Topology, glycosylation and conformational changes in the membrane domain of the vacuolar H+-ATPase a subunit†
The authors declare no conflicts of interest.
Abstract
Published topological models of the integral membrane a subunit of the vacuolar proton-translocating ATPase complex have not been in agreement with respect to either the number of transmembrane helices within the integral membrane domain, or their limits and orientations within the lipid bilayer. In the present work we have constructed a predictive model of the membrane insertion of the yeast a subunit, Vph1p, from a consensus of seven topology prediction algorithms. The model was tested experimentally using epitope tagging, green fluorescent protein fusion, and protease accessibility analysis in purified yeast vacuoles. Results suggest that a consensus prediction of eight transmembrane helices with both the amino-terminus and carboxyl-terminus in the cytoplasm is correct. Characterization of two glycosylation sites within the homologous mouse a subunit membrane domain further corroborates this topology. Moreover, the model takes into account published data on cytoplasmic and luminal accessibility of specific amino acids. Changes in the degree of protease accessibility in response to the V-ATPase substrate, MgATP, and the V-ATPase-specific inhibitor, concanamycin A, suggest that functional conformational changes occur in the large cytoplasmic loop between TM6 and TM7 of Vph1p. These data substantially confirm one topological model of the V-ATPase a subunit and support the notion that conformational changes occur within the membrane domain, possibly involving previously proposed axial rotation and/or linear displacement of TM7 in the proton transport cycle. J. Cell. Biochem. 114: 1474–1487, 2013. © 2013 Wiley Periodicals, Inc.
Vacuolar-type H+-ATPases (V-ATPases) are highly conserved multi-subunit proton pumps. They are ubiquitous in eukaryotic cells where they perform essential housekeeping functions by acidifying endomembrane organelles [Beyenbach and Wieczorek, 2006; Saroussi and Nelson, 2009]. In specialized mammalian cells V-ATPases can be targeted to the plasma membrane to translocate protons out of the cell. This can be for the purpose of intracellular pH regulation [Nordström et al., 1995], or to maintain organismal acid–base balance [Wagner et al., 2009]. In some cases, proton secretion is a means to achieve low luminal pH in specialized extracellular compartments, such as the lumina of the epididymis [Brown et al., 1997], or osteoclast resorption lacunae [Manolson et al., 2003].
V-ATPases consist of two major subcomplexes, the V0 sector, which is membrane-bound and involved in proton translocation, and the V1 sector, which hydrolyzes ATP. V0 consists of subunits a, c, c′, c″, d, and e, and V1 is composed of subunits A through H [Zhang et al., 2008]. The energy of ATP hydrolysis is coupled to proton translocation by physical rotation of a central stalk structure that drives a hydrophobic c-barrel composed of (in yeast) four c subunits, one c′ subunit and one c″ subunit. The physical rotation of the c-barrel in contact with the membrane domain of the adjacent a subunit (Vph1p in yeast vacuoles) has been proposed to drive proton translocation by a mechanism that is analogous to F-ATPases “running in reverse.”
The a subunit is c. 100 kDa in size and consists of a cytoplasmic N-terminal half (NTa domain), and an integral membrane, hydrophobic C-terminal half (CTa domain). The NTa domain interacts with the V1 sector and provides a stabilizing stator function [Landolt-Marticorena et al., 2000; Kartner et al., 2010; Muench et al., 2011]. The CTa domain plays a major role in proton translocation by forming the transmembrane proton channel [Toei et al., 2011]. Protons are thought to diffuse into a cytoplasmically accessible hemichannel, resulting in protonation of a passing glutamate residue situated in a transmembrane α-helix (TM) of one of the six c-type subunits within the rotating c-barrel. As a consequence of coupled ATP hydrolysis, the c-barrel rotates 360° in three 120° steps, each consuming one ATP molecule, to deliver the same proton to a putative second hemichannel with aqueous access to the lumen. Dissociation of the proton and diffusion into the lumen completes its translocation across the lipid bilayer [Toei et al., 2011]. Because there are six proton-carrying sites evenly spaced at the periphery of the c-barrel, two protons can be translocated for every molecule of ATP hydrolyzed. A critical arginine residue (R735 in yeast) is thought to interact with the c-barrel glutamic acid at the luminal hemichannel, possibly providing the necessary alignment and environment for protonation and deprotonation phases of the transport cycle. The general postulate is derived from one proposed originally to describe F-ATPase proton translocation [Vik et al., 2000].
The CTa domain of the a subunit has been reported as having six [Manolson et al., 1992], six or eight [Jackson and Stevens, 1997], seven [Leng et al., 1996; Leng et al., 1998; Peng et al., 1999; Ma et al., 2011], eight [Landolt-Marticorena et al., 1999; Wang et al., 2008; Toei et al., 2011], or nine [Leng et al., 1999] TMs. In addition to these discrepancies, topology models of the V-ATPase a subunit based on experimental accessibility of membrane-permeable and impermeable reagents, or protease accessibility, have not been unequivocal in their assignments of TM or inter-helical loop sequences [Jackson and Stevens, 1997; Leng et al., 1999; Wang et al., 2008], although a better defined model has emerged recently [Toei et al., 2011]. The present work seeks to clarify features of topology and function of the a subunit, using approaches complementary to those of previously published works, and in so doing add to its potential as a therapeutic target.
MATERIALS AND METHODS
Enzymes and Antibodies
Specialty enzymes were from: zymolyase, BioLynx (Zymolyase 100T, Seikagaku, catalog no. SK1204931); endoproteinase Glu-C, Roche (sequencing grade Staphylococcus aureus V8 protease; 1420399); proteinase K, Promega (V3021); enterokinase, GenScript (enteropeptidase, porcine light chain; Z01003); bovine pancreatic trypsin, BioShop (3× crystallized; TRP 003); PNGase F, New England Biolabs (P0704S). Antibodies were from: anti-Influenza hemagglutinin (HA) epitope, Santa Cruz (HA-Probe Y-11, rabbit polyclonal IgG, 200 µg/ml; SC-805); anti-green fluorescent protein (GFP), Santa Cruz (GFP (FL), rabbit polyclonal IgG, 200 µg/ml; SC-8334); anti-Vph1p, Invitrogen (10D7, mouse monoclonal IgG, directed against yeast V-ATPase a subunit NTa domain (epitope accessible only in dissociated V0 complexes); A-6426); anti-B, Invitrogen (13D11, mouse monoclonal IgG directed against yeast V-ATPase B subunit; A-6427); anti-CPY, Invitrogen (10A5, mouse monoclonal IgG directed against yeast carboxypeptidase Y; A-6428).
Cell Culture, Media, and Reagents
Yeast cells were typically grown in YPD medium (pH 6.5) consisting of 10 g/l yeast extract (Difco), 20 g/l bacto-peptone (Difco), and 20 g/l dextrose. In some assays, as indicated, ZnCl2 was added to 4 mM to YPD (pH 6.5). The Δvma2 strains were maintained in YPD adjusted to pH 5.0 with succinic acid (cells lacking vacuolar V-ATPase activity require acidified medium for optimal growth). Human HeLa cells were grown in DMEM supplemented with 10% FBS and antibiotics (100 units/ml penicillin, 100 µg/ml streptomycin; Invitrogen). FBS was heat inactivated in-house at 56°C for 1 h. Prior to transfection, HeLa cells were grown in serum-free Opti-MEM (Invitrogen), as noted. Bone marrow mononuclear (BMM) cell-derived osteoclasts were obtained by harvesting bone marrow cells from femurs of 2-month-old male mice. These were plated in 100 mm tissue culture dishes at 1.0 × 105 cells/ml in α-MEM without nucleosides (Invitrogen; 12561) supplemented with 10% FBS, 50 ng/ml macrophage colony stimulating factor (M-CSF) and antibiotics for 2 days. The medium was then replaced with one containing 50 ng/ml M-CSF and 200 ng/ml soluble, recombinant receptor activator of NF-κB ligand (RANKL) and growth was allowed for another 4 days prior to cell extraction. Mammalian cells were grown at 37°C in a 5% CO2 humidified incubator.
G418 was from GIBCO BRL (11811-031); concanamycin A was from Kamiya Biomedical (MT-803); protease inhibitor cocktail (P8215), mammalian protease inhibitors (P8340), and phosphatase inhibitor cocktail (P5726) were from Sigma-Aldrich; ATP was from Amersham Biosciences (27-2056); M-CSF was from Calbiochem; and Fugene HD was from Roche, Inc.
Yeast Strains
All strains used in this work are listed in Table I and were derived from the protease deficient Saccharomyces cerevisiae strain, BJ1991 (Matα leu2 trp1 ura3-52 pep4-3 prb1-1122) [Brachmann et al., 1998]. MM525 is a VPH1 deletion strain (Δvph1) that was derived from MM476 by deletion of a major portion of the coding region of the VPH1 gene and disruptive LEU2 insertion [Manolson et al., 1992]. MM725 was derived by integration of the plasmid pMM719 into strain MM525 and selection on Ura− medium; MM799 was similarly derived by integration of pMM798. As all strains were derived from the parental MM476, they are all deficient in Pep4p and Prb1p (yeast proteinases A and B).
| Straina | Phenotypeb (ref.) | Genotype (background)c |
|---|---|---|
| MM476 | Protease deficient (ref. 22) | Matα leu2 trp1 ura3-52 pep4-3 prb1-1122 |
| MM525 | Vph1p deletion | Δvph1::LEU2 (on MM476) |
| MM725 | Vph1p-3HA | VPH1-3HA (on MM525) |
| MM799 | Vph1p-GFP | VPH1-GFP (on MM525) |
| Plasmid | Constructd | Expression product |
|---|---|---|
| pMM126 | pRS316 (ref. 23) | None (empty vector) |
| pMM122 | pRS306 (ref. 23) | None (empty vector) |
| pMM509 | pRS316-VPH1-3HA | Vph1p-3HA |
| pMM719 | pRS306-VPH1-3HA | Vph1p-3HA |
| pMM787 | pRS316-VPH1-GFP | Vph1p-GFP |
| pMM798 | pRS306-VPH1-GFP | Vph1p-GFP |
| pMM789e | pEGFP-N1 (Genbank U55762) | GFP |
| pMM919e | pEGFP-N1-a3 | a3-GFP |
| pMM963e | pEGFP-N1-a3-N41Q | a3N41Q-GFP |
| pMM964e | pEGFP-N1-a3-N484Q/N504Q | a3N484Q/N504Q-GFP |
- a Designations refer to the strains collection of the communicating author (M.F.M.).
- b Experimentally relevant phenotype is shown. All strains are protease deficient. Unreferenced plasmids were developed in the present work (see Materials and Methods Section).
- c Genotype added to the parental background is shown (background strain in parentheses). For derivation of strains, see Materials and Methods Section.
- d See Materials and Methods Section for details of plasmid construction.
- e Plasmid for mammalian expression.
Plasmids and Homologous Recombination
All site-directed mutagenesis was performed using the Quickchange Site-Directed Mutagenesis kit (Agilent Biotechnologies), in accordance with supplier instructions. Plasmid constructs were prepared in the shuttle vector pRS316 (pMM126) for transient plasmid expression, or pRS306 (pMM122) for yeast genomic integration [Sikorski and Hieter, 1989], and are listed in Table I. The plasmid pMM509 was originally pDJ65, a gift from Dr. Tom Stevens, University of Oregon. It was constructed with VPH1 followed by three C-terminal Influenza virus hemagglutinin (HA) epitope tags, YPYDVPDYA, in series after S837 of Vph1p. The plasmid pMM719 was constructed in pRS306 (pMM122) by transferring the VPH1-3HA insert (NotI/SalI fragment) from pMM509 to pRS306 (pMM122). The plasmid pMM787, with VPH1 ligated with in-frame, C-terminal enhanced green fluorescent protein (GFP) after codon 840, was a gift from Dr. Min Zhao, University of Toronto. The plasmid pMM798 was constructed in pRS306 (pMM122) by transferring the insert from pMM787. In this case, the NotI–BamHI fragment containing the VPH1 gene and a BamHI–EcoRI fragment containing the GFP gene from pMM787 were ligated in-frame into pRS306, between NotI and EcoRI sites. All plasmid inserts were fully sequenced, including their junctions, to confirm the expected DNA sequence.
To obtain stable yeast expression strains, pRS306-derived plasmids were linearized with PstI and integrated into the receptor yeast strain genome (see Table I) using standard lithium acetate transformation [Sherman et al., 1986] followed by selection on SD − Ura agar. After growth of selected colonies in non-selective YPD medium, the stable transformants were validated for appropriate protein expression, using SDS–PAGE and immunoblotting with appropriate anti-HA, anti-GFP, and anti-Vph1p antibodies. Similar methods were used to select transiently expressing constructs in pRS316.
Functional Testing of V-ATPase Assembled With Tagged Vph1p
Recombinant yeast strains were grown on YPD plates in the presence or absence of 4 mM ZnCl2 at 30°C. Heavy metal resistance requires functional V-ATPase in the yeast vacuole [Iwaki et al., 2004]. Growth was compared (see Results Section) among strains bearing VPH1 (wild type), Δvph1 (VPH1 deletion mutant), VPH1-GFP (C-terminal GFP-tagged Vph1p), and VPH1-3HA (C-terminal HA-tagged Vph1p).
Mammalian Constructs and Expression
The mouse a3 subunit cDNA in pcDNA3.1-a3 was a gift from Dr. Beth S. Lee, Ohio State University. The pcDNA3.1-a3 plasmid was used as a template to obtain the a3 coding sequence, flanked by EcoRI and SacII restriction sites, as a PCR product. The cut insert DNA was cloned into the pEGFP-N1 plasmid (Clontech), also cut with EcoRI and SacII. Expression from this construct produces the mouse a3 protein with a C-terminal fusion to the N-terminus of GFP. A short, random linker region, ARDPPVAT, exists between the two polypeptides. The following three mutations were introduced into this construct by site-directed mutagenesis, using the primers indicated (paired with complementary antisense primers): N41Q, 5′-gagttcagagacctccaggaatccgtgagcgcc-3′; N484Q, 5′-ggctgccatggcccagcagtcaggctgga-3′; and N504Q, 5′-gctcaccctgaaccctcagatcactggtgtcttcc-3′. All plasmid inserts were fully sequenced, including their junctions, to confirm the expected DNA sequence. Wild type a3, a3N41Q and the double-mutant, a3N484Q/N504Q constructs were used for exogenous expression of C-terminally GFP-tagged a3 proteins. HeLa cells were seeded at 1.0 × 106 cells/well in 100 mm tissue culture dishes and maintained in DMEM, supplemented with 10% FBS, until 85% confluent. The medium was replaced with Opti-MEM after washing cells with PBS, 24 h prior to transfection. Cells were transfected with 10 µg of DNA, using Fugene HD at a Fugene/DNA ratio of 3:1.
Cells were harvested 48 h post-transfection, after washing twice in ice-cold PBS, by scraping on ice in the presence of hypotonic lysis buffer (10 mM KCl, 1.5 mM MgCl2, 2 mM dithiothreitol, 10 mM Tris–HCl, pH 7.4 at 25°C, with the addition of mammalian protease inhibitor cocktail (1:200, v/v) and 1 mM phenylmethylsulfonyl fluoride (PMSF) from 100 mM stock in anhydrous ethanol). The cell suspension was homogenized by passage through a 0.5 in., 27.5 gauge syringe needle 15–20 times and was then centrifuged at 5,000g for 10 min. The supernatant was centrifuged at 100,000g for 1 h and the microsomal membrane pellet was resuspended in 50 µl hypotonic lysis buffer.
To observe native a3 subunit with and without PNGase F treatment, cell extracts were prepared from mouse BMM-derived osteoclasts. Cells were washed twice with ice cold PBS and lysed in lysis buffer (0.1% Triton X-100, 300 mM NaCl, 5 mM EDTA, 50 mM Tris–HCl, pH 7.4 at 25°C, with addition of protease inhibitor cocktail, phosphatase inhibitor cocktail and PMSF, as above). Microsomal membrane and lysate protein concentrations were determined using the Pierce 660 nm Protein Assay Reagent (Thermo Scientific).
Deglycosylation and SDS–PAGE of a3-GFP
Glycoproteins were deglycosylated with PNGase F according to a modification of the supplier protocol. Microsomal membranes (20 µg total protein) were diluted to a final volume of 30 µl in glycoprotein denaturing buffer (manufacturer supplied). Proteins were denatured at 65°C for 10 min, then 1/10th vol. each of 10% Nonidet P-40 and 10× G7 reaction buffer (manufacturer supplied) were added, followed by 1,000 units PNGase F. The final volume was adjusted to 40 µl with distilled water. The reaction mixture was incubated for 1 h at 37°C. Proteins were then solubilized by addition of 5× SDS–PAGE gel-loading buffer and resolved on 7% polyacrylamide gels. The a3-GFP fusion proteins were visualized by immunoblotting with anti-GFP antibody.
Isolation of Intact Yeast Vacuoles
Vacuoles were isolated essentially as described by Manolson et al. [1992]. Vacuole protein concentration was quantified, as above, and the integrity of purified vacuoles to exogenously added proteases was assayed by observing the ability of proteinase K to cleave luminal procarboxypeptidase Y (pro-CPY) to its mature form (CPY) in the presence or absence of detergent. Intact vacuoles purified from exponentially-growing yeast strains were resuspended to 0.5 mg/ml protein concentration in 10 mM MES–Tris, pH 6.9 at 25°C, with 30 µg/ml proteinase K in the presence or absence, of 0.1% or 0.5% (w/v) Triton X-100 for 2 or 10 min. Proteolytic digests were terminated at designated times by the addition of PMSF to a final concentration of 22 mM, followed immediately by protein precipitation with ice cold 20% trichloroacetic acid (TCA) and centrifugation. Protein pellets were washed in ice-cold acetone to remove residual TCA, and immunoblotted with anti-CPY antibody.
Glu-C Digestion of Intact Vacuoles
Purified intact vacuoles from MM725 were subjected to endoproteinase Glu-C proteolysis. Briefly, an aliquot of intact vacuoles (20 µg protein) in 7% Ficoll-400, 10 mM MES–Tris, pH 7.8 at 25°C, was diluted with ammonium carbonate buffer, pH 7.8, to a final concentration of 25 mM and incubated with varying concentrations of endoproteinase Glu-C at 25°C, for designated times. In addition, 5 mM ATP, 5 mM MgCl2, or both, and 1 µM or 10 µM concanamycin A were included during proteolysis, as indicated for each experiment. The final reaction volume was 100 µl. Endoproteinase Glu-C proteolysis was inhibited by the addition of PMSF to a final concentration of 1 mM, tosyl-L-lysine chloromethylketone (TLCK) to a final concentration of 5 mM, and protease inhibitor cocktail to a 1:100 dilution, followed by incubation on ice for 15 min. Samples were denatured by the addition of 5× SDS sample-loading buffer and incubation at 65°C for 3 min. Western blotting was performed by standard methods and immunoblots were probed with anti-HA, or anti-Vph1p, antibodies.
Tryptic Digestion of Intact Vacuoles
Following digestion with 1 mg trypsin/mg vacuole protein at 0°C for 5 min, proteins were resolved on tricine gels and transferred to 0.025 µm pore-size nitrocellulose sheets in 25 mM sodium phosphate buffer, pH 4.5, for 2 h. Transfers were blocked with 1% bovine serum albumin (BSA) in Tris-buffered saline (TBS) for 1 h and developed using the same detection reagents, buffers and dilutions specified below for ELISA, with the exception that 3,3′-diaminobenzidine (DAB; Sigma D5637) was used to visualize bound peroxidase activity.
Proteinase K Proteolysis of Vph1p-GFP
Aliquots containing purified, intact vacuoles (50 µg protein) from strain MM799 (expressing the Vph1p-GFP fusion protein) were treated with varying amounts of proteinase K in 7% Ficoll-400, 10 mM MES–Tris, pH 6.9, with a final volume of 200 µl, at 0°C for 20 min. The reaction was stopped by addition of protease inhibitor cocktail (1:100) and PMSF (5 mM final). Samples were imaged by fluorescence microscopy (Leica DM IRE 2 with GFP filter) and then were precipitated with ice-cold 20% TCA. Protein pellets were washed in ice-cold acetone to remove residual TCA and analyzed by SDS–PAGE and Western blotting, using anti-GFP and anti-CPY antibody probes, followed by HRP-conjugated second-antibody and enhanced chemiluminescence.
Enterokinase Cleavage of Flag-Tags
Freshly prepared intact yeast vacuoles (15–20 µg protein) were suspended in 50 µl of enterokinase cleavage buffer (5.6% Ficol-400, 8 mM MES–Tris, pH 6.9, and 0.4 mM MgCl2 final) and were digested with 8 units of enterokinase in the presence or absence of 0.1% and 0.5% (w/v) Triton X-100 at 22°C for 2 h. Vacuole samples treated in exactly the same way, but without addition of protease, were later subjected to proteinase K digestion on ice to determine vacuolar integrity. Proteolytic digests were terminated by addition of 5 µl of 0.1 M PMSF in anhydrous isopropanol and 2 µl of protease inhibitor cocktail, followed by mixing with 11 µl of 6× SDS–PAGE sample buffer and heating at 65°C for 5 min.
Competitive Anti-HA ELISA
The topological orientation of the C-terminus of Vph1p was determined by competitive ELISA, using freshly isolated intact vacuoles, from cells expressing Vph1p-3HA, or from the isogenic wild type strain. A peptide corresponding to the HA epitope (plus an N-terminal cysteine) was conjugated to BSA using m-maleimidobenzoyl-N-hyroxysuccinimide ester (MBS; Pierce 22311) [Kitagawa and Aikawa, 1976]. The BSA-HA conjugate (0.5 µg/well) was covalently coupled to microtiter plates using 1-cyclohexyl-3-(2-morpholinoethyl)-carbodiimide-metho-p-toluene sulfonate (CMC; Sigma–Aldrich C1011). Anti-HA antibody (1:100,000 dilution) was preincubated in 1% BSA in TBS for 2 h at 4°C with various amounts of either intact, or freeze-thawed and sonicated (permeable), vacuoles. Aliquots (100 µl) of the mixtures were added to HA/BSA-coated microtiter-plate wells and incubated for 1 h at 4°C. Wells were washed twice with 200 ml each of cold TBS, and bound H7 antibody was detected with a 1:5,000 dilution of biotinylated anti-mouse IgG followed by Elite VectaStain A/B (Vector Laboratories PK-6102) in 100 µl/well of 1% BSA in TBS. Peroxidase activity was developed colorimetrically using 1-Step Turbo TMB-ELISA (Pierce 34022).
Computed Topology Prediction
A topology model of Vph1p was derived by applying seven different, commonly used predictive algorithms to determine putative TM helices (Fig. 1). These were HMMTOP [Tusnády and Simon, 1998, 2001], TMHMM [Krogh et al., 2001], SOSUI [Hirokawa et al., 1998], TMpred [Hofmann and Stoffel, 1993], TopPred [von Heijne, 1992; Claros and von Heijne, 1994], DAS [Cserzo et al., 1997], and PHDhtm [Rost et al., 1995]. All were available on the Swiss Institute of Bioinformatics ExPASy Proteomics Server at the time that this work was done and were generally used with default settings. A description of the consensus model computation is given in Results Section.
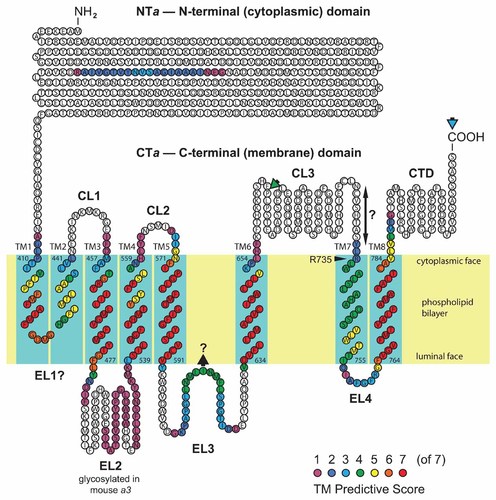
Consensus computed topology model for Vph1p. In this model, N- and C-termini are both cytoplasmic; NTa is depicted en bloc and CTa with 8 TM α-helices (blue rectangles), designated TM1–8 (start and end residues numbered). R735 is essential for proton translocation within predicted TM7, which may be mobile perpendicular to the plane of the membrane bilayer (as suggested by double-ended black arrow with question mark). EL3 could possibly form a reentrant loop (black arrowhead with question mark). C-terminal HA and GFP tags were engineered onto the C-terminus (blue arrowhead) and Glu-C protease cleavage of CL3 was observed (green arrowhead) in supporting evidence for the model.
RESULTS
Questions of Vph1p topology were addressed by first deriving computed models using seven commonly available TM prediction algorithms (see Materials and Methods Section). Their methodologies are similar; however, these algorithms employ somewhat different amino acid properties tables, are derived from various partitioning data, and comparisons are made with various surveys of both globular and membrane proteins, and some use hidden Markov models [von Heijne, 1992; Hofmann and Stoffel, 1993; Claros and von Heijne, 1994; Rost et al., 1995; Cserzo et al., 1997; Hirokawa et al., 1998; Tusnády and Simon, 1998, 2001; Krogh et al., 2001]. Consequently, there were considerable differences in the numbers, locations and spans of the predicted TM segments for Vph1p, depending on the algorithm used. Thus, it was felt that by building a consensus of results the most reliable features of this panel of TM-prediction tools could be incorporated into a model of greater accuracy than would be predicted by any single method.
To build a consensus, the seven different predictions were mapped to the primary sequence of Vph1p, and each amino acid in the sequence was given an unbiased score numerically equivalent to the number of predictive algorithms that flagged it as being within a TM. Stretches of flagged amino acids were identified as likely TMs if there were at least 18 contiguous amino acids with a score ≥4 (i.e., predicted by at least half of the algorithms used). Stretches of amino acids flagged by ≤3 of the seven algorithms over <18 contiguous residues were rejected as unreliable predictors of TM helices.
The resulting consensus computed topology prediction for Vph1p (see Fig. 1) was grossly defined by its well-established features: two nearly equally-sized domains, the largely hydrophilic, cytoplasmically oriented, amino-terminal domain (NTa) comprising residues M1 to N406, and the largely hydrophobic, integral membrane, carboxyl-terminal domain (CTa) comprising residues A407 to the C-terminus, S840. Furthermore, CTa was comprised of seven to eight predicted TM helices. There was ambiguity in the first hydrophobic stretch, which should have been a single TM by our stated criteria; however, further topological considerations, discussed below, require that it be treated as two TMs. The model also predicts putative cytoplasmic inter-helical loops, CL1–3; putative extracellular loops, EL1–4 (EL1 may not exist if TM1 and TM2 form a reentrant loop that does not penetrate to the lumen, as is suggested in Fig. 1); and the C-terminal tail domain, CTD. In Figure 1, the color-coded predictive scores (color legend, lower right) are mapped to the topology diagram, and TMs have been positioned within the phospholipid bilayer to maximize the predictive score within a 21 amino acid span.
There is abundant evidence that the NTa domain has a cytoplasmic orientation, in part because of its known interactions with a number of cytoplasmic V1 subunits, such as A, B, and H [Jackson and Stevens, 1997; Landolt-Marticorena et al., 1999, 2000; Leng et al., 1999; Kartner et al., 2010], and there is evidence that the hydrophilic C-terminal “tail” domain (CTD) is oriented to the cytoplasm. The latter evidence comes from scanning cysteine mutagenesis and sulfhydryl reagent accessibility studies [Wang et al., 2008; Toei et al., 2011], but past investigations with similar methods have yielded contradictory results [Leng et al., 1998]. Additional support for a cytoplasmic orientation comes from the observation that the CTD interacts with the cytoplasmic glycolytic enzyme, phosphofructokinase-1 [Su et al., 2003, 2008]. With both the N-terminus and C-terminus in the cytoplasm, the number of TMs must be even. Thus, with a default of eight TMs, the first contiguous stretch of hydrophobic amino acids necessarily has to be divided into two TMs. Alternatively, since these two putative TMs are relatively short spans that do not fully meet our criteria for being transmembrane, it seems plausible that this segment forms a reentrant loop, as shown speculatively in Figure 1, rather than spanning the membrane twice; the implications for orientation of other TMs, however, remain the same. A third alternative is that the low-scoring hydrophobic region between TM5 and TM6 is a TM (while keeping the first hydrophobic stretch as one TM to satisfy the even-number requirement). This, however, would invert TMs 3–5 (as labeled in Fig. 1), drastically altering the topology. To determine which model is correct required further experimental evidence.
To test the model of Figure 1, it was necessary first to dispel any remaining doubts regarding the disposition of the C-terminus. Two constructs were prepared where the C-terminus of Vph1p was tagged, either with the 3HA epitope tag to yield Vph1p-3HA, or with GFP to yield Vph1p-GFP (see Materials and Methods Section and Table I). These constructs were integrated separately into the genome of a yeast strain with a VPH1 deletion, Δvph1. Since these tags are polypeptide additions to the Vph1p structure, and GFP has a substantial size (27 kDa), it was necessary to verify that the tagged Vph1p subunits were functionally expressed within the V-ATPase complex. This was accomplished with a growth assay in the presence of 4 mM ZnCl2 (Fig. 2A). Zinc is toxic to yeast, but can be detoxified, possibly by sequestration within the vacuole, requiring a cation-exchange mechanism dependent upon the proton gradient generated by V-ATPase, or some other V-ATPase-dependent mechanism [Iwaki et al., 2004; Kane, 2006]. In yeast where Vph1p is not functional, V-ATPase activity is lacking in the vacuolar membrane and, consequently, the cells are sensitive to zinc toxicity and will not grow on media containing 4 mM ZnCl2. This was observed for the Vph1p-deletion strain Δvph1 in a 4-day growth assay (Fig. 2A). In a dilution series, it was seen that Δvph1 grew poorly compared with the wild type VPH1 strain in standard medium (that it grows at all may be due to low expression of the paralogous subunit, Stv1p, which can partially complement the Δvph1 phenotype, though this remains speculative [Kane, 2006]), but in the presence of Zn2+, growth was completely eliminated. On the other hand, in the strain expressing the Vph1p-GFP fusion protein, growth was restored to a level indistinguishable from that of the wild type. Similar results were obtained with the Vph1p-3HA construct (not shown). Thus, neither the large C-terminal GFP fusion, nor the short 3HA epitope tag, significantly compromised Vph1p function.
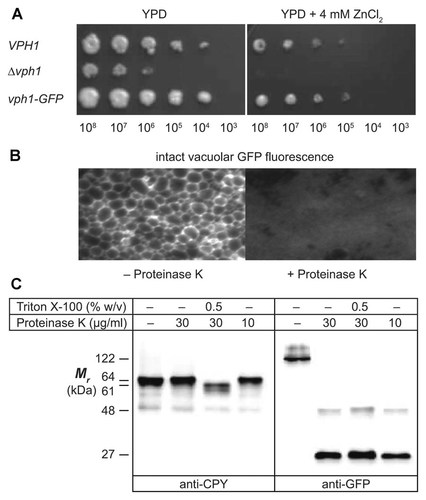
C-terminal GFP-tag protease accessibility evidence that the CTD of Vph1p is cytoplasmic. A: A yeast strain expressing wild type Vph1p (VPH1), a Vph1p-deletion strain (Δvph1), or a strain expressing Vph1p-GFP fusion protein (vph1-GFP) in a Δvph1 background, were 10-fold serially diluted and spotted onto either standard YPD agar medium or YPD agar containing Zn2+ (both pH 6.5). B: Tightly packed vacuoles from VPH1-GFP yeast were observed by epifluorescence microscopy before (left) and after (right) treatment with proteinase K (5 s exposures with FITC filter and 100× objective). C: Vacuole proteins separated by SDS–PAGE were immunoblotted with anti-CPY (left blot), or anti-GFP (right blot), antibodies. For the right-most lanes of each panel, a range of proteinase K was used for digestion without detergent; results were identical over the range of protease concentrations, but only the 10 µg/ml protease lane is shown.
To determine the orientation of the hydrophilic CTD of Vph1p, accessibility of either the 3HA epitope tag, or the GFP fusion tag, on the cytoplasmic surface of purified, intact vacuoles was tested. Upon purification of intact vacuoles from the Vph1p-GFP-expressing strain, GFP fluorescence was observed in densely packed vacuoles by fluorescence microscopy (Fig. 2B), revealing the characteristic ring-staining expected for a membrane-bound fluorescent marker. Treatment of vacuoles with 30 µg/ml proteinase K at 0°C for 20 min resulted in complete proteolytic quenching of GFP fluorescence. This suggested that the GFP-tag was accessible on the outer (cytoplasmic) surface of the vacuolar membrane and, therefore, that the CTD of Vph1p, to which it is fused, must reside in the cytoplasmic compartment; however, this conclusion depends entirely upon the integrity of the vacuoles to protease penetration.
To show that the vacuole preparation consisted of vacuoles that were impermeable to proteinase K, accessibility of the protease to pro-CPY, a luminal marker of the vacuolar compartment, was tested. Figure 2C (left panel) shows vacuolar proteins immunoblotted with anti-CPY antibody. It demonstrates that pro-CPY (66 kDa band) was not digested to mature CPY (61 kDa) in the vacuole preparation even under conditions of high protease concentration and extended digestion time (up to 40 µg/ml proteinase K, 20 min at 0°C); however, with the addition of 0.5% (w/v) of the membrane-disrupting, non-ionic detergent, Triton X-100, complete digestion of pro-CPY to CPY was observed. Furthermore, when probed with anti-GFP antibody, the blot of Figure 2C (right panel) showed that Vph1p-GFP was as completely digested in the absence of Triton-X100 as in its presence, even at low protease concentrations (right-most lane). These data support the notion that the GFP-tagged CTD of Vph1p is accessible at the cytoplasmic surface of the vacuole membrane.
Further evidence of the orientation of the C-terminus of Vph1p was sought using the HA epitope tag. To do this, a competitive ELISA was performed, where intact vacuoles were used to compete for anti-HA antibodies targeted to antigen-coated microtiter plate wells, as described in Materials and Methods section. Figure 3A shows results of competition with vacuoles from different sources. While intact and detergent solubilized vacuoles expressing Vph1p-3HA strongly competed for anti-HA antibodies at between 0.1 and 1 µg vacuole protein per assay, neither intact nor detergent-solubilized vacuoles expressing wild type Vph1p had any effect at up to 2 µg protein added.
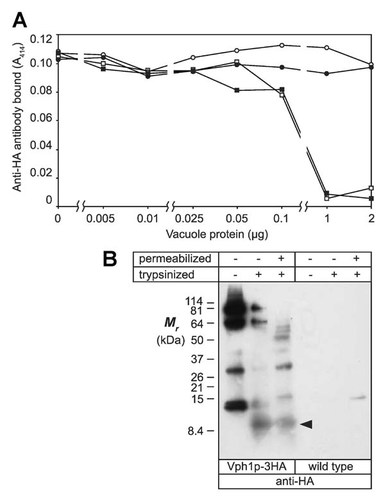
C-terminal epitope tag (3HA) protease accessibility evidence that the CTD of Vph1p is cytoplasmic. A: Competitive ELISA shows that the C-terminal 3HA-tag on Vph1p-3HA (expressed in yeast strain MM725) is accessible to anti-HA antibody from the cytoplasmic side of intact vacuoles. Open circles: intact wild type Vph1p-containing vacuoles; open squares, intact vacuoles from yeast expressing Vph1p-3HA; solid circles, detergent-solubilized wild type vacuoles; solid squares, detergent-solubilized Vph1p-3HA vacuoles. Abscissa is a discontinuous logarithmic scale. B: Right panel, intact vacuoles (100 µg protein per lane) were control digested without trypsin (autodigestion by vacuolar proteases is evident; left lane), with 100 µg trypsin (middle lane), or with trypsin and 0.5% Triton X-100 (right lane). Polypeptide fragments were resolved on tricine gels and immunoblotted with anti-HA antibody. This revealed a 10 kDa tryptic peptide (arrowhead) bearing the C-terminal 3HA epitope tag (itself 5 kDa) of Vph1p-3HA, which is of approximately equal intensity when derived from either intact or detergent permeabilized vacuoles (and is not seen in the absence of trypsin). Left panel: control, wild type vacuoles showed no significant background bands.
Figure 3B shows vacuolar proteins resolved by SDS–PAGE and immunoblotted using anti-HA antibody. Vph1p-3HA was observed at its native size of c. 100 kDa in untreated vacuoles (left-most lane). Although vacuoles were “untreated” with respect to trypsinization in this lane, incubation was the same as in other lanes, but without addition of trypsin or detergent. Smaller bands were observed presumably due to autodigestion by vacuolar proteases during this incubation. While all lanes were of 100 µg initial vacuolar protein, this left-most lane appears overloaded relative to adjacent lanes where trypsin digestion resulted in significant loss of the HA epitope tag signal. When either intact vacuoles, or detergent-solubilized vacuoles, were treated with trypsin, similar digestion of Vph1p-3HA occurred, with the smallest HA-tagged band having a size of c. 10 kDa (arrowhead). There was no evidence of the 10 kDa band in the untreated control lane, indicating that this band was generated by trypsin digestion. No significant background bands were seen with wild type Vph1p (not HA-tagged). The 10-kDa band corresponds approximately to the size of the CTD, from its proposed junction with TM8 to, and including, the C-terminal 3HA-tag, which itself adds c. 5 kDa to the fragment size. That the fragment was released equally upon trypsinization in the presence or absence of detergent suggests strongly that it is accessible on the cytoplasmic face of the intact, trypsin-impermeable vacuoles. All of the above data, taken together, demonstrate unequivocally that the orientation of the hydrophilic CTD of Vph1p is cytoplasmic and is accurately depicted in the topology model shown in Figure 1.
In order to provide further evidence in support of the model presented in Figure 1, additional protease digestion experiments were done with endoproteinase Glu-C and intact vacuoles from yeast expressing Vph1p-3HA. Under the buffer conditions employed, endoproteinase Glu-C cleaves polypeptides on the carboxyl side of a glutamate residue [Sørensen et al., 1991] and, therefore, its protease activity is more selective than that of trypsin or proteinase K, making it a relatively infrequent cutter. Figure 4A shows an immunoblot probed with anti-HA antibodies after digestion of intact vacuoles with a range of Glu-C concentrations. A single major HA-positive band was released, with a size of c. 23 kDa (23.4 ± 0.3 kDa, n = 3; note that this includes the C-terminal 3HA epitope tag, with a predicted size of 5.0 kDa). This places the site of digestion near the N-terminal end of the large hydrophilic loop (green arrowhead in CL3 of Fig. 1). The indicated site would yield a predicted fragment of 23.7 kDa in size; the closest potential cleavage sites on the luminal side of the membrane would yield a fragment c. 10 kDa greater than the observed size. Figure 4B shows an immunoblot identical to that in Figure 4A, but probed instead with an antibody directed to the NTa domain of Vph1p. This yielded a single major band of c. 80 kDa (79.6 ± 1.7 kDa, n = 3), which complements the 23-kDa, HA-positive band of Figure 4A to give empirically a native protein size of 103.0 kDa ± 2.0 kDa, in good agreement with the apparent size observed in SDS–PAGE.
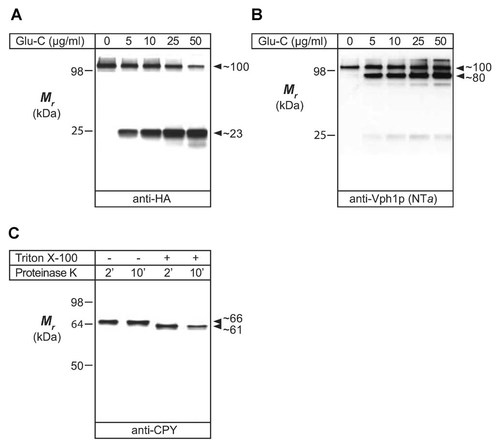
Endoproteinase Glu-C accessibility evidence that the loop between TM6 and TM7 (CL3) of Vph1p is cytoplasmic. Purified, intact vacuoles (4 µg protein) were digested with Glu-C concentrations indicated, then resolved by SDS–PAGE and immunoblotted. A: Anti-HA probe revealed undigested Vph1p-3HA at 100 kDa; band at 23 kDa is C-terminal HA-tagged fragment of Vph1p-3HA. B: Anti-Vph1p (NTa domain) probe also identified 100 kDa undigested Vph1p-3HA; the band at 80 kDa is a proteolytic fragment of Vph1p-3HA retaining the NTa domain. C: Intact vacuoles were treated with proteinase K (30 µg/ml) in the presence or absence of 0.5% (w/v) Triton X-100 for 2 and 10 min on ice, as indicated. Digestion of pro-CPY to CPY was seen only upon detergent permeabilization of vacuoles, indicating that purified vacuoles were intact and impermeable to protease. Full-length pro-polypeptide bands (c. 66 kDa) and the processed form (c. 61 kDa) of CPY are indicated (arrowheads).
Further elucidation of Vph1p topology comes by inference from studies of V-ATPase a subunits in mammalian systems. Apps et al. [1989] showed over 20 years ago that a subunits of V-ATPases in bovine adrenal medullary chromaffin granules were sensitive to neuraminidase and PNGase F digestion. For the latter enzyme, half of the “120 kDa band” was reduced in size on SDS–PAGE gels when digested in the presence of ocytl-β-glucoside, but the other half could be digested only with the addition of 0.1% SDS, suggesting two distinct oligosaccharide chains (our interpretation). This was the first publication clearly to show N-linked glycosylation in any V-ATPase a subunit. Surprisingly, no further glycosylation studies appeared until recently, when Lee et al. [2010] published work on mouse neuronal autophagy that showed that the lysosomal V-ATPase a1 subunit, a highly conserved ortholog of Vph1p, was glycosylated.
The mammalian a3 subunit has often been referred to as the “116 kDa subunit” because of its relative mobility in SDS–PAGE, when in fact its predicted polypeptide size (in mouse) is 93.4 kDa. Many membrane proteins have anomalous mobility in SDS–PAGE, but our observations on native mouse a3 banding patterns strongly suggested that it had the characteristics of a glycoprotein with complex N-linked glycosylation, showing a diffuse band that was more than 20 kDa in excess of its predicted size. On examination of potential glycosylation signals (N-X-S/T, where X ≠ P) in the mouse a3 polypeptide sequence, we found that only two, at N484 and N504, within the putative second extracellular loop (EL2) were highly conserved in vertebrate species (Fig. 5A). Only one other site, at N41 in NTa could be informative regarding topology, but NTa was already clearly established to be in the cytoplasm, as discussed above. We felt that if we could precisely identify the glycosylation site, which must be luminal, it would help to clarify the orientations of TMs in the N-terminal half of CTa.
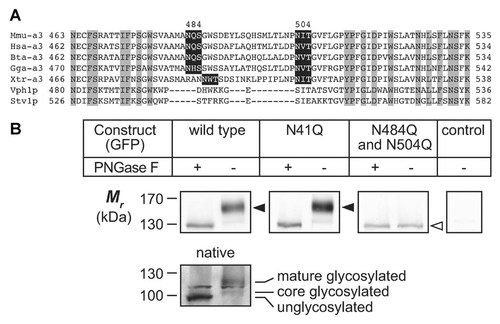
Evidence that the loop between TM2 and TM3 (EL2) of Vph1p is luminal. A: Alignments of putative EL2 polypeptide sequences from mouse (Mmu), human (Hsa), bovine (Bta), chicken (Gga), and Xenopus (Xtr) a3 subunit, and yeast Vph1p and Stv1p. Gray highlights, residues identical over all species; black rectangles, glycosylation sites. Numbers above top line indicate glycosylated Asn residues in mouse a3. B: HeLa cells were transfected with constructs for transient expression and microsomal membrane proteins (20 µg per lane) were treated with PNGase F. Upper row of panels, immunoblots probed with anti-GFP antibody, indicating: wild type, unmodified a3 polypeptide with C-terminal GFP fusion (arrowhead indicates 152 kDa band [right lane] that is absent after PNGase F treatment [left lane]); N41Q, knockout of glycosylation site in NTa domain had no effect; N484Q and N504Q, double knockout eliminated the 152 kDa band (open arrowhead, 128 kDa unglycosylated band); control, untransfected HeLa cells. Bottom inset panel: native a3 from mouse BMM-derived osteoclasts with, or without, PNGase F treatment. This shows (right lane) a diffuse glycosylated band at c. 116 kDa, a core-glycosylated band at c. 102 kDa and (left lane) an unglycosylated band at c. 94 kDa. The sharp intermediate band is a common, non-specific band. For upper row of panels, n = 3; lower inset panel, n = 2.
Figure 5B, shows immunoblots of a3-GFP constructs expressed in HeLa cells, with and without PNGase F treatment. The wild type a3 fusion protein was observed largely as an intense band at 152 kDa. The 152 kDa band was diffuse, which is typical of complex-glycosylated glycoproteins, and it was reduced to 128 kDa in size upon deglycosylation by PNGase F treatment. A faint band was observed additionally at 134 kDa, which we have recently characterized as the core-glycosylated protein [Bhargava et al., 2012], present at low steady-state levels in the ER prior to trafficking and processing in the Golgi (to the complex-glycosylated 152 kDa band). Similar observations were made with native a3 protein, endogenously expressed in BMM-derived mouse osteoclasts, but since this protein has no C-terminal GFP fusion, molecular weights of the identified bands were: complex glycosylated glycoprotein, 116 kDa; core-glycosylated glycoprotein, 102 kDa; and deglycosylated protein, 94 kDa. These latter observations, along with previously published work [Bhargava et al., 2012], support the notion that heterologous GFP-tagged a3 expression is identical, in terms of biosynthesis and trafficking, to that of the native protein. Site-directed mutagenesis (N → Q conversion) of the N41 site did not alter glycosylation of mouse a3-GFP relative to wild type, as was expected, and the a3N41Q-GFP could be deglycosylated with PNGase-F in the same manner as the wild type protein. Individual mutagenesis of N484 and N504 also did not abrogate expression of the glycosylated band (data not shown); however, the double mutation N484Q/N504Q resulted in only a smaller-sized, unglycosylated band being observed at 128 kDa, similar to that seen after enzymatic deglycosylation of wild type a3-GFP (Fig. 5B). This demonstrated for the first time that the mouse a3 EL2 loop is glycosylated at two sites, N484 and N504. While there is no evidence that yeast Vph1p is N-glycosylated, nor are any glycosylation sites predicted in its EL2 sequence (Fig. 5A), the otherwise high degree of structural homology between mammalian and yeast a subunits suggests that the Vph1p EL2 loop must also be luminal. The further implications of this finding are that it supports the validity of dividing the first hydrophobic stretch of amino acids into TM1 and TM2 (or representing it as a reentrant loop, as shown in Fig. 1) and requiring an even number of TMs between EL2 and CL3, eliminating the possibility of an additional membrane-spanning TM between TM5 and TM6. As is speculatively indicated in Figure 1, however, this does not rule out the possibility that EL3 could also be a reentrant loop similar to what has been indicated for TM1 and TM2.
While performing experiments with Glu-C, it was observed that the cleavage of Vph1p with the protease appeared to be sensitive to Mg2+ concentration. Since the endoproteinase Glu-C itself is not Mg2+-dependent, this phenomenon was investigated further. Figure 6A shows results of a series of experiments where endoproteinase Glu-C cleavage of Vph1p (in vacuolar V-ATPase) was performed in the presence of 5 mM Mg2+, MgATP, or ATP. These data are further quantified and shown graphically in Figure 6B. At 5 µg/ml Glu-C concentration the yield of the 23-kDa, HA-tagged cleavage product was more than doubled in the presence of 5 mM Mg2+ and approximately doubled in the presence of 5 mM MgATP. ATP alone increased cleavage by c. 40%. This suggests that binding of substrate to the catalytic subunits (Vma1p and/or Vma2p; i.e., the A and B subunits) of the V-ATPase complex is able to effect an inter-subunit conformational change, transmitted from Vma1p/Vma2p to the Vph1p subunit, that affects the accessibility of the hydrophilic loop between TM6 and TM7 to endoproteinase Glu-C. Whether this is by direct interaction of the hydrophilic loop with the catalytic subunits, or by transmission through some intermediary subunit between the two, or allosterically by transmission through some other domain of Vph1p (likely NTa), is presently unknown.
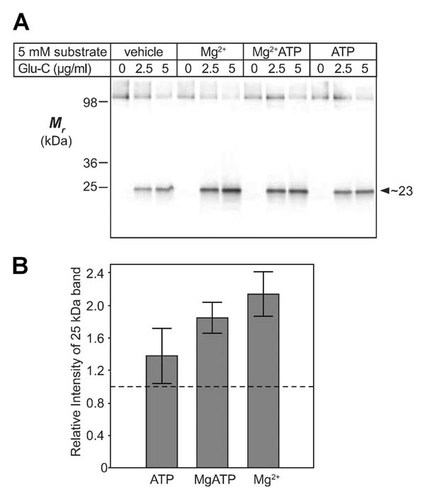
Mg2+ and MgATP (V-ATPase substrates) enhance endoproteinase Glu-C proteolysis between TM6 and TM7. A: Vacuolar Glu-C digests (0.7 µg protein per lane) were immunoblotted as in Figure 4. Glu-C concentrations were as indicated, with control vehicle, Mg2+, MgATP, or ATP (5 mM each). Arrowhead indicates cleaved, C-terminal HA-tagged fragment of Vph1p-3HA at c. 23 kDa. B: Quantification of substrate-enhanced accessibility of protease to the hydrophilic loop between TM6 and TM7 of Vph1p-3HA. Chemiluminescence of the 23-kDa proteolytic fragment of panel A was quantified using a Molecular Imager Gel Doc XR+ system (Bio-Rad). Data were normalized to vehicle control value (dashed line). Error bars, ±1 SD (ATP, n = 3; MgATP, n = 2; Mg2+, n = 4).
The above results encouraged additional experiments with concanamycin A, a known inhibitor of V-ATPase function which is thought to interact directly with Vph1p at the junction between it and the c subunits of the c-barrel structure of the V-ATPase rotor [Huss et al., 2002; Bowman et al., 2004]. It was of interest to determine if this binding could also effect apparent conformational changes within Vph1p. Figure 7A shows results of a series of experiments, similar in execution to those of Figure 6A, but where endoproteinase Glu-C cleavage of Vph1p was performed in the presence of 1 or 10 µM concanamycin A. These data are further quantified and shown graphically in Figure 7B. Concanamycin A at 10 µM reduced the yield of the 23-kDa HA-tagged cleavage product of Vph1p to c. 70% of that seen in its absence, while at 1 µM the effect was somewhat less, at c. 80% of control. Concanamycin A at 10 µM concentration did not have any significant effect on Glu-C digestion of control protein (bovine serum albumin; data not shown). These data suggest that binding of concanamycin in the membrane domain of Vph1p has some allosteric effect on the accessibility of the hydrophilic CL3 loop between TM6 and TM7 to endoproteinase Glu-C. The apparent conformational changes observed here are consistent with the hypothesis that TM7 may undergo some physical displacement within the phospholipid bilayer during the catalytic transport cycle of the V-ATPase complex [Kawasaki-Nishi et al., 2003; Toei et al., 2011].
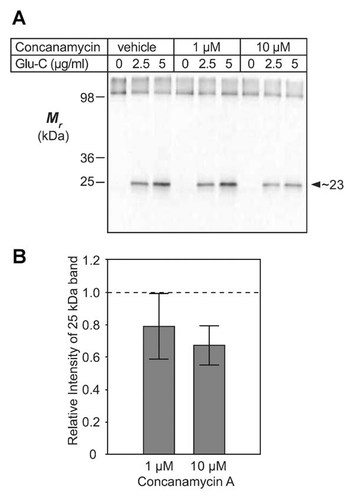
Concanamycin A (V-ATPase inhibitor) decreases endoproteinase Glu-C accessibility between TM6 and TM7. A: Glu-C digests were immunoblotted, as in Figure 6 (4 µg vacuolar protein per lane). Glu-C concentrations were as indicated, in the presence of control vehicle, 1 µM, or 10 µM concanamycin A. Arrowhead, cleaved, HA-tagged, C-terminal fragment of Vph1p-3HA (23 kDa). B: Quantification (as in Fig. 6) of the concanamycin-mediated decrease in accessibility of protease to the hydrophilic loop, CL3, between TM6 and TM7 of Vph1p-3HA. Data were normalized to the vehicle control value (dashed line). Error bars, ±1 SD (1 µM concanamycin A, n = 5; 10 µM concanamycin A, n = 3).
DISCUSSION
In the present work we attempted to refine the structural model for the Vph1p membrane domain by starting with a consensus of topology prediction algorithms. This approach yielded a model that was confirmed experimentally and is similar in organization to a recently published model that was also confirmed experimentally [Toei et al., 2011]. This supports the notion that using a consensus predictive approach is preferable to the use of any single algorithm. In contrast, a recent V-ATPase review features a Vph1p topology prediction using the TMHMM algorithm alone, which yields a model having seven TMs, with the CL3 and CTD domains (our designations) in the lumen [Ma et al., 2011]. Thus, the single-algorithm prediction has misplaced important features of V-ATPase a subunit topology that are now unequivocally established through many lines of experimental evidence.
Validation of the present topology model was performed using epitope-tagged Vph1p, and C-terminal fusion proteins. These experiments provided strong confirmation that the C-terminus of Vph1p is in the cytoplasm. Similarly, the orientation of the CL3 loop was verified by demonstrating its accessibility to endoproteinase Glu-C on the cytoplasmic surface of intact vacuoles. Additionally, homology modeling, using data from mouse a3 subunit glycosylation studies placed Vph1p EL2 in the lumen. Taking all these data into account supports both the number of predicted TM segments (totaling 8) and the dispositions of the hydrophilic loops and termini of the Vph1p topology model presented in Figure 1.
The current working hypothesis for V-ATPase proton translocation proposes that TMs of the a subunit form separate cytoplasmic and luminal hemichannels in the membrane bilayer. Evidence to support this comes from site-directed mutagenesis of polar and charged amino acids within potentially aqueous channel-forming segments that are part of, or are immediately peripheral to, TMs. These mutations block proton translocation and ATP hydrolysis without affecting V-ATPase assembly [Leng et al., 1996, 1998; Kawasaki-Nishi et al., 2001; Toei et al., 2011]. Physical evidence is also seen in 21 Å resolution single-particle electron microscopy of V-ATPase, where two openings are evident on the luminal side and one on the cytoplasmic side of the V0 complex [Wilkens and Forgac, 2001]. A more detailed, dynamic model suggests that proton translocation requires not only rotation of the rotor c-barrel, but also intra-molecular, axial rotation of TM7, and/or its linear displacement, within the membrane domain of the a subunit [Kawasaki-Nishi et al., 2003; Toei et al., 2011].
In the present work, Glu-C protease accessibility experiments revealed conformational changes in the Vph1p cytoplasmic loop, CL3, in response to V-ATPase substrate or inhibitor binding. These data may lend support to the dynamic model for TM7 involvement in proton transport, as TM7 is contiguous with CL3. Thus, we speculate that conformational changes in the V-ATPase catalytic headpiece might be physically coupled to Vph1p CL3 through the NTa domain, which is in contact with the V-ATPase V1 sector [Landolt-Marticorena et al., 1999, 2000; Kartner et al., 2010]. This could account for changes in the rate of CL3 proteolytic digestion in response to MgATP and suggests a physical transduction of conformational change from the V1 sector to Vph1p CL3 that ultimately might be involved in displacement of TM7 in concert with the catalytic events of the proton transport cycle. Response to concanamycin A suggests that if the displacement of TM7 is inhibited (at the a subunit to c-barrel interface), then CL3 is constrained to a less protease-accessible conformational state. Consistent with this, Wang et al. [2005] found that bafilomycin and concanamycin resistance could be conferred by the mutations E721K, L724A, or N725F in Vph1p. These mutations are clustered at the CL3–TM7 interface, so they proposed that bafilomycin and concanamycin may be inhibitory by “locking” the V0 domain into a conformation that prevents the dynamic association of TM7 and the c-barrel.
While the topology model presented here is in overall agreement with that of Toei et al. [2011] there are some notable differences. One surprising feature of the predicted topology shown in Figure 1 is that the critical R735 residue of TM7 is very near the cytoplasmic membrane interface, not near the middle of the TM, as has been proposed to accommodate its hypothetical interaction with the centrally located glutamate residues of the c-barrel, or even nearer the luminal membrane interface, as has been suggested in some models [Duarte et al., 2007; Wang et al., 2008; Toei et al., 2011]. TM7 has a surprisingly amphipathic sequence for a TM; nevertheless this is consistent with its proposed function as an aqueous channel-forming domain. Because of its amphipathic nature, in some orthologs of other species, TM7 is not detected as a TM by any of the TM-predicting algorithms used in the present work (data not shown). Thus, while we show the unrevised results of predictive modeling in Figure 1, it seems likely that the limits of TM7 cannot be accurately identified by hydropathy mapping alone. Cysteine accessibility scanning suggests that TM7 may include sequences as far upstream as C723, with a luminal border corresponding to A744, shifting the proposed TM7 α-helix upstream by a dozen amino acids with respect to the assignments shown in Figure 1 [Toei et al., 2011].
The possibility of axial rotation of TM7 to open alternately cytoplasmic and luminal hemichannels has been discussed by Toei et al. [2011]. We speculate that an alternative possibility might be that TM7 goes through a linear displacement, perpendicular to the membrane plane, in the direction of the lumen, allowing the hypothetical alignment of R735 with the rotating glutamic acid proton carriers of the c-barrel. The amphipathic nature of TM7 (including the upstream CL3 sequence) may allow such “bobbing” to occur (see Fig. 1), taking R735 from a position near the cytoplasmic interface to near mid-membrane and back again, in synchrony with the catalytic cycle, opening and closing aqueous channels, or promoting influx and efflux of protons in the lipid bilayer. This notion is in part supported by cross-linking studies suggesting that linear displacement of TM7 by more than one α-helical turn (at least four amino acids) may occur relative to the c-barrel [Kawasaki-Nishi et al., 2003]. A mechanism for this may be provided by the proposed conformational changes in CL3.
It is notable that many of the charged and polar amino acids suggested to form cytoplasmic or luminal hemichannels in the model of Toei et al. [2011] are actually in the cytoplasm (e.g., in the CTD domain), or in the lumen (in EL2), respectively, rather than in the TMs identified in the model of Figure 1. These charged and polar amino acids are thought to form hydrophilic segments of the proton hemichannels. To some extent the model of Figure 1 can accommodate these discrepancies, as the limits of TMs are arbitrarily constrained to a 21 amino acid window. Clearly some TMs will be somewhat longer, or shorter, but extending TM3, TM4, and especially TM8 to accommodate the working model proposed by Toei et al. would likely require significant tilting of α-helices to accommodate their length in the lipid bilayer. This is not an unusual feature of membrane proteins, but the three dimensional modeling of tilted helices would certainly complicate the predictions of dynamic helical rotation of TM7 in relation to other channel-forming helices.
Finally, we have suggested diagrammatically in Figure 1 that there is the possibility that EL3 could form a reentrant loop within the membrane. Reentrant loops are not common, but are seen in some transport proteins and may play regulatory roles. In the AE2 anion exchanger, for example, a reentrant loop is thought to act in acute regulation of anion transport by pH [Stewart et al., 2009]. The central hydrophobic segment shown in Figure 1 is flanked by charged residues, and Leng et al. [1999] have shown that the introduction of a factor Xa site (IEGR) into EL3 before P606 significantly reduced proton transport activity compared with wild type Vph1p. These observations are consistent with a possible role in proton transport for the EL3 domain.
CONCLUSION
In the present work we have demonstrated the utility of consensus predictive modeling, and have confirmed features of the predicted Vph1p topology model with experimental evidence. The amino and carboxyl termini of Vph1p and the CL3 loop domain have been unequivocally shown to be situated in the cytoplasm. We have shown for the first time that the mouse V-ATPase a3 subunit is N-glycosylated at two sites and by homology modeling deduced that EL2 of Vph1p is luminal. These observations and the constraints they place on topology resulted in the model of Figure 1, which is largely in agreement with the recently published experimentally determined topology of Toei et al. [2011]. Determination of CL3 orientation led to the observation that the conformation of CL3 is affected by V-ATPase substrate and inhibitor binding. This, and questions concerning the limits of TM7 suggest considering alternate models of TM7 dynamics in proton transport. Further experimental work will be required to resolve these possibilities.
With its important roles in housekeeping functions and disease processes, further elucidating the structure and function of the V-ATPase a subunit would be of benefit to both a greater understanding of V-ATPase cell biology and the potential development of novel V-ATPase-targeted therapeutics.
Acknowledgements
We thank: Dr. Jeffrey Charuk of Larial Proteomics, Toronto, Canada, for helpful discussion and services on ELISA quantification of Vph1p; Dr. Mark S. Longtine, Biochemistry and Molecular Biology Dept., Oklahoma State University, Stillwater, OK, for his gift of the pFA6a-KanMX6 plasmid; Dr. Beth S. Lee, Physiology and Cell Biology Dept., Ohio State University, Columbus, OH, for her gift of the cDNA clone for mouse a3, and anti-mouse a3 antibodies; Dr. Patricia M. Kane, Biochemistry and Molecular Biology Dept., State University of New York, Syracuse, NY, for her gift of anti-Vph1p monoclonal antibodies; Dr. Tom Stevens, Institute of Molecular Biology, University of Oregon, Eugene, OR, for his gift of pDJ65 plasmid; and Dr. Min Zhao, Visiting Professor, University of Toronto, Toronto, Canada, for his gift of Vph1p-GFP plasmid.




