LIGHT regulates the adipogenic differentiation of mesenchymal stem cells†
Jianrong Xu and Gang-Ming Zou have shared senior authorships.
Abstract
LIGHT is a cytokine belonging to the TNF family. This cytokine has been extensively defined in its role on T-cell regulation and dendritic cell maturation. It also exhibits the role in liver regeneration. We recently identified its role in regulation of hematopoietic stem cell differentiation. However, the question whether this cytokine regulates mesenchymal stem cells (MSCs) proliferation and/or differentiation remains unknown. In this study, we observed that MSCs express LT-βR but not HVEM. PCR analysis show LIGHT mRNA is undectable in MSCs. LIGHT did promote neither MSCs proliferation nor migration. However, LIGHT promoted MSCs differentiation into adipocyte which was confirmed by Oil Red O Staining Assay. Since either MSCs or adipocytes are the major cell population in bone marrow niche, we then suggest that LIGHT regulate bone marrow niche, such as MSCs differentiation. J. Cell. Biochem. 114: 346–353, 2013. © 2012 Wiley Periodicals, Inc.
Stem cells are characterized by the ability to self-renew and differentiate into various functional cell types. Mesenchymal stem cells (MSCs) are multipotent stromal cells that have the capacity to differentiate into chondrocytes, osteoblasts, and adipocytes as well as have immunosuppressive effects on lymphocytes. MSCs have been isolated from bone marrow [Ame-Thomas et al., 2007], adipose tissue, peripheral blood, fetal liver, lung, amniotic fluid, chorionic villi of the placenta, and umbilical cord blood. MSCs are easy to isolate from the bone marrow and umbilical cord blood and to expand in vitro. MSCs are promising tools for regenerative medicine.
MSCs proliferation or differentiation can be regulated by various cytokine. For instance, FGF18 induced osteogenic differentiation of MSCs [Hamidouche et al., 2010]. Cytokines of TNF family member regulate various cellular responses, including proliferation, migration, inflammation, and cell death. For example, TWEAK has been reported to induce proliferation and migration of human umbilical vein endothelial cells in vitro. Human cord blood MSC and bone marrow MSCs express TNFR1, the receptor of TNFα, and TNFα stimulation lead to cytokine IL-8 expression. Bone marrow MSCs are incapable of producing TNFα under the circumstance tested in their investigations [van den Berk et al., 2010]. TNFα might synergize with IFN-γ in regulating gene expression in MSCs [Xu et al., 2009]. TRAIL is another member of TNF family and it induces significantly human MSC migration and TNF-α may modulate this course [Corallini et al., 2010]. However, the question whether or not other members of TNF family, such as LIGHT, activate MSCs remains unclear currently.
LIGHT (HVEM-L, TNFSF-14, or CD258) is a member of TNF superfamily. It is expressed as a homotrimer on activated T cells [Mauri et al., 1998], and also on immune dendritic cells [Tamada et al., 2000]. So far, three receptors with distinct cellular expression patterns are described to interact with LIGHT. LT-βR, found on follicular DCs and stromal cells binds LIGHT. LT-βR is also expressed on hepatocytes which contribute to liver regeneration (Anders et al., 2005). HVEM, however, in contrast, is detected on immune DCs, T and B cells, NK cells, monocytes, and endothelial cells [Scheu et al., 2002]. LIGHT also binds to decoy receptor 3 (DcR3) [Wang et al., 2009]. LIGHT is produced by activated T cells, monocytes, granulocytes, immature dendritic cells, and platelets [Otterdal et al., 2006]. Due to monocyte/macrophage has been listed as one of cell members in bone marrow niche [Ehninger and Trumpp, 2011], we then hypothesis that LIGHT secreted by one niche cells, such as monocyte/macrophage, may regulate other niche cells, like MSCs.
LIGHT was originally identified as a weak inducer of apoptosis. It could induce both apoptotic and non-apoptotic events in the cells. Normally, caspase signal transduction is involved in apoptosis mediated by LIGHT. This cytokine has been extensively defined in its role on T-cell regulation. It also regulates liver regeneration through its receptor LT-βR [Anders et al., 2005]. A recent study demonstrated it inhibit adipose differentiation suggesting its potential use as an anti-obesity agent [Tiller et al., 2011]. Our previous report demonstrated that LIGHT could induce monocyte differentiation toward dendritic cells [Zou et al., 2004]. However, the question regarding whether this cytokine regulates MSCs proliferation and or differentiation remains unknown. In the present study, we identified the novel role of LIGHT in regulating MSC differentiation.
MATERIALS AND METHODS
Cell Culture
Mouse BM-MSCs (CD29+, CD44+, Sca-1+, CD34+, and CD117−) were purchased from Cyagen Biosciences (Sunnyvale, CA). BM-MSCs were maintained in MSCs basal medium. BM-MSCs were expanded to passage 8 before experiments. Sub-confluent or confluent cultures of cells were treated and maintained with LIGHT cytokine and BSA as control, or in adipogenic induction medium until RNA extraction or microscopic evaluation. MSCs were stimulated with Dex, insulin, Indo, or IBMX alone using the same concentration as found in the adipogenic induction medium.
Quantitative RT-PCR
cDNA was prepared using the PrimeScript kit. In each case, random and oligo(dT) primers were used. Quantitative PCR analysis was performed with a 7300 (ABI), using SYBR Premix EX Taq as the reaction reagent. The relative quantities of the genes were calculated using β-actin as a reference, using the formula: 2 (−[CtGene−Ctβ-actin]). Primer sequences are listed in Table I.
| Primers | Sequence | Product (bp) | Annealing (°C) |
|---|---|---|---|
| Pparg_Fw | ACCACTCGCATTCCTTTGAC | 100 | 60 |
| Pparg_Rv | TGGGTCAGCTCTTGTGAATG | 60 | |
| Cfd_Fw | TGCATCAACTCAGAGTGTCAATCA | 50 | 60 |
| Cfd_Rv | TGCGCAGATTGCAGGTTGT | 60 | |
| β-actin_Fw | GCTTCTTTGCAGCTCCTTCGT | 62 | 60 |
| β-actin_Rv | ATCGTCATCCATGGCGAACT | 60 |
Bisulfite Modification and methylation-specific PCR (MSP)
DNA from mouse MSCs were subjected to bisulfite treatment, The EpiTect Bisulfite Kit was used to converting unmethylated cytosine residues in genomic DNA to uracil according the manufacturer's protocol. The modified DNA was used as a template for MSP. The primers for the methylated reaction were: LIGHT_M_SF: (sense) 5′-TAGAGGTTTTTGTTTGTTAGCGC, and LIGHT_M_SR (anti sense), 5′-AATAAATAAAACAACCCCTACTCCG, which amplify a 130-bp product. The primers for the unmethylated reaction were: LIGHT_U_SF (sense), 5′-TAGAGGTTTTTGTTTGTTAGTGTGT, and LIGHT_U_SR (anti sense), 5′-TAAATAAAACAACCCCTACTCCAAT, which amplify a 128-bp product. The PCR amplification of the modified DNA samples consisted of 1 cycle at 95°C for 5 min; 35 cycles at 95°C for 30 s, 69°C for 30 s and 72°C for 30s; 1 cycle at 72°C for 10 min. PCR product was directly loaded onto non-denaturing 2% polyacrylamide gels, stained with ethidium bromide and visualized under UV illumination. Each MSP was repeated at least three times. We considered that the presence of a visible PCR product in Lane U or M indicated the presence of unmethylated or methylated genes, respectively.
Flow Cytometry Analysis
Cell surface staining involved direct immunofluorescence, and the samples were analyzed using Cell Quest software. Staining was performed with the following FITC-conjugated isotype control antibody, HVEM mAb or LT-β receptor antibody. The cells were washed and resuspended in PBS containing 2% FCS and 2 mM EDTA then read by flow cytometry. To perform HVEM expression analysis, the cells were stained with PE conjugate Rat-anti-mouse HVEM antibody, then analyzed using FACScan analyzer.
In vitro Migration Assay
Chemotactic migration of MSCs was performed using transwell assays (diameter, 6.5 mm; pore size, 8 µm; Millipore, USA). Briefly, 200 µl serum-free IMDM, containing 1 × 105 MSCs was added to the upper chamber, while 1,300 µl serum-free IMDM, with SDF1 (100 ng/ml), was added to the bottom chamber. LIGHT (50, 100, and 200 ng/ml) were added to the bottom chamber, in order to evaluate their chemotactic activity. After 20 h of incubation (37°C, 5% CO2), MSCs in the upper surface of the chambers were eliminated using of a cotton bud and those in the lower surface were fixed with 95% ethanol, stained with hematoxylin, and counted using a 200× microscope. Five visual fields were observed in each chamber. All experimental steps were repeated three times under the same conditions and settings. All samples were prepared in duplicate.
Western Blot Analysis
Western blot analysis was performed as previously reported [Zou et al., 2003]. In summary, whole cell extracts were prepared using protein extract buffer from both LIGHT-treated or BSA-treated cells. A volume of 30 µg of protein was separated by SDS-polyacrylamide electrophoresis using a 15% (w/v)polyacrylamide resolving gel and transferred electrophoretically to a nitrocellulose membrane. The membrane was then incubated with 5% non-fat dry milk, subsequently stained with antibodies against IkBα, etc. After overnight incubation, the blots were washed three times with a wash buffer PBST for 10 min each time at room temperature, then, incubated for 45 min with a secondary horseradish peroxidase-conjugated Rabbit-anti-mouse antibody.
Oil Red O Staining Assay
MSCs were plated to a six-well plate (5,000/cm2), the cells grew to 80% confluence. The adipogenic differentiation ability was induced in adipogenic medium for 10 days (15% normal horse serum and 100 nM dexamethasone in basal DMEM medium). First, the density of Oil Red O positive cells were calculated using Image Pro Plus software (Media Cybernetics Inc., Silver Spring, MD); Second, the intracellular lipid droplets were extracted and quantified. The cells were fixed with 10% neutral buffered formalin followed by incubating with 60% propylene glycol, then incubated with a newly filtered Oil Red O staining solution. After staining, the cells were rinsed with distilled water, and 1 ml of isopropyl alcohol was added to the stained dish. Aliquots of the extracted Oil Red O were measured at 510 nm with spectrophotometer (Ultrospec 3000, Pharmacia Biotech, USA).
NF-κB Transcription Factor DNA-Binding ELISA Assay
The transcription factor NF-κB family activation assay was measured using the TransAM NF-κB family kit (Active motif, Carlsbad, USA), according to the instructions of manufacturers.
Microarray Screening
Each RNA sample was labeled with Cy3, and hybridized to an Illumina Whole Murine Genome Microarray chip consisting of over 30,850 probes according to the manufacturer's protocol. Briefly, biotinylated cRNA was prepared using the Illumina RNA Amplification Kit according to the manufacturer's directions starting with ∼100 ng total RNA. Samples were purified using the RNeasy kit. Hybridization to the Whole Murine Genome Microarray chip, washing and scanning was performed according to the Illumina Bead Station. After image processing, background-subtracted intensities were normalized using the quantile method [Bolstad et al., 2003]. DAVID TOOL 6.7 (National Institute of Allergy and Infectious Diseases, http://david.abcc.ncifcrf.gov/) was used to examine selected lists of genes in order to identify overrepresentation of functional classes accordingly with gene-ontology classification.
Statistical Analysis
With the SPSS 9.0 statistical software package, a Student's test was used to test the probability of significant differences between samples. Statistical significance was set at P < 0.05.
RESULT
Mouse BM-MSCs Constitutively Express LT-βR But Not HVEM
Three receptors, LT-β receptor, HVEM/TR2 and TR6, bind LIGHT. We examined whether BM-MSCs express LIGHT receptor and LIGHT cytokine. The FACS analysis result shown that LT-β receptor is expressed on BM-MSC, and HVEM is not or at very low levels (Fig. 1A and B). However, the mRNA of LIGHT was not detectable in our RT-PCR, and qRT-PCR assay and LPS stimulation did not induce LIGHT expression in MSCs (Fig. 1C and D). Moreover, LIGHT treat MSC caused a number of gene expression has been upregulated (Table II).
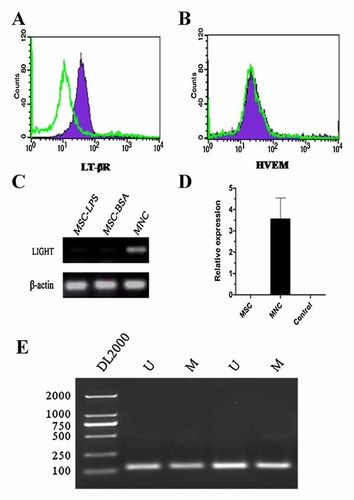
LIGHT and its receptor expression on mouse BM-MSCs. A and B: LT-βR and HVEM proteins on the cell surface of mouse bone marrow-derived MSCs were determined by FACS analysis. BM-MSCs were stained with specific antibodies against LT-βR and HVEM, and each receptor expression level was determined by FACS analysis. Cells treated solely with isotype control were used. C and D: Intracellular LIHGT expression in BM-MSCs and BM-MNCs (born marrow-derived mononuclear cells as a positive control) were estimated by RT-PCR and qRT-PCR. E: Methylation status of LIGHT gene promoter as determined by MSP. The gene studied is given on the left of each panel. Lane U: amplified product with primers recognizing unmethylated sequence. Lane M: amplified product with primers recognizing methylated sequence. The LIGHT promoter showed both M- and U-signal, due to partial methylation of the DNA.
| Category | Gene number | P-value |
|---|---|---|
| Biological process | ||
| Translation | 19 | 1.12 × 10−1 |
| Macromolecular complex subunit organization | 10 | 0.003307 |
| Macromolecular complex assembly | 9 | 0.00683 |
| Negative regulation of macromolecule biosynthetic process | 9 | 0.02219 |
| Negative regulation of macromolecule metabolic process | 10 | 0.023869 |
| Negative regulation of cellular biosynthetic process | 9 | 0.02573 |
| Negative regulation of biosynthetic process | 9 | 0.026994 |
| Hatching | 2 | 0.032568 |
| Blastocyst hatching | 2 | 0.032568 |
| Regulation of cell proliferation | 10 | 0.033555 |
| Induction of apoptosis by intracellular signals | 3 | 0.033574 |
| Negative regulation of transcription | 8 | 0.034153 |
| Transcription initiation | 3 | 0.037184 |
| Negative regulation of nucleobase, nucleoside, nucleotide, and nucleic acid metabolic process | 8 | 0.045837 |
| Negative regulation of nitrogen compound metabolic process | 8 | 0.047918 |
| Translational elongation | 3 | 0.048843 |
| Molecular function | ||
| Structural constituent of ribosome | 13 | 1.33 × 10−9 |
| Structural molecule activity | 14 | 3.15 × 10−5 |
| Translation factor activity and nucleic acid binding | 6 | 8.53 × 10−4 |
| Ribosome binding | 3 | 0.007084 |
| Transcription regulator activity | 17 | 0.017753 |
| Nuclear hormone receptor binding | 3 | 0.026903 |
| Ribonucleo protein binding | 3 | 0.026903 |
| Hormone receptor binding | 3 | 0.034687 |
| Cellular component | ||
| Ribosome | 14 | 1.78 × 10−9 |
| Ribonucleo protein complex | 17 | 3.00 × 10−7 |
| Non-membrane-bounded organelle | 26 | 0.003348837 |
| Intracellular non-membrane-bounded organelle | 26 | 0.003348837 |
| Golgi apparatus part | 7 | 0.007239565 |
| Ribosomal subunit | 4 | 0.013381826 |
| Endomembrane system | 10 | 0.019023994 |
| Small ribosomal subunit | 3 | 0.019898892 |
| Intracellular organelle lumen | 16 | 0.021428928 |
| Organelle lumen | 16 | 0.021891024 |
| Membrane-enclosed lumen | 16 | 0.028425775 |
| Golgi apparatus | 11 | 0.031864932 |
| Nuclear lumen | 13 | 0.032480924 |
| Nucleoplasm part | 9 | 0.038971919 |
| Proton-transporting V-type ATPase, V0 domain | 2 | 0.044274989 |
| Trans-Golgi network | 3 | 0.04858062 |
- GO analysis was performed using DAVID Bioinformatics Resources. Shown are GO biological process terms that significantly overrepresented (a modified Fisher Exact P-value < 0.05) for the LIGHT-responsive genes.
LIGHT Promoter is Methylated in Mouse BM-MSC
As LIGHT protein in the supernatants of LPS-stimulated MSC was not detected (Fig. 1C), we hypothesize that the promoter of LIGHT is silenced by methylation in these cells. To prove this hypothesis, methylation-specific primers were developed that flank the region in the LIGHT promoter. Using the MSP, we established the methylation status of the LIGHT gene promoter in the BM-MSCs. we found that the LIGHT gene promoter was partially methylated (Fig. 1E).
Proliferation Potential and Migration of Mouse BM-MSCs is Not Affected by LIGHT In vitro
We investigated whether or not LIGHT regulate MSC proliferation. In our study, we observed that MSC growth was not regulated by LIGHT (Fig. 2A); LIGHT regulate monocyte migration in other reports [Heo et al., 2010]. However, in our study, the regulatory role of LIGHT in SDF-1 dependent MSC migration was not observed (Fig. 2B).
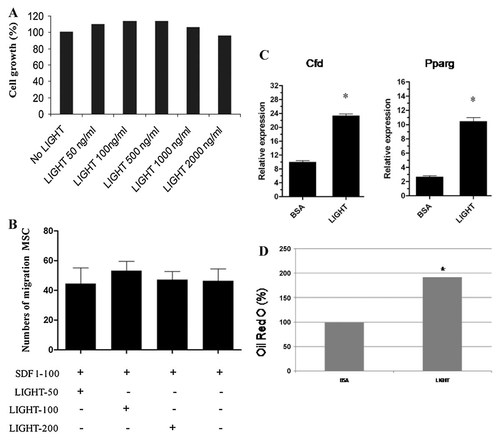
Mouse BM-MSC expansion, migration, and differentiation with LIGHT stimulation. A: The percentage of MSC expansion after LIGHT treatment with different concentration compared with LIHGT untreated control. B: The effect of LIGHT on MSC migration. MSC were treated with various concentration of LIGHT as described under the figure and cell numbers that migrate from the upper chamber to the lower chamber was counted on a Zeiss Axiovert 200 inverted fluorescence microscope. C: LIGHT promote the adipogenic differentiation potential of mouse BM-MSCs. Gene expression of adipocytic (Cfd: adipsin; Pparg: peroxisome proliferator-activated receptor gamma 2) markers in mouse MSC cultures after 3 days of differentiation procedures as measured by qRT-PCR. Mouse MSCs were cultured in the induction medium (15% normal horse serum and 100 nM dexamethasone in [Justesen et al., 2002] basal DMEM medium) [Justesen et al., 2002] together with LIGHT (100 µg/ml). Data are mean ± standard error (SEM). *P < 0.05, when compared with control (BSA). Differences between two groups were analyzed by the two-tailed Student's t-test and of more than two groups by one-way ANOVA with post hoc Dunnett's and Tukey's Multiple Comparison test. D: Oil Red O Staining Assay: quantification result showed much more lipid droplets in LIGHT group as compared with BSA (control) group (*P < 0.05).
LIGHT Promote Mouse BM-MSC Differentiate into Adipocyte
MSCs exhibit the potentials to differentiate into various cell lineages, including adipocyte [Justesen et al., 2002; Kanda et al., 2011]. The characteristics of mouse bone marrow MSCs during adipogenic differentiation are different from those of murine cells [Qian et al., 2010]. Our studies found that differentiation was further enhanced when LIGHT was applied in the cell differentiation culture. Cfd and Pparggenes are the specific markers of adipocyte. Addition of LIGHT in the culture increased Cfd and Pparg expression examined by real-time PCR (Fig. 2C). Moreover, Oil Red O Staining Assay showed much more adipocyte cell formation in LIGHT-treated MSCs than control groups which MSCs were treated by BSA (Fig. 2D).
NF-kB is Not Activated by LIGHT in Mouse BM-MSC
LIGHT stimulates NF-kB-dependent transcriptional activity in human hepatocyte [Matsui et al., 2002]. To investigate whether the downstream of LIGHT signal pathway is also affected in mouse BM-MSCs, we then examined NF-κB activation in mouse BM-MSC treated by LIGHT. Though we found that LIGHT activated NF-κB in immune cells in our previous report [Zou and Hu, 2005]; in the present study, we did not observe NF-κB was activated by LIGHT in mouse BM-MSCs (Fig. 3).
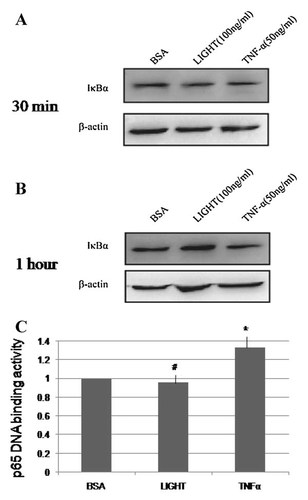
LIGHT and NF-κB activation in mouse BM-MSC cells. NF-κB is not activated by LIGHT in mouse BM-MSCs. Mouse BM-MSCs were exposed to LIGHT (100 ng/ml) for 30 min (A) or 1 h (B) after which samples were removed and analyzed by Western blot with anti-Iκ-Bα antibody. The blot showed that LIHGT did not induce the degradation of Iκ-Bα in mouse MSCs. Similar results were obtained in three separate experiments. p65NF-κB binding activity assay. The transcription factor NF-κB family activation assay was measured using the TransAM NF-κB family kit (C).
LIGHT Upregulates Gene Expression in Mouse BM-MSCs
GeneChip technology has proven to be an effective way to measure the co-expression of tens of thousands of genes [Upton et al., 2009]. GO analysis indicated genes differentially expressed when MSCs were treated by LIGHT (Table II). By gene chip assay, we identified a number of gene expression has been upregulated in mouse BM-MSCs (Fig. 4A); Meanwhile, a number of other genes expression has been downregulated (Fig. 4B). These downregulated genes include c-Jun and c-Myc. LIGHT regulating c-Jun and c-Myc expression in mouse BM-MSCs was also confirmed by real-time PCR assay (Fig. 4C).
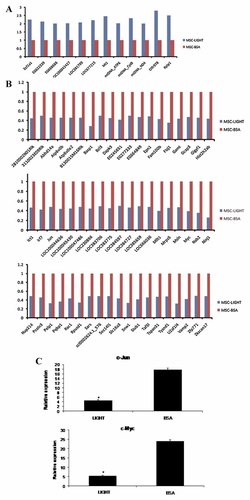
The gene expression up-regulation and down-regulation by LIGHT in mouse BM-MSCs. A: The up-regulation gene expression by LIGHT. B: The down-regulation gene expression by LIGHT. C: Real-time PCR for c-Myc and Jun.
DISCUSSION
The concept of the hematopoietic stem cell (HSC) niche was formulated by Schofield in the 1970s, as a region within the bone marrow containing functional cell types that can maintain HSC potency throughout life [Schofield, 1978]. This niche is composed of a specialized population of cells that plays an essential role in regulating adult stem cell self-renewal and differentiation. Osteoblasts function as a key component of the HSC niche (namely, the osteoblastic niche), controlling HSC numbers. HSCs also interact other stromal cells, including endothelial cells. In adults, osteoblasts, and hematopoietic cells, are closely associated in the bone marrow, suggesting a reciprocal relationship between the two. Sinusoidal endothelial cells in bone marrow have been revealed as an alternative HSC niche called the vascular niche [Yin and Li, 2006]. It was unclear before whether or not MSCs regulate HSCs directly; however, a recent report shows that MSCs co-localized with HSCs in the bone marrow, and maintains HSCs in bone marrow. These MSCs also regulate HSC/progenitor homing [Mendez-Ferrer et al., 2010]. So MSCs play an important role in hematopoietic regulations.
MSCs are heterogeneous subsets of stromal stem cells which can differentiate into of the mesodermal lineage, such as adipocytes, osteocytes, and chondrocytes [Uccelli et al., 2008]. Osteoblasts and endothelium constitute functional niches that support HSCs in mammalian bone marrow [Zhang et al., 2003]. Osteoblast derived from MSC promotes HSC expansion [Mishima et al., 2010]. Recently, it has been also suggested that bone marrow adipocyte may also regulate hematopoiesis which function as a negative regulator of the hematopoietic microenvironments; consequently, influence hematopoiesis [Naveiras et al., 2009]. Therefore, one of pattern of MSC in hematopoietic regulation might be through differentiation into adipocyte. In the present study, we identified that MSC differentiation into adipocyte can be regulated by LIGHT, a cytokine of TNF superfamily. This study then offers the evidence of LIGHT in hematopoietic regulation.
LIGHT has been well documented about its immune regulatory role. However, the knowledge about its role in stem cell regulation is limited. Our previous study showed its regulatory role in embryonic stem cell differentiation [Zou et al., 2006]. In the present study, we identified its regulatory role in adult stem cell differentiation. Due to MSC functions as stromal stem cell in bone marrow. This study then suggests that LIGHT may regulate hematopoiesis by action of MSC in bone marrow. So, we suggest that LIGHT is a regulator of hematopoietic microenvironment.
LIGHT protected liver injury induced by TNF-α [Matsui et al., 2002]. Moreover, primary hepatocytes express LT-ß receptor. LIGHT activates NF-κB in these cells which contributed to liver regeneration [Anders et al., 2005]. NF-κB can be activated by LIGHT in monocyte [Heo et al., 2008]. Our early work also shows that in dendritic cells NF-κB can be activated by LIGHT [Zou and Hu, 2005]. However, in the present study, we found NF-κB activation was not induced by LIGHT in MSC. The mechanism of this phenomenon is unclear and further studies are necessary to clarify it. It has been suggested that adult human MSC differentiation to the adipogenic lineage was regulated by MAPK [Jaiswal et al., 2000]. We identified c-Jun and c-Myc were upregulated in LIGHT-treated MSCs. We then suggested that LIGHT may regulate adipogenic differentiation of BM-MSCs through modulation of c-Myc and Jun. It is unclear whether MSCs autocrine LIGHT. Human MSCs secret TGF-ß1 but TNF-α was not detectable in the supernatant [Oh et al., 2009]. In the present study, we examined whether or not MSC express LIGHT. Our study demonstrated MSCs did not express LIGHT, even they were stimulated by LPS. Further analysis identified that LIGHT promoter was methylated in MSC; this may explained the results that LIGHT gene was not transcripted in MSCs. MSCs do not autocrine LIGHT which may potentially promote their differentiation provide the homeostasis of MSCs in bone marrow.
Adipocyte was recently identified on its role in hematopoietic regulation. The class dogma that adipocyte act as passive space filler in the marrow, but a recent study using fatless mice model confirm its role as a negative regulator of hematopoiesis. These adipocytes play as a negative role in bone marrow microenvironment [Naveiras et al., 2009]. We then suggest LIGHT may regulate hematopoiesis through action on MSCs in HSC niche.
In summary, our study described that MSCs express LIGHT receptor, named LT-ßR, a TNF receptor family member, where expression of the LIGHT receptor make the cells responsive to LIGHT.




