plays pivotal role in the disease development. This study was designed to investigate the pathological role of variants of the ISL1 gene promoter region in ASD patients.
Methods
Total DNA extracted from 625 subjects, including 332 ASD patients and 293 healthy controls, was sequenced to identify variants in the promoter region of ISL1 gene. Further functional analyses of the variants were performed with dual luciferase reporter assay and electrophoretic mobility shift assay (EMSA). All possible binding sites of transcription factor affected by the identified variants were predicted using the JASPAR database.
Results
Four variants in the ISL1 gene promoter were found only in patients with ASD by sequencing. Three of the four variants [g.4923 G > C (rs541081886), g.5079 A > G (rs1371835943) and g.5309 G > A (rs116222082)] significantly decreased the transcriptional activities compared with the wild-type ISL1 gene promoter (p < 0.05). The EMSA revealed that these variants [g.4923 G > C (rs541081886), g.5079 A > G (rs1371835943) and g.5309 G > A (rs116222082)] in the ISL1 gene promoter affected the number and affinity of binding sites of transcription factors. Further analysis with the online JASPAR database demonstrated that a cluster of putative binding sites for transcription factors may be altered by these variants.
Conclusions
These sequence variants identified from the promoter region of ISL1 gene in ASD patients are probably involved in the development of ASD by affecting the transcriptional activity and altering ISL1 levels. Therefore, these findings may provide new insights into the molecular etiology and potential therapeutic strategy of ASD.
Abbreviations
-
- ASD
-
- atrial septal defect
-
- CHD
-
- congenital heart defect
-
- EMSA
-
- electrophoretic mobility shift assay
-
- PCR
-
- polymerase chain reaction
-
- TFBS
-
- transcription factor binding sites
1 INTRODUCTION
Congenital heart disease (CHD) has been recognized as the most common type of birth defect, with a total birth prevalence of six to 13 per 1,000 newborn babies,1 which is considered to be a substantial cause of early fetal morbidity and mortality.2 With the tremendous advances in antenatal diagnosis, medical care and surgical treatment in childhood, mortality in infants and children has notably declined, causing a rapid increase in the number of adults with CHD.3-5 Life-long CHD and multisystem health issues may impose enormous physical, emotional and social economic burdens on patients and families, affecting their quality of life.2 Therefore, it is imperative to understand the etiology and mechanisms of CHD for precision medicine and genetic counseling.
Although genomic techniques have dramatically changed our understanding of the causes of CHD, the mechanisms underlying the development of CHD are complex and remain incompletely understood.6, 7 Accumulating evidence has indicated that genetic defects play a pivotal role in the pathogenesis of CHD,8-10 and has identified numerous genes associated with CHD. The majority of these genes code for transcription factors that modulate specific events in cardiac development.8, 11 Atrial septal defect (ASD) is one of the most common types of CHD12, accounting for 10% of CHD.13 ASD might not be diagnosed until it appears in adulthood and therefore the incidence of ASD is usually higher than estimated.14 Although studies have found that cardiac core transcription factor genes, encompassing NKX2-5, GATA4 and TBX5, are involved in ASD,15 to date the genetic causes of ASD remain largely unknown.
As a key LIM homeobox transcription factor localized on chromosome 5q11.1, ISL1 (OMIM: 600366) plays a crucial role in marking cardiac progenitor cells and cardiac differentiation, especially in generating diverse multipotent cardiovascular cell lineages.16, 17 Animal experiments have shown that ISL1-deficiency mice developed severe cardiac deformities, including loss of structures derived from the second heart field such as the right ventricle, outflow tract and large portions of the atria.18 Importantly, there is growing evidence that mutation and deletion in the ISL1 gene can cause diverse types of CHD, including ventricular septal defect, congenital double outlet right ventricle and d-transposition of the great arteries.17, 19-21 The promoter region of a gene may interrupt transcriptional regulation, which alters the gene expression, leading to disease.22, 23 In our previous study, novel variants in the ISL1 gene promoter region have been identified in ventricular septal defect.19 However, the role of genetic variants within the ISL1 gene promoter in the development of ASD has not been reported. Thus, we hypothesized that variants in the ISL1 gene promoter region may alter the expression of ISL1 gene, leading to the formation of ASD. To test this hypothesis, we studied the variants of the promoter region of ISL1 gene in ASD patients compared with healthy controls. Further, functional studies at the cellular level were performed to exam the possible functional role of the variants.
2 MATERIALS AND METHODS
2.1 Study subjects
This study enrolled 625 subjects. Up to March, 2021, 332 unrelated patients with clinically confirmed ASD at the Department of Cardiovascular Surgery, TEDA International Cardiovascular Hospital, Tianjin University (Tianjin, China) were recruited in this study. A total of 293 healthy control subjects from routine check-up or CHD screening program were also enrolled (Figure 1). All healthy subjects were confirmed to have no diseases by clinical screening including echocardiography. The study was conducted according to the principles of the Declaration of Helsinki. Written informed consents were signed by the legal guardians of participants prior to the study. This research protocol was approved by the Ethics Committee of TEDA international Cardiovascular Hospital.
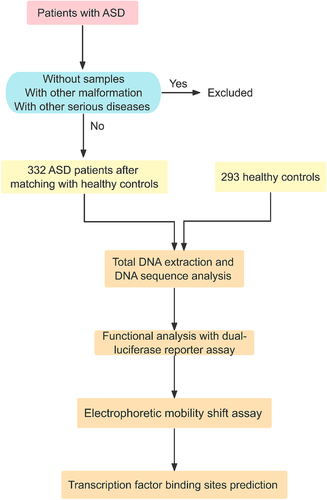
2.2 DNA sequencing analysis in the promoter region of the ISL1 gene
Peripheral blood leukocytes were isolated. Genomic DNA was prepared with the TIANamp Blood DNA kit (TIANGEN, Beijing, China) according to the manufacturer’s instructions. The polymerase chain reaction (PCR) primers were designed from the genomic sequence of the human ISL1 gene (NCBI: NG_023040.1) as previously reported.19 The ISL1 gene promoter was generated by PCR and bidirectionally sequenced (Table 1). The sequences were aligned with the wild-type sequence of the ISL1 gene promoter to compare and identify variants.
| Primers name | DNA sequences 5′–3′ | Location | Position |
|---|---|---|---|
| PCR and sequencing primers | |||
| ISL1-F1 | 5′-CTGTCTTTGGGAGACCGTAACA-3′ | 4,039 | −1,206 |
| ISL1-R1 | 5′-TGCCAATGCTGAAAGAGCCG-3′ | 5,436 | +111 |
| The double-stranded biotinylated oligonucleotides for the EMSA | |||
| Sequence variants | Oligonucleotides sequences | ||
| g.4923G > C | 5′-TGCGGGGACCCCAG(G/C)AGCGCAGGGCGGAG-3′ | ||
| g.5079A > G | 5′-GGGAGAACGGCCTG(A/G)GCCCCGAGCAAGTTG-3′ | ||
| g.5247A > G | 5′-CGCGCGCTGCGTC(A/G)GACCAATGGCGATGG-3′ | ||
| g.5309G > A | 5′-AGAGATAAGGAAGAGAG(G/A)TGCCCGAGCCGCGC-3′ | ||
- Notes: The PCR primers are designed based on the genomic DNA sequence of the ISL1 gene (NG_023040.1). The transcription start site is at the position of 5,325 (+1).
- Abbreviations: F, forward; R, reverse; PCR, polymerase chain reaction; EMSA, electrophoretic mobility shift assay.
2.3 Construction of expression plasmids and cellular transfection
To functionally analyze the activity of ISL1 promoter affected by variants, DNA fragments of the ISL1 gene promoter region with or without variants were generated by PCR and subcloned into the KpnI and BglII sites of pGL3-basic, a reporter vector expressed by the firefly luciferase (Figure 2). These insert fragments of expression vectors were then confirmed by direct Sanger sequencing.
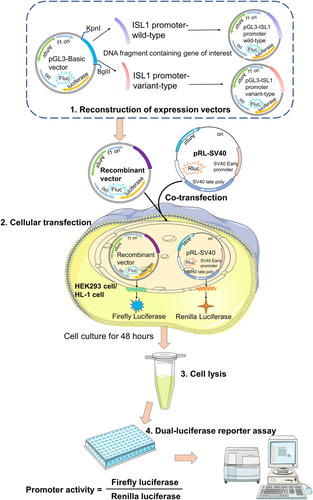
The HEK293 cells (Cell Resource Center, IBMS, CAMS/PUMC) and HL-1 cells were regularly cultivated in minium essential medium (Gibco)/Dullbecco's modified Eagle's medium (Gibco), containing 10% fetal bovine serum (Thermofisher) and 1% penicillin/streptomycin in an incubator with 5% CO2 at 37°C. Cells were seeded into six-well plates at a density of 6 × 105 cells per well and grown to 70–80% confluence before transfection. Designated expression plasmids (2.5 ng) together with pRL-SV40 (0.35 ng) were co-transfected into the HEK293 and HL-1 cells, respectively. pRL-SV40, a renilla luciferase reporter plasmid, was used as the internal control. The empty pGL3-basic plasmid without the ISL1 promoter sequence was used as a negative control.
2.4 Dual-luciferase reporter assay
The transfected cells were harvested and lysed after 48 hours. Subsequently, the firefly and renilla luciferase activities of the cell lysates were measured with the dual-luciferase reporter assay system (Beyotime Biotechnology, Beijing, China) following the manufacturer’s instructions. The transcriptional activities of ISL1 gene promoter were assessed by relative luciferase activity, which were calculated as ratios of firefly luciferase activity to renilla luciferase activity (Figure 2). Wild-type ISL1 gene promoter activity was designated as 100%. The whole experiments were performed three times independently, each in triplicate.
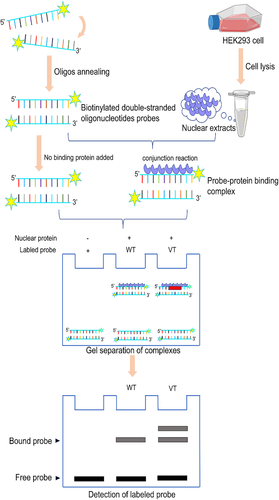
2.5 Preparation of nuclear extracts and electrophoretic mobility shift assay
Nuclear extracts of HEK293 cells were prepared with a nuclear and cytoplasmic protein extraction kit (Beyotime, Beijing, China). The protein concentration was determined using an enhanced BCA protein assay kit (Beyotime, Beijing, China). Biotinylated double-stranded oligonucleotides including wild-type or variant-type sequences (Table 1) in the ISL1 gene promoter were used as probes. A chemiluminescent electrophoretic mobility shift assay (EMSA) kit (Beyotime, Beijing, China) was used to detect the binding reaction between transcription factors and the ISL1 promoter with equal amounts of biotinylated oligonucleotide probes and nuclear extracts (3.0 μg) following the protocol (Figure 3).
2.6 Transcription factor binding site prediction
To analyze whether variants identified above affected transcription factor binding sites (TFBS) of ISL1 promoter region, the JASPAR database was used for predicting all possible TFBS destroyed or newly generated by variants. The relative profile score threshold was set as 85%.
2.7 Statistical analysis
All statistical analyses were performed with SPSS 25.0. A standard Student’s test was used to compare the quantitative experimental data. The data were expressed as means ± SEM. Values of p < 0.05 were taken as statistically significant.
3 RESULTS
3.1 The variants of the promoter region of ISL1 gene identified in ASD patients and healthy controls
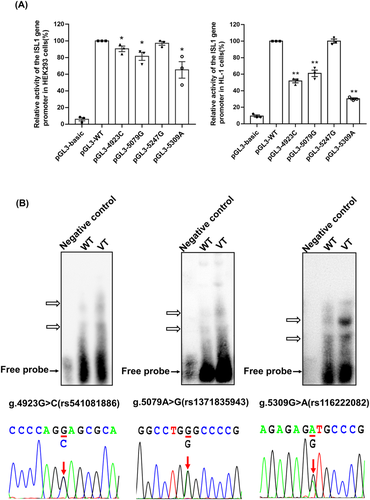
In the 625 subjects (332 patients with ASD and 293 healthy controls), 11 variants were identified by Sanger sequencing (Table 2). The variants were named according to their locations in the ISL1 genomic sequences (Figure 4A). Among the variants identified, four were only found in patients with ASD [g.4923 G > C (rs541081886), g.5079 A > G (rs1371835943), g.5247 A > G (rs36216897), g.5309 G > A (rs116222082); Figure 4B]. Importantly, of these four variants, the allele frequency of the variant [g.5079 A > G (rs1371835943)] is less than 0.0001 in the NCBI dbSNP database and GnomAD database (Accession: PRJNA398795), and another one [g.4923 G > C (rs541081886)] is without allele frequency reports. Interestingly, one of these four variants [g.5309G > A (rs116222082)] was found in two patients (Table 2). Also important, in East Asian all four of these variants have allele frequencies equal to 0.00 in the ALFA database and equal to 0.0003 or 0.0000 in GnomAD database (Table 2). Finally, these four variants were the objectives for further functional validations at the cellular level.
| Variants | ASD | Controls | Positiona | Genotypes | Allele frequency | |
|---|---|---|---|---|---|---|
| Frequency in control = 0 (further validation) | GnomAD | East-Asian | ||||
| g.4923G > C (rs541081886) | 1 | 0 | −402 | GC | None | C = 0.00* |
| g.5079A > G (rs1371835943) | 1 | 0 | −246 | AG | G = 0.00004 |
G = 0.00* G = 0.0000# |
| g.5247A > G (rs36216897) | 1 | 0 | −78 | AG | G = 0.00953 |
G = 0.00* G = 0.0000# |
| g.5309G > A (rs116222082) | 2 | 0 | −16 | GA | A = 0.00906 |
A = 0.00* A = 0.0003# |
| Frequency in control ≠ 0 (no further validation) | ||||||
| g.4184 T > C | 0 | 1 | −1,141 | TC | None | |
| g.4213C > T (rs36216895) | 19 | 24 | −1,112 | CT | T = 0.13 | |
| g.4457C > G (rs6899279) | 19 | 24 | −868 | CG | G = 0.174 | |
| g.4613G > A (rs142427249) | 1 | 1 | −712 | GA | A = 0.00013 | |
| g.4720A > G (rs1200176972) | 0 | 1 | −605 | AG | None | |
| g.5006A > C | 0 | 1 | −319 | AC | None | |
| g.5057A > G (rs3762977) | 19 | 24 | −268 | AG | G = 0.149 | |
- Abbreviations: ASD, atrial septal defects.
- a Variants are located upstream (−) to the transcription start site of the ISL1 gene at 5325 of NG_023040.1.
- * The allele frequency of East Asian in ALFA database (version 20201027095038).
- # The allele frequency of East Asian in GnomAD database (accession no. PRJNA398795).

In addition, three variants were only found in healthy controls [g.4184 T > C, g.4720 A > G (rs1200176972) and g.5006 A > C]. Four variants were found in both ASD patients and controls [g.4213 C > T (rs36216895), g.4457 C > G (rs6899279), g.4613 G > A (rs142427249) and g.5057 A > G (rs3762977)]. These seven variants were excluded from the subsequent study.
3.2 Functional analysis of the variants by dual-luciferase reporter assay
To further analyze the effect of ISL1 gene promoter variants on transcriptional activity, the reporter gene expression vectors were constructed by subcloning wild-type and variants of the ISL1 gene promoter into the luciferase reporter vector (pGL3-basic), including empty pGL3-basic (negative control), pGL3-WT (wild-type ISL1 gene promoter), pGL3-4923C, pGL3-5079G, pGL3-5247G and pGL3-5309A. These expression vectors were transfected into cultured HEK293 and HL-1 cells, respectively. After the predetermined time had elapsed, a dual-luciferase activity assay was performed.
Three of the four variants [g.4923 G > C (rs541081886), g.5079 A > G (rs1371835943) and g.5309 G > A (rs116222082)] significantly decreased the activity of the ISL1 gene promoter compared with the wild-type (p < 0.05; Figure 5A). In contrast, the transcriptional activity was not significantly altered by the other variants [g.5247 G > A (rs36216897); p > 0.05].
3.3 Variant-affected binding sites of transcription factors
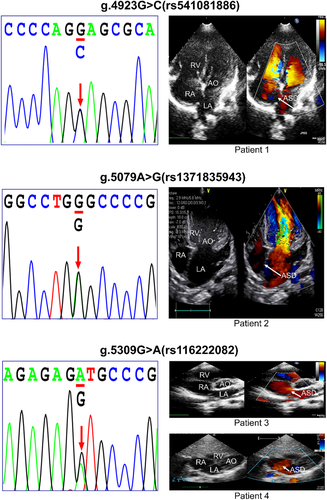
To examine whether the variants affect the binding ability of transcription factors, the EMSA experiment was performed with wild-type or variant-type oligonucleotides. The above three significant variants [g.4923 G > C (rs541081886), g.5079 A > G (rs1371835943) and g.5309 G > A (rs116222082)] in the ISL1 gene promoter affected the affinity of binding sites of transcription factors, resulting in alteration of the transcription of ISL1 gene promoter (Figure 5B). In addition, the other variant [g.5247 A > G (rs36216897)] did not affect the ISL1 gene promoter activity in cultured cells, so it was not further tested with EMSA. The color Doppler echocardiography of the parasternal short-axis view at aortic valve level showed ASD (arrow) in the patients with the discovered variants at the promoter region of the ISL1 gene (Figure 6).
3.4 Putative binding sites for transcription factors affected by genetic variants
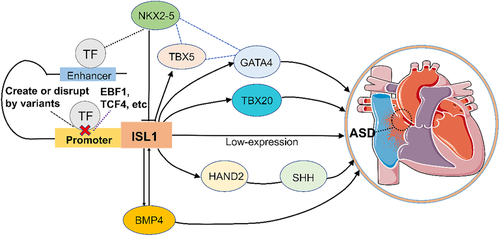
We analyzed the ISL1 gene promoter with the JASPAR database to further investigate the putative binding sites for transcription factors affected by variants identified in the ISL1 gene promoter (Table 3). The variant [g.4923 G > C (rs541081886)] may create three binding sites for basic helix–loop–helix A15 (BHLHA15), specificity protein 1 (SP1) and zinc-finger protein of unknown function (ZNF462), and disrupt the binding sites for the Early B-cell family 1 (EBF1), transcription factor T cell factor 4 (TCF4), SP4, ZNF337 and ZNF786. Five binding sites for transcription factor AP-2C (TFAP2C), nuclear factor IX (NFIX), regulatory factor X-5 (RFX5), PLAGL2 and ZFP64 may be created by the variant [g.5079A > G (rs1371835943)]. Further, it may disrupt the binding sites for androgen receptor (AR), Rhox homeobox family member 1 (RHOXF1), TFAP2A, TFAP2B, zinc finger and BTB domain containing 6 (ZBTB6), ZNF793 and ZNF750. In addition, the variant [g.5309 G > A (rs116222082)] may create three binding sites for ETS homologous factor (EHF), ZNF594 and HAND2, and disrupt the binding sites for GATA1, SNAI2, SNAI1, TBX3 and ZNF28. To illustrate the possible influence of ISL1 gene promoter variants on the development of ASD, the results from the JASPAR database analysis, cellular functional experiments and previously reported studies were combined to establish a schema (Figure 7) that contains genes and pathways including TBX5, TBX20, NKX2-5, GATA4, BMP4, HAND2 and SHH.
| Variants | Binding sites for transcription factors | Promoter activity | |
|---|---|---|---|
| Create | Disrupt | ||
| g.4923G > C | BHLHA15, SP1, ZNF462 | EBF1, TCF4, SP4, ZNF337, ZNF786 | ↓ |
| g.5079A > G | TFAP2C, NFIX, RFX5, PLAGL2, ZFP64 |
AR, RHOXF1, TFAP2A, TFAP2B, ZBTB6, ZNF793, ZNF750 |
↓ |
| g.5247A > G | NFYA, MSGN1 | MEIS1, NR2C2 | No change |
| g.5309G > A | EHF, ZNF594, HAND2 |
GATA1, SNAI2, SNAI1, TBX3, ZNF28 |
↓ |
4 DISCUSSION
The current study for the first time identified that: (1) there are four variants within the ISL1 gene promoter that are only found in the ASD patients with zero incidence in the control; (2) three of these four variants (g.4923 G > C, g.5079 A > G and g.5309 G > A) significantly decreased the transcriptional activity of ISL1 promoter and therefore have functional significance in the development of ASD; and (3) these variants (g.4923 G > C, g.5079 A > G, g.5309 G > A) affected the binding of transcription factors demonstrated by EMSA experiments. From these results the variants (g.4923 G > C, g.5079 A > G, g.5309 G > A) probably contribute to the development of ASD.
Human ISL1 gene, a transcription factor consisting of six exons and five introns and comprising 349 amino acids, codes members of a family of transcription factors containing homologous domains. ISL1 has been used as a marker for cardiac progenitors, it is highly expressed in animal and human embryonic hearts and plays an important role in their normal development.16, 18 Animal studies showed that almost all mice with reduced ISL1 expression presented ASD and ventricular septal defects.24 Several common genetic variants of ISL1 have been associated with the genetic susceptibility of human CHD.17, 25 In addition, sequence variants of ISL1 have also been reported in the cardiac conduction system including atrial fibrillation.26
Growing evidence implies that variants in the gene promoter region, which regulates genes mainly at the transcriptional level, may create or destroy TFBS and change the normal gene promoter activation process, leading to the occurrence of disease.22, 27, 28 In the present study, combined with EMSA and TFBS prediction, we analyzed the promoter of the ISL1 gene and revealed that the variants (g.4923 G > C) may disrupt the potential binding sites for EBF1. As reported, EBF1 is a transcription factor that is the upstream of ISL1. Knockdown of EBF1 results in the downregulation of ISL1 expression.29 In addition, g.4923 G > C may also disrupt the potential binding sites for TCF4, a CHD-related gene interaction with β-catenin, directly regulating the ISL1 gene promoter.30, 31 These transcription factors need to be further identified and investigated.
Based on the above, taken together with the results from previous studies, we established a schema (Figure 7). The low ISL1 gene promoter activity caused by the variants contributes to the low expression of ISL1, as shown in our study. Consequently, the low expression of ISL1 may be directly involved in the formation of ASD.24 Interestingly, a large number of downstream target genes and interacting partners for ISL1 may be involved in the development of ASD. Firstly, NKX2-5, a pivotal regulator of the cardiac lineage, directly binds to an enhancer of ISL1, repressing the transcriptional activity of ISL1.32 In addition, NKX2-5 interacts with GATA4 and TBX5; both are known downstream targets of ISL1.24, 33 Low expression of ISL1 may reduce the expression of TBX5 and GATA4, which are associated with ASD.34, 35 Furthermore, TBX20 is also a downstream gene of ISL1 and its decreased expression may promote the development of ASD.36 Figure 7 also shows that ISL1 is a crucial upstream regulator of the HAND2-SHH pathway.37 The low expression of ISL1 probably contributes to the decrease in SHH signaling related to ASD.38 Finally, BMP4 and ISL1 interact and regulate each other and therefore the low level of BMP4 may increase the risk of ASD.37, 39, 40
The precise role of the discovered ISL1 promoter variants in the development of ASD and CHD requires further in vivo animal experiments. However, often, one or few variants in a particular gene may not show CHD in an in vivo animal model because most CHDs are not caused by single gene mutations but developed with the interaction of related genes as shown in Figure 7. Further studies would be necessary to reveal the role of possible interactions among those genes in the development of CHD.
There are several limitations in our study. The exact role of the genotype–phenotype relationship needs to be further established. In addition, to confirm the interaction between ISL1 and other genes mentioned in the schema, further verification would be the next step. These considerations will be taken into account in our future investigations.
5 CONCLUSIONS
In summary, this study for the first time discovered that variants in the promoter region of ISL1 gene are associated with ASD. Functional analysis and EMSA revealed that these variants significantly altered the transcriptional activity of the ISL1 gene by affecting the binding sites of the transcription factor, contributing to the development of ASD as risk factors. Therefore, the present study may provide new insights into the molecular etiology and potential therapeutic strategy of ASD.
ACKNOWLEDGEMENTS
We thank the patients and their family members for their collaboration. The assistance of nursing staff at the Division of Pediatric Cardiac Surgery, Department of Cardiovascular Surgery is gratefully acknowledged.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
ETHICS STATEMENT
This work was performed in strict compliance with the regulations of the Ethics Committee of TEDA international Cardiovascular Hospital.
AUTHOR CONTRIBUTIONS
GWH and HXC conceived and designed research; XYY and HXC performed experiments; XYY, HXC, ZC, QY and GWH analyzed data; XYY, HXC, ZC, JH, QY, and GWH interpreted results of experiments; XYY prepared figures; XYY, HXC, and GWH drafted manuscript; GWH edited and revised manuscript; XYY, HXC, ZC, JH, QY and GWH approved final version of manuscript.
Open Research
DATA AVAILABILITY STATEMENT
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.




