Myostatin inactivation increases myotube size through regulation of translational initiation machinery
Abstract
Myostatin deficiency leads in skeletal muscle overgrowth but the precise molecular mechanisms underlying this hypertrophy are not well understood. In this study, to gain insight into the role of endogenous myostatin in the translational regulation, we used an in vitro model of cultured satellite cells derived from myostatin knock-out mice. Our results show that myostatin knock-out myotubes are larger than control myotubes and that this phenotype is associated with an increased activation of the Akt/mTOR signaling pathway, a known regulator of muscle hypertrophy. These results demonstrate that hypertrophy due to myostatin deficiency is preserved in vitro and suggest that myostatin deletion results in an increased protein synthesis. Accordingly, the rates of global RNA content, polysome formation and protein synthesis are all increased in myostatin-deficient myotubes while they are counteracted by the addition of recombinant myostatin. We furthermore demonstrated that genetic deletion of myostatin stimulates cap-dependent translation by positively regulating assembly of the translation preinitiation complex. Together the data indicate that myostatin controls muscle hypertrophy in part by regulating protein synthesis initiation rates, that is, translational efficiency. J. Cell. Biochem. 112: 3531–3542, 2011. © 2011 Wiley Periodicals, Inc.
The control of muscle size is essential for proper development and homeostasis of adult musculature. This control involves in large part signaling molecules that promote the synthesis or breakdown of skeletal muscle cell proteins. One of the growth factor regulating the development and growth of skeletal muscle is myostatin. Myostatin, also called growth differentiation factor 8 (GDF-8), is a TGF-β family member and a negative regulator of muscle mass. Naturally occurring mutations as well as experimental knock-out of the myostatin gene lead to a hypermuscular phenotype in mice, sheep, dogs, cattle, and human [Grobet et al., 1997; Kambadur et al., 1997; McPherron et al., 1997; Schuelke et al., 2004; Clop et al., 2006; Mosher et al., 2007] resulting from both hypertrophy and hyperplasia. The fundamental role of myostatin in adult muscle homeostasis is demonstrated by the increased muscle mass of mice carrying a postnatal deletion of the myostatin gene or adult mice injected with various inhibitors of myostatin [Grobet et al., 2003; Whittemore et al., 2003; Wolfman et al., 2003; Lee et al., 2005]. Also myostatin inhibition demonstrates promising potential for clinical applications, a better understanding of the molecular mechanisms driving myostatin loss-induced hypertrophy is an important question to tackle when potential therapeutic approaches are investigated.
Although it is clear that myostatin regulates skeletal muscle hypertrophy, the mechanisms by which this regulation occurs are not fully elucidated. Myostatin can inhibit the activation and self renewal of satellite cells [McCroskery et al., 2003]. These adult stem cells are resident of the skeletal muscle fiber and are responsible for postnatal muscle cell growth and new muscle protein production [Moss and Leblond, 1971], supporting the relevance of satellite cell contribution to muscle hypertrophy consecutive to myostatin loss. However, contradictory observations suggest that the muscle hypertrophic phenotype of transgenic myostatin knockout mice involves little or no input from satellite cells [Amthor et al., 2009].
Increased protein synthesis is another mechanism important for hypertrophy. A number of studies have shown that skeletal muscle protein synthesis is sensitive to myostatin. In vivo increased rate of muscle protein synthesis (determined by measuring tracer incorporation after a systemic flooding dose of L-[ring-2H5] phenylalanined) was reported in neonatal rats receiving chronic infusion of the myostatin inhibitor follistatin [Suryawan et al., 2006], in constitutive myostatin knock-out mice as well as in mature mice treated with anti-myostatin antibodies [Welle et al., 2006, 2009]. Additionally, a decreased of protein synthesis was seen in C2C12 myotubes after the addition of recombinant myostatin protein [Ma et al., 2001].
Recent studies have indicated that a potential mechanism for increasing rate of protein is the activation of the mammalian Target of rapamycin (mTOR) pathway. mTOR resides in two large complexes (mTORCs) associated with either raptor protein, mTORC1, or rictor protein, mTORC2. It is well known that mTORC1 directly regulates protein synthesis and is sensitive to rapamycin [Wullschleger et al., 2006]. mTOR is an important regulator of translational initiation through at least two distinct but integrated pathways [Hay and Sonenberg, 2004; Tee and Blenis, 2005; Ma and Blenis, 2009]. On the one hand mTORC1 controls phosphorylation of 4E-binding protein 1 (4EBP1), releasing its inhibitory interaction with eukaryotic translation initiation factor 4E (eIF4E), allowing eIF4E to associate with eIF4G to form the active eIF4F complex, a necessary component of the 40S initiation complex controlling cap-dependent translation. On the other hand, mTORC1 regulates ribosome biogenesis by at least two different mechanisms: phophorylation of ribosomal protein S6 by the ribosomal S6 kinase 1 (S6K1) that stimulates the translation of mRNA with a 5′-oligopyrimidine tract (5′-TOP) [Meyuhas, 2000] and modulation of the synthesis of ribosomal RNA (rRNA) [Hannan et al., 2003].
Several lines of evidence have highlighted the cross-talk between myostatin and the mTOR pathway. Indeed, hypertrophied skeletal muscle of myostatin-deficient mice showed a higher activation status of the Akt/mTOR signaling components [Morissette et al., 2009; Lipina et al., 2010]. Transfection of a dominant negative myostatin receptor into adult skeletal muscle leads to an increase of muscle mass associated with increased phosphorylation of S6 a downstream effectors of Akt in the mTOR pathway [Sartori et al., 2009]. In contrast, down-regulation of the Akt/mTOR signaling pathway was seen in muscles after myostatin gene electrotransfer [Amirouche et al., 2009]. Few in vitro studies have further demonstrated that modulation of Akt/mTOR pathway is important for myostatin anti-hypertrophic effects [McFarlane et al., 2006; Morissette et al., 2009; Trendelenburg et al., 2009]. Most of these in vitro experiments were designed in “a gain- or loss-of-function approach” using the recombinant myostatin protein or pharmacological inhibitor of a myostatin receptor (ALK) or forced expression of a dominant negative form of ALK. However, these studies (both in vivo and in vitro) never addressed the effect of myostatin on the translational initiation machinery. In order to gain insight into the role of endogenous myostatin in the translational regulation, we used an in vitro model of cultured satellite cells derived from myostatin knock-out mice. Here, we demonstrate that myostatin-deficient myotubes exhibit an hypertrophic phenotype associated with a higher activation of the Akt/mTOR pathway, which validates this cellular model to study the effects of myostatin on the translational machinery. We further showed that genetic deletion of myostatin not only activates translational effectors but also increases the formation of the translational initiation complex and activates cap-dependent translation. This study documents for the first time that myostatin controls translational efficiency in skeletal muscle cells.
MATERIALS AND METHODS
Cell Culture Products
Dulbecco's modified Eagle's medium (DMEM), nutrient mixture F-12 (Ham), nutrient mixture F-10 (Ham) were purchased from Sigma (Saint Quentin Fallavier, France); horse serum and foetal calf serum (FCS) were purchased from Hyclone (Thermo Fisher Scientific, Waltham, MA).
Reagents
Recombinant Myostatin was purchased from R&D Systems (Mineapolis, MN). Insulin was purchased from SIGMA, Rapamycin from Tebu (Le Perray en Yvelines, France). The 35Stranslabel was obtained from Perkin Elmer (MA).
C2 Cell Culture
C2.7, a subclone of C2 myoblasts [Yaffe and Saxel, 1977; Pinset et al., 1988], maintained in a humidified incubator (37°C, 5% CO2), was grown in DMEM/Hamf12 containing 10% (v/v) FCS: growth medium (GM). For differentiation, confluent cells were cultured in DMEM containing 2% FCS (DM). Cells were kept in differentiation medium (DM) for 4 days.
Primary Mouse Muscle Cell Culture
Myostatin KO mice used in this study have been described previously [Grobet et al., 2003] and were generously provided by L. Grobet (Faculty of Veterinary Medicine, University of Liège, Belgium). These mice harbor a constitutive deletion of the third myostatin exon leading to the deletion of the entire mature C-terminal region of myostatin. Therefore, these animals are null for myostatin function. Animals were maintained on a 12:12 h light/dark cycle with free access to food. For our experiments mice were genotyped by polymerase chain reaction analysis of tail DNA to ensure the production of homozygous null for myostatin. In vitro studies were performed using male mice between 4 and 6 weeks of age. Briefly, murine myoblasts were isolated from the whole mice muscles of the paw after enzymatic digestion by pronase [Levin et al., 2001; Descamps et al., 2008]. Cells were plated at a density of 2 × 104 cells/cm2 on Matrigel®-coated Petri dishes (BD Biosciences, Francklin Lakes, NJ), in 80% Ham's F10 supplemented with 20% horse serum (HS) (complete medium). They were maintained at 37°C in a water-saturated atmosphere containing 5% CO2 in air. After 2 days, cells were washed with Ham's F10 and placed in complete medium supplemented with 5 ng/ml of basic fibroblast growth factor (bFGF). After 2 days, to achieve differentiation, satellite cells were switched to DM consisting of Ham's F10, and 20% HS. Cells were kept in DM for 3 days. This study was approved by the regional committee for Ethics in Animal experimentation in accordance with the international guidelines for animal welfare.
Immunofluorescence Assay and Analysis
Murine myotubes were fixed in phosphate buffer saline (PBS) containing 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) and permeabilized with PBS 0.5% triton X-100. Expression of myosin heavy chain was analyzed using mouse monoclonal antibody against monoclonal anti-fast MHC diluted 1/200 (Sigma). Primary antibody diluted in PBS/BSA was incubated for 1 h at 37°C, then washed in PBS, followed by a 30 min incubation with fluorescein-conjugated anti-mouse (1/100; Fluoprobes 488, Interchim, Montluçon, France). DNA was stained with Hoechst (0.1 mg/ml; Sigma).
The number of nuclei per myotube was counted. For each experimental situation, the number of nuclei in at least 200 myotubes were counted in three independent cultures. To estimate myotube size, the diameter of at least 200 myotubes was measured using PerfectImage software. The average diameter per myotube was calculated as the mean of three measurements taken along the length of the myotube.
Image Processing
Series of stack of fluorescently stained KO and WT myotubes were scanned using a confocal laser. Morphometry of myotubes was analyzed using extensive three-dimensional reconstructing software IMARIS. Confocal optical sectioning was performed with an LSM510 META confocal laser scanning (CLS) microscopy system (Carl Zeiss, Oberkochen, Germany) coupled to an inverted 200 M microscope (Zeiss) with Plan-NeoFluar objective (40× oil, N.A. = 1.3) at the UMR866 Laboratory, INRA Montpellier. The refraction index of immersion media (Zeiss 518F) was 1.518. Theoretical xy- and z-axes resolutions were 0.095 and 0.3 µm, respectively. Frame size of the image was 193.9 × 193.9 µm with an 12-bit color depth. Confocal images were taken with 0.3 µm Z-step size. The 3D structure of the myotubes was reconstructed from CLS images using IMARIS software (Bitplane). CLS images from 6.6 to 18.9 µm in depth were reconstructed for visualizing the myotubes. Isosurface of a selected myotube was performed by IMARIS software which allowed us to calculate the volume.
Western Blot
For Western blot analysis, protein lysates were prepared using lysis buffer (20 mM Tris pH 7.5, 10 mM NaCl, 1 mM DTT, 50 mM NaF, 1 mM Na3VO4, and protease inhibitor cocktail set [Réf: P2714 Sigma)] and protein concentrations were determined using the BioRad DC kit (BioRad, Hercules, CA). Fifty microgram of total proteins were denaturated by mixing with an equal volume of Laemmli buffer (100 mM Tris–HCl, pH 6.8, 4% SDS, 20% glycerol, 200 mM DTT, bromophenol blue). Linearization was achieved by heating the samples for 5 min at 100°C. The samples were loaded with pre-stained molecular mass markers (BioRad). The proteins were separated on SDS–polyacrylamide gels. The gels were run in an electrophoresis unit at constant voltage (i.e., 140 V). Proteins were then transferred from the gels onto nitrocellulose membranes (Protran BA 85, 0.45 µm; Whatman, Dassel, Germany) using a transfer buffer containing 25 mM Tris Base, 192 mM glycine and 200 mM methanol at constant electric charge (i.e., 300 mA) for 1 h. The membranes were incubated on a shaker with a blocking buffer (5% non-fat dry milk in TBS-Tween: 50 mM Tris Base, 150 mM NaCl, HCl, adjusted to pH 7.5, 0.1% v/v Tween 20). The membranes were then incubated with the following primary antibodies overnight in the following blocking buffer (TBS, 0.1% Tween, 5% BSA) at 4°C: monoclonal anti-α-tubulin (1/5,000) from Sigma; monoclonal anti-Raptor (10E10) (1/200) from Santa-Cruz (Santa Cruz, CA); anti-Akt, anti-phospho-Akt (Ser473), anti-mTOR, anti-phospho-mTOR (Ser24/48), anti-S6 ribosomal protein, anti-phospho-S6 ribosomal protein (Ser235/236), anti-4E-BP1, anti-phospho-4E-BP1 (Thr37/46), and anti-eIF4G were from Cell Signaling Technology (Danvers, MA) and were all used at a dilution of 1/1,000. After washing, primary antibodies were detected with peroxidase-conjugated secondary antibodies (Amersham, GE Healthcare, Buckinghamshire, UK) at a dilution of 1/5,000 and an ECL kit. Blots were exposed with Amersham Hyperfilm ECL (GE Healthcare) films. Signals were quantified by gel scan and with the ImageJ software.
Monitoring Cellular Translation Efficiency
Measurement of protein synthesis by 35S-labeling
Primary skeletal muscle myotubes (from wild-type and myostatin KO mice) at 3 days of differentiation were washed once with DMEM lacking cysteine and methionine, and the medium was replaced with DMEM lacking cysteine and methionine supplemented with 20% HS. After incubation for 1 h, 50 µCi of 35S (Perkin Elmer) was added to the cells for 4 h. Myotubes were washed once with ice-cold PBS and lysed as described above for Western blotting. 35S-labeled proteins were quantified by scintillation counting with a PACKARD TRICARB 1600 counter. Following separation by SDS–PAGE and treatment with an amplifier fluorographic reagent (GE Healthcare), 35S-labeled proteins were visualized by autoradiography.
Determination of protein, DNA, and RNA content
This experiment was done in parallel with the analysis of protein synthesis by 35S methionine incorporation. Primary skeletal muscle myotubes (from wild-type and myostatin KO mice) were collected at 3 days of differentiation. For DNA, RNA, and protein analysis cells were cultured in six-well plates, of which three wells were used for RNA or protein and the other three were used for DNA quantitation. The experiments were performed in triplicate (n = 3/plate). DNA was isolated using the DNA isolation Genelute kit (Sigma) according to the manufacturer's guidelines and quantified on spectrophotometer at 260 nm. The protein quantitation was evaluated using the BioRad DC kit (BioRad). RNA was prepared using the RNeasy kit (Qiagen, Hilden, Germany). RNA concentration was determined on spectrophotometer at 260 nm. Quality of the RNA was controlled measuring A260/A280 ratio. The integrity of the RNA was assessed visually using agarose gel fractionation of the 28S and 18S rRNA subunits.
Polysome analysis
For polysome analysis, cells were washed with PBS and treated with cycloheximide (100 µg/ml in PBS) for 15 min at 37°C immediately before collection for lysis. Cells were scraped in 5 mM Tris–HCl (pH 7.5), 1.5 mM KCl, 2.5 mM MgCl2, 2 mM DTT, 100 µg/ml cycloheximide, and 80 U/ml RNase inhibitor (Promega Corporation, Madison, WI). Samples were incubated on ice for 5 min before addition of 0.5% Triton X-100 and 0.5% sodium deoxycholate. Following a 5 min incubation on ice, lysates were spun at 14,000 rpm for 5 min at 4°C. An aliquot of the supernatant was removed to measure protein concentration. Polysomes were fractionated through a 10–50% continuous sucrose gradient (10 mM Tris–HCl, pH7.5, 100 mM KCl, 5 mM MgCl2) and centrifuged at 35,000 rpm in a SW41 rotor at 4°C for 120 min. Following centrifugation, fractions were collected and monitored by UV spectroscopy at 260 nm.
Cap pull-down assay
Primary skeletal muscle myotubes (day 3 of differentiation) were lysed in cap lysis buffer (140 mM KCl, 10 mM Tris pH 7.5, 1 mM EDTA, 4 mM MgCl2, 1 mM DTT, 1% NP-40, 1 mM sodium orthovanate, 50 mM β-glycerophosphate, 10 mM NaF, and proteases inhibitors). Seventy microliter of detergent-free cap lysis buffer and 20 µl of pre-washed cap beads (m7GTP Sepharose 4B from GE Healthcare) were added to 300 µg of cleared lysate and incubated at 4°C overnight with tumbling. The beads were washed twice with 400 µl of cap wash buffer (cap lysis buffer with 0.5% NP-40 instead of 1%) and twice with 400 µl of ice-cold PBS. The beads were boiled at 100°C in SDS–PAGE sample buffer and the retained proteins were analyzed by Western blot.
Transient transfections and luciferase activity measurement
Primary murine myoblasts were plated at a density of 2 × 104 cells/cm2 (in 12-multiwell plates) and of respectively in GM. After 3 days myoblasts were transfected using a turbofect reagent as described by the supplier (Thermo Fisher Scientific) with 1 µg of the luciferase reporter construct (pHRluc-C2). Twenty-four hours after transfection myoblasts were switched to DM. Two days after, myotubes were treated for 24 h with insulin (10 nM) or pretreated with rapamycin (40 nM) and stimulated with insulin for and additional 24 h. The myostatin KO myotubes transfected with luciferase reporter construct were incubated in the presence or absence of recombinant myostatin (2 µg/ml) for 24 h before harvesting. Forty-eight hours after transfection, cells were harvested for luciferase assay. Luciferase activity was measured using the luciferase assay kit reagent as described by the supplier (Promega Corporation) and corrected with respect to protein content of the extracts. Duplicate samples were subjected to qRT-PCR to assess levels of Luc reporter RNA to normalize translation results.
For bicistronic Luciferase assay, primary murine myoblasts (WT and myostatin KO) and C2.7 myoblasts were transfected with a bicistronic luciferase reporter plasmid pRL-HCV-FL, which directs cap-dependent translation of the Renilla luciferase (RL) gene and cap-independent HCV IRES-mediated translation of the firefly (FL) gene [Roux et al., 2007]. Twenty-four hours after transfection myoblasts were switched to DM for 2 days. The C2.7 myotubes and the myostatin KO myotubes were incubated in the presence or absence of recombinant myostatin (2 µg/ml) for 24 h. Three days after transfection cells were harvested, and the Renilla and Firefly luciferase activity was measured using the Dual-luciferase kit (Promega Corporation). Cap-dependent translation levels in transiently transfected cells were calculated by normalizing Renilla luciferase levels to control (IRES-directed) firefly luciferase levels.
Real-Time PCR
Total RNA was extracted from primary skeletal muscle myotubes using standard manufacturer's protocols (RNeasy plus mini kit; Qiagen). First strand cDNAs were synthesized on 1 µg of total RNA using SuperScript first-strand synthesis system (Invitrogen, Carlsbad, CA) with oligo(dT) and random primers. Reverse transcription (RT) products were then 1/5-fold diluted in nuclease-free water and kept at −20°C until use. qPCR was performed using the SYBR Green chemistry on a MiniOpticon thermal cycler (BioRad). qPCR mixture contained the following: 1 µl of diluted RT product, 250 nM of each specific primers, 1× IQ SYBR Green Supermix (Bio-Rad; containing iTaq DNA polymerase, reaction buffer 2×, dNTPs, 6 mM MgCl2, SYBR Green I, fluorescein, and stabilizers) and nuclease-free water to obtain a final volume of 15 µl. PCR program was the following: initial Taq polymerase activation at 95°C for 3 min, followed by 40 cycles at 95°C for 10 s and 60°C (Akt1/PKBα, Akt2/PKBβ, mTOR, RpS6, Luc) or 58°C (Rplp0) for 25 s. Melting temperatures were measured by gradually heating from 65 to 95°C. Each sample was tested in duplicate, and negative controls containing water in place of cDNA template were included in each run.
Sizes of amplicons were verified by agarose gel electrophoresis and specificity of the target by comparing the melting temperature obtain by qPCR to the predicted one calculated by Poland's algorithm. Crossing threshold values (Ct) were measured and converted into mRNA starting quantities by CFX analysis software (BioRad) using standard curves. Expression levels of genes of interest were evaluated by qPCR and data were normalized to the expression level of the reference gene (Rplp0) [Stern-Straeter et al., 2009]. Fold difference in genes of interest expression were then calculated relatively to an untreated control.
PCR primers (see table below) were designed using the LightCycler probe design software 2.0 (Roche Diagnostics, Meylan, France) and tested for homology with other sequences at the NCBI gene BLAST website.
Primer Table
| Gene | Primer sequences (position on mRNA) | Accession numbers | Amplicon sizes (bp) |
|---|---|---|---|
| Akt1 | F: ACTTCCTCCTCAAGAACGAT (447-466) | NM_001165894.1 | 159 |
| R: GCAGGCAGCGGATGATAA (588-605) | |||
| Akt2 | F: CTTCCTTACAGCCCTCAAGT (793-812) | NM_007434.3 | 155 |
| R: CTGACACAATCTCTGCTCCATAAA (924-947) | |||
| mTOR | F: CAGACTGGCTCTTGCTCATAA (5112-5132) | NM_020009.2 | 155 |
| R: GCTGGAAGGCGTCAATC (5250-5266) | |||
| RpS6 | F: CGTCGGTTGGGACCTAA (432-448) | NM_009096.3 | 163 |
| R: GGACACGAGGAGTAACAAGT (575-594) | |||
| hRLuc | F: ACTACAACGCCTACCTGAGA(4029-4048) | AY995140.1 | 155 |
| R: GTCCTCCTGGCTGAAGTG (4166-4183) | |||
| Rplp0 | F: GGGCATCACCACGAAAATC (544-562) | NM_007475.5 | 168 |
| R: GCCGTTGTCAAACACCTG (692-709) |
Statistical Analysis
The results are expressed as mean ± SEM. Statistical analysis was performed by Student's t-test using Instat 3.01 software (GraphPad, San Diego, CA). Statistical significance was set at P-values < 0.05.
RESULTS
Hypertrophy of Myostatin-Deficient Myotubes
To examine the role of myostatin in the control of muscle size, we set up primary cultures of skeletal muscles isolated from myostatin KO (KO) and wild-type (WT) mice. Briefly, we differentiated confluent myoblasts to multinucleated cells (myotubes) that express muscle-specific structural proteins by mitogen removal. The resulting cultures were examined for myotube size measurement by immunofluorescence analysis using anti-myosin heavy chain antibody. Three days after switching confluent myoblast to differentiation media, both KO and WT primary cell cultures were fully differentiated, as assessed by morphology (Fig. 1A). However, KO myotubes displayed a 15% increased diameter when compared to WT myotubes (Fig. 1B). We further explored the morphology and morphometry of reconstructed three dimensional (3D) of KO and WT myotubes (Fig. 1C). We found that the deletion of myostatin induced a 88% increase in myotube volume consistent with the crucial role of myostatin on adaptation to muscle hypertrophy (Fig. 1D). Moreover, the number of nuclei per myotube did not significantly differ between WT and KO cells supporting the fact that KO muscle fiber hypertrophy was due to a better cytoplasmic growth rather than to improved cell fusion (Fig. 1E,F).
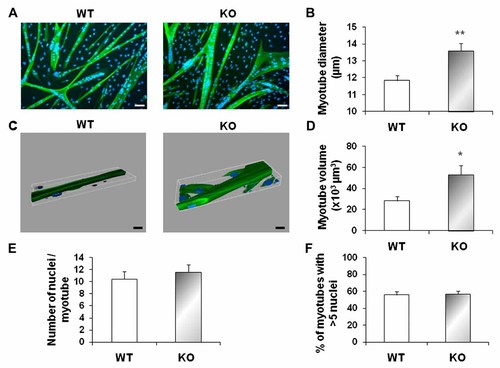
Cell size control in primary muscle cultures lacking myostatin. A: Primary muscle cells from wild-type (WT) and myostatin KO mice (KO) were allowed to differentiate for 3 days and then immunostained with anti-myosin heavy chain (MHC). Nuclei were revealed by Hoescht staining. Scale bar, 20 µm. B: Myotube diameter of myostatin-deficient cells is increased when compared to wild-type cells. Histograms are means ± SEM for six assays. Independent cell cultures were obtained from at least five mice of the indicated genotype. At least 300 myotubes were analyzed for WT and KO mice. **P < 0.01; n = 6. C: Three-dimensional reconstruction of WT and KO myotubes by IMARIS software. We used confocal light scanning images from 6.6 to 18.9 µm for the reconstruction. Scale bar, 20 µm. D: Myotube volume of myostatin-deficient cells is increased compared to myotube volume of wild-type cells. Histograms are means ± SEM for at least six independent cell cultures. Scale bar, 20 µm. At least 12 myotubes were analyzed for WT and KO mice. *P < 0.05; n = 6. E: Wild-type (WT) and myostatin KO cultures (KO) have a similar number of nuclei per myotube. The average number of nuclei per myotube was determined by counting 1,000 nuclei as described in Materials and Methods section. Histograms are means ± SEM for at least three assays. F: The percent of myotubes having more than five nuclei is indicated. At least 200 myotubes were analyzed for each culture.
Myostatin Deletion Up-Regulates Akt/mTOR Signaling in Skeletal Muscle Cells
We examined the effect of myostatin deficiency on Akt/mTOR signaling pathway known to play a prominent role in muscle hypertrophy [Bodine et al., 2001]. For this purpose, WT and KO myotubes were stimulated or not with 10 nM of insulin for 15 min and cell lysates were examined by Western blotting analysis for expression and phosphorylation of Akt signaling proteins. As shown on Figure 2A and quantified in 2B, Akt, mTOR, and rpS6 protein abundance was higher in KO myotubes when compared to WT myotubes. To further investigate the mechanism responsible for the increase in Akt, mTOR, and rpS6 protein level, all three were examined at the mRNA level. No significant variation in Akt, mTOR, and rpS6 mRNA were found between KO and WT myotubes (Fig. 2C) suggesting that myostatin deletion regulates Akt, mTOR, and rpS6 expression at the translational level.
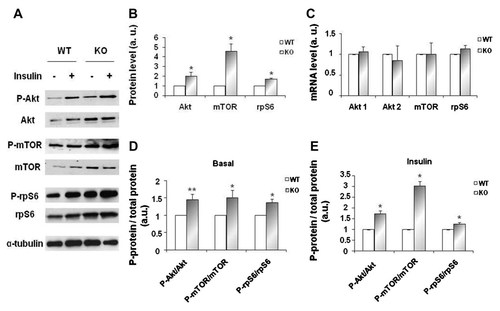
Increased expression and activation of Akt/mTOR signaling pathway in myostatin KO myotube. A: Western blot analysis of protein extracts from wild-type (WT) and myostatin KO myotubes (KO) with (+) or without (−) insulin stimulation (10 nM for 15 min). Cells extracts were then prepared and immunoblotted for both the expression and phosphorylation of Akt, mTOR, rpS6 and for α-tubulin expression (as a loading control). B: Akt, mTOR, and rpS6 protein levels are increased in myostatin KO myotubes. Quantification of Akt, mTOR, and rpS6 protein levels expressed relative to α-tubulin levels in WT and KO myotubes in basal conditions. Histogram shows the average ± SEM for at least five independent cultures. *P < 0.05 versus wild-type culture (a.u., arbitrary units). C: Akt1, Akt2, mTOR, and rpS6 mRNA levels are not changed upon myostatin deletion. Transcriptional expression of Akt 1, Akt 2, mTOR, and rpS6 were measured by qRT-PCR on total RNA extracted from WT and KO myotubes in basal conditions as in (B). Histogram shows the average ± SEM for at least four independent cultures (a.u. arbitrary units). D: Effect of myostatin deletion on the activation of Akt, mTOR, and ribosomal protein S6 in basal conditions. Quantification of phospho-protein to total protein ratio in basal conditions, from five cultures established from WT and KO mice and conducted in parallel. The total protein level was first expressed to α-tubulin protein level. Histogram shows the average ± SEM for at least five independent cultures. *P < 0.05 and **P < 0.01 versus wild-type culture (a.u. arbitrary units). E: Effect of myostatin deletion on the activation of Akt, mTOR, and ribosomal protein S6 upon insulin stimulation. Quantification of phospho-protein to total protein ratio upon insulin stimulation, from three independent cultures established from WT and KO mice and conducted in parallel. The total protein level was first expressed to α-tubulin protein level. Histogram shows the average ± SEM for at least three independent cultures. *P < 0.05 versus wild-type culture (a.u. arbitrary units).
Consistently, KO myotubes exhibited a significant increase in baseline phosphorylation of Akt, mTOR, and rpS6 when compared to WT myotubes. Insulin stimulated the phosphorylation of Akt as well as mTOR and rpS6. Accordingly, insulin stimulation resulted in a more robust increase of Akt, mTOR, and rpS6 protein phosphorylation in KO versus WT myotubes as determined by the measurement of Phospho versus total protein ratio (Fig. 2D,E). These results indicate that the observed increase in Akt/mTOR pathway activation may be due in part by a change in the ratio between phosphorylated species to total protein rather than to the enhanced signaling protein levels.
Myostatin Deletion Increases Translation Initiation and Protein Synthesis
Since myostatin deletion results in Akt/mTOR pathway up-regulation we decided to measure the rate of protein synthesis in KO cells. KO myotubes showed a 21% increased protein synthesis rates as demonstrated by 35S methionine incorporation when compared to WT myotubes (Fig. 3A,B). Total amount of protein per DNA was increased in myostatin KO myotubes (Fig. 3C). Similarly to the protein changes, cellular content in RNA towards DNA was increased in KO myotubes (Fig. 3D). Of note, the average DNA content was not different between WT and KO myotubes, and the values were 4.13 ± 0.46 for WT myotubes and 4.46 ± 0.64 for KO myotubes (P > 0.05). The protein synthesis efficiency appeared to be sensitive to myostatin re-introduction. Indeed, addition of exogenous recombinant myostatin (2 µg/ml, 24 h) decreased the protein synthesis rate in KO myotubes to the level of WT myotubes in basal conditions (Fig. 3A,B). The addition of recombinant myostatin also decreased both total cellular protein and RNA content (Fig. 3C,D). We next used polysome profiling to determine the step if any of translation that is improved as a result of myostatin deletion. We analyzed polysome seggregation in WT and KO cells using sucrose gradient centrifugation which allows to separate polysomes (P) from the 40S/60S free ribosomal subunits and 80S ribosomes, that is, the subpolysomal fraction (SP). As shown in Figure 3E,F, the P/S ratio was higher in KO cells compared to WT cells. This increase was mirrored by a decrease in 80S monomers (Fig. 3E) indicative of a better initiation of translation. Interestingly, myostatin addition tended to decrease the ratio of P/S in KO cells (Fig. 3F) (P = 0.06). Collectively, our data suggest that translation initiation is improved in the absence of myostatin.
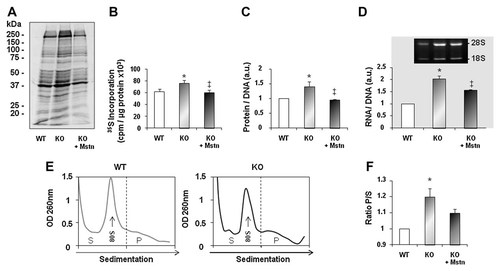
Myostatin-deficient muscle cells display increased protein synthesis rate, increased polysome formation as well as RNA content. A: Mouse primary cultured satellite cells from wild-type (WT) or myostatin KO mice (KO) were allowed to differentiate for 3 days prior to labeling new protein synthesis with 35S Methionine. KO myotubes were treated or not with recombinant myostatin (2 µg/ml) (+Mstn) for 24 h before labeling. Newly synthesized proteins were separated by SDS–PAGE and visualized by autoradiography. B: Protein synthesis rate was measured as the incorporation of 35S methionine into protein. The results are presented as CPM of 35S methionine incorporated into protein corrected for the total amount of protein in cell homogenates. Each experiment was done in triplicate, and histograms are mean ± SEM for four independent cell cultures. *P < 0.05 versus WT culture, ‡P < 0.05 versus untreated KO cultures. C: Protein quantity was evaluated by measuring protein and DNA concentrations and expressed as the ratio of protein to DNA. The ratio is expressed relative to control wild-type cells (arbitrary taken as 1). Each experiment was done in triplicate, and histograms are mean ± SEM for four independent cell cultures. *P < 0.05 versus WT culture, ‡P < 0.05 versus untreated KO cultures (a.u. arbitrary units). D: Total RNA content was evaluated by measuring RNA and DNA concentrations and expressed as the ratio of RNA to DNA. The ratio is expressed relative to control wild-type cells (arbitrary taken as 1). Each experiment was done in triplicate, and histograms are mean ± SEM for four independent cell cultures. *P < 0.05 versus WT culture, ‡P < 0.05 versus untreated KO cultures (a.u. arbitrary units). Total RNA of cell cultures from WT and KO mice cultured as described in (A) were analyzed on agarose gel to visualize the 18S and 28S rRNAs. Inset: Typical agarose gel fractionation of total RNA is shown. E: Cellular extracts from WT and KO cells were size-fractionated by centrifugation through sucrose gradients (10–50%) as described in Materials and Methods section. The absorbance of polysomes and subpolysomal particles was monitored at 260 nm and the vertical dashed line denotes the separation between the polysomal (P) and subpolysomal fractions (S). A representative absorbance profile from the WT and KO gradients is shown. F: Quantification of the ratio of P/S in WT cells and in KO cells treated or not with recombinant myostatin (+Mstn) cultured as in (A). The assay was performed by calculating the area under the curves using ImageJ (NIH). The ratio P/S is expressed relative to control WT cells (arbitrary taken as 1). Data are shown as mean ± SEM and represent an average of three independent experiments for each culture. *P < 0.05 versus WT culture.
Myostatin Controls the Recruitment of Regulatory Factors to the Translational Pre-Initiation Complex
The above data predict that myostatin (through changes in the activities of downstream effectors of mTOR signaling) could modulate the translational machinery. Actually, activation of mTOR leads to sequential hyperphosphorylation of 4E-BP1 and to its release from eIF4E, allowing eIF4E to bind eIF4G and form active initiation complexes. To assess the impact of myostatin on the formation of the initiation complexes, we performed pull-down experiments with methyl-GTP (m7-GTP) linked to Sepharose beads which mimic the 5′ mRNA cap to precipitate cap-interacting proteins. We observed that endogenous eIF4G, raptor (core component of mTORC1 complex) and P-rpS6 were highly bound to the m7-GTP cap complexes in KO myotubes compared to WT myotubes (Fig. 4A,C). In addition, the association of 4E-BP1 to the cap was less sustained in KO myotubes than in WT. The impact of myostatin on the regulation of the translational machinery was further demonstrated by reconstitution experiments (Fig. 4B). Two days after differentiation, KO cells were treated for 24 h with recombinant myostatin (2 µg/ml). We found that treatment with recombinant myostatin of KO myotubes significantly reduced the recruitment of eIF4G to the m7-GTP cap complex, abolished the recruitment of raptor and P-rpS6 to the m7-GTP cap complex while it increased the retention of 4E-BP1 compared to untreated KO myotubes (Fig. 4B,C). These results show that myostatin can regulate the recruitment and consequently the assembly of the translational initiation complex in skeletal muscle cells.
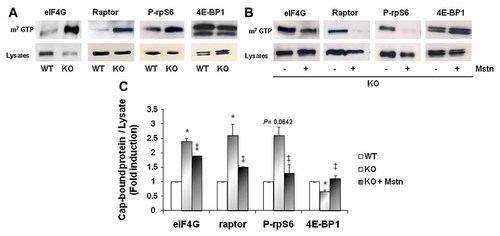
Myostatin regulates the recruitment of translational components to the 7-methylguanosine cap. A: Binding of translational components to the 7-methylguanosine cap is increased in KO myotubes. Mouse primary cultured satellite cells from wild-type (WT) or myostatin KO mice (KO) were allowed to differentiate for 3 days. Cap-binding proteins in lysates were precipitated using m7GTP-Sepharose-coated beads and analyzed by Western blot with the indicated specific antibodies. One representative experiment out of three is shown. B: Myostatin decreases the binding of translational components to the 7-methylguanosine cap. The same experiment as in (A) is presented with lysates from KO cells treated or not with recombinant myostatin (2 µg/ml) (+Mstn) for 24 h before harvesting cells. One representative experiment out of three is shown. C: Quantification of cap-bound protein to total protein in lysate ratio. The results are presented as a mean ± SEM (n = 3). *P < 0.05 KO versus WT cultures, ‡P < 0.05 myostatin-treated versus untreated KO cultures.
Myostatin Deletion Enhances Cap-Dependent Translation
Also our experiments suggested a role for myostatin in translation initiation, we next performed transient transfection experiments to investigate the consequences of myostatin inhibition on cap-dependent translation using luciferase reporter gene. To address this question we first transfected the luciferase reporter plasmid (containing the renilla luciferase cDNA downstream of a CMV promoter: phRLUC-C2 which allows cap-dependent expression of luciferase as previously described [Yang et al., 2004; Xu et al., 2010]) in wild-type and in myostatin KO cells supplemented or not with recombinant myostatin. The effect of insulin and rapamycin on cap-dependent translation efficiency was measured as well. As expected, the luciferase activity relative to luciferase mRNA levels was increased upon insulin stimulation and decreased in the presence of rapamycin (Fig. 5A). It is of note that both baseline and insulin-stimulated translation initiation were found to be higher in KO cells when compared to WT cells which is consistent with the enhanced mTOR activation in KO cells versus WT cells. Luciferase mRNA levels did not show any significant variations altered in all tested conditions (Fig. 5B). Interestingly, we observed that addition of recombinant myostatin led to a decrease in translation rates in KO cells. Secondly, we used a bicistronic luciferase reporter system that distinguishes cap versus cap-independent translation by separating Renilla luciferase from firefly with the HCV IRES as previously described [Roux et al., 2007] (Fig. 5C). Therefore, the Renilla/Firefly ratio would determine the cap-dependent translation ratio in cells. Using this system, we confirmed that cap-dependent translation was increased in KO cells when compared to WT cells (Fig. 5D). Accordingly, addition of recombinant myostatin on KO cells down-regulated the cap-dependent translation. The addition of recombinant myostatin on C2.7 cells also resulted in a decrease of the cap-dependent translation (Fig. 5E). Therefore, these results indicate that myostatin expression modulates cap-dependent translation.
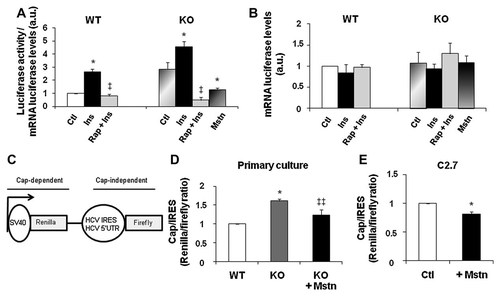
Myostatin regulates cap-dependant translation of luciferase gene. A,B: Primary muscle cells from wild-type mice (WT) and myostatin KO mice (KO) were transfected with the phRLUC-C2 plasmid. The cells were harvested 48 h after transfection. Twenty-four after transfection cells were stimulated with insulin (Ins) or pretreated with rapamycin (Rap) and stimulated with insulin for an additional 24 h. The KO cells transfected with luciferase cDNA gene reporter were treated or not with recombinant myostatin (2 µg/ml) (+Mstn) for 24 h before harvesting. Luciferase activities were expressed relative to that in control WT cells (arbitrary taken as 1). All luciferase values were normalized by qRT-PCR analysis of actual LUCmRNA present in lysates (B). Independent cell cultures were obtained from at least five mice of the indicated genotype. Results are presented as the mean ± SEM of data from three independent experiments, each of which was conducted in duplicate. *P < 0.05 versus control culture, ‡P < 0.05 versus insulin-treated cultures (a.u. arbitrary units). C: Structure of the bicistronic reporter plasmid allowing cap-dependent expression of renilla luciferase and expression of firefly luciferase dependent on HCV IRES. D: Primary muscle cells from wild-type mice (WT) and myostatin KO mice (KO) were transfected with the bicistronic reporter vector. The cells were harvested 48 h after transfection. The KO cells transfected with luciferase bicistronic gene reporter were treated or not with recombinant myostatin (2 µg/ml) (+Mstn) for 24 h before harvesting. The ratio of Renilla (Cap-dependent) to Firefly (IRES-dependent) luciferase activity was calculated. Data are presented as the mean ± SEM from three independent experiments carried out in duplicate. *P < 0.05 versus WT culture, ‡‡P < 0.01 versus untreated KO cultures. E: C2.7 myoblasts were transfected with the bicistronic reporter vector. Twenty-four hours after transfection, cells were incubated in presence (+Mstn) or in absence (Ctl) of recombinant myostatin for 24 h. Luciferase activities were measured as in (D). Data are presented as the mean ± SEM from four independent experiments carried out in duplicate. *P < 0.05 versus untreated C2.7 cells.
DISCUSSION
In this study, we explored the mechanism by which myostatin controls skeletal muscle hypertrophy. Using an in vitro model of cultured satellite cells derived from myostatin knock-out mice we demonstrate: (1) that myostatin deficiency leads to myotube hypertrophy associated with an enhancement of Akt/mTOR pathway; (2) that myostatin deficiency leads to increased protein synthesis and translation initiation; (3) and finally that the cap-dependent translation as well as the recruitment of translational components to the 7-methylguanosine cap complexes were more efficient in myostatin-deficient cells while myostatin restoration in these cells decreases the assembly of the translation initiation complexes as well as protein synthesis. These findings provide evidence that at least part of the mechanism by which myostatin regulates muscle hypertrophy is by modulating the rate of protein synthesis initiation, an important limiting step in the translational efficiency. Together, these findings highlight an important link between myostatin and the translational machinery, and extend our current understanding of the mechanisms by which myostatin deficiency promotes muscle cell hypertrophy.
Gain and loss of function approaches have reported a connection between myostatin and Akt/mTOR signaling. To address this question, most of the in vitro experiments used transfection technique to perturb the pathway (systemic overexpression of myostatin or inhibition of myostatin by expressing inactive activin receptor dnActRIIB). These studies never addressed the effect of myostatin downstream the Akt/mTOR pathway, that is, on translational machinery. In addition, the existence of a differential response to exogenous and endogenous myostatin has been demonstrated in cultured myoblasts suggesting that myostatin actions are exerted in vivo in an autocrine fashion [Rios et al., 2004]. This led us to explore the role of endogenous myostatin in the control of translation. Using an in vitro model of cultured satellite cells derived from myostatin KO mice we established that genetic deletion of myostatin increased translational efficiency by regulating protein synthesis initiation rates. We emphasize that this is the first demonstration of the role of endogenous myostatin in translation.
Extensive investigations pinpoint the central role of Akt/mTOR signaling pathway for both in vitro and in vivo skeletal muscle growth [Bodine et al., 2001; Rommel et al., 2001]. Previous in vivo studies have suggested that inhibition of myostatin activity may lead to increased myofibrillar protein synthesis and skeletal muscle overgrowth. In order to gain insight into the role of myostatin in these processes we used primary skeletal muscle cells isolated from myostatin KO mice. Our results show that ex vivo myostatin KO myotubes display a 88% volume increase when compared to WT myotubes demonstrating that hypertrophy due to myostatin deficiency is preserved in vitro. We also observed that the myostatin inhibition causes an 88% increase in the volume of the myotubes while there is no difference in the number of nuclei per myotube nor in the % of myotubes with more than five nuclei. Since the volume is increased but the number of nuclei remains constant then the myonuclear domain size (i.e., cytoplasmic volume-to-myonucleus ratio) would be larger in KO myotubes compared to WT myotubes. This suggests a dysregulation of the myonuclear domain size. Further studies should be designed to address this point.
At the molecular level, the myotube hypertrophy due to myostatin deficiency is associated with an increase in the expression and activation of Akt signaling protein already in basal culture conditions. In response to insulin-stimulation, activation of the Akt, mTOR, and rpS6 pathway activation is more robust in the myostatin KO myotubes. The myostatin KO myotubes are thus characterized by an upregulation of Akt/mTOR signaling pathway. These data are in agreement with previous studies showing an activation of this anabolic pathway in skeletal muscle where myostatin is inhibited [Welle et al., 2006, 2009; Morissette et al., 2009; Lipina et al., 2010]. We next showed that deletion of myostatin is associated with an increased expression of Akt, mTOR, and rpS6 proteins likely to result from activation of the translational but not the transcriptional machinery in myotubes. Morrissette et al. recently reported an increase of Akt protein secondary to an increase of Akt mRNA level in myostatin KO mice, whereas no change in total Akt protein was observed in C2C12 infected with adenoviral constructs expressing myostatin or the inhibitory myostatin propeptide [Morissette et al., 2009]. Such discrepancies may reflect (i) different adaptation of signaling pathway in muscles and muscle cells to the chronic loss of myostatin and (ii) different cell lines used in these studies. In the present study, we report that in muscle cells the effect of chronic myostatin loss on expression of Akt/mTOR signaling components is most likely to occur at the translational level.
Previous observations already try to establish a link between myostatin and protein synthesis. Reports where the activation state of the Akt/mTOR signaling pathway—the major regulator of protein synthesis in muscle cells—was investigated showed a decreased activation both in vivo, upon ectopic expression of myostatin in adult muscle [Amirouche et al., 2009] and in cellulo [Trendelenburg et al., 2009]. A more direct effect was reported in C2C12 myotubes where myostatin treatments were shown to decrease total protein synthesis [Taylor et al., 2001]. Here, we demonstrate that the polysomal association and the global protein synthesis rates are increased in myostatin KO muscle cells while this effect is counteracted by the addition of recombinant myostatin. However, whether this difference may as well involve a change in total protein degradation remains to be determined. In this line, Mendias et al. [2011] recently reported that the absence of myostatin in muscles of myostatin null mice resulted in lower atrogin-1 protein levels and lower levels of ubiquitinated myosin heavy chain. Although we cannot exclude an inhibition of the proteolytic systems in absence of myostatin, our findings reveal the important contribution of myostatin to the protein synthesis and support a role for global translational control in the overgrowth due to myostatin deletion.
Our results shed a light on an important link between myostatin and the translational machinery. We show that myostatin gene deletion results in an increased phosphorylation status of the Akt/mTOR-dependent translation factors. In addition we demonstrate that translational factors are recruited to the 7-methyl guanosine cap binding complexes more efficiently in cells lacking myostatin while myostatin disrupted this process indicating that assembly of the translation initiation complex may be regulated by myostatin. Our results also indicate that myostatin inhibition positively influences polysome formation. Collectively, these data suggest that an interaction between Akt/mTOR and myostatin signaling cascade is likely to play a role in the control of translation initiation. Cooperation between these pathways was previously shown by the group of M. Sandri. The authors demonstrated that inhibition of myostatin signaling mediated by overexpression of dnActRIIB promotes hypertrophy via mTOR signaling [Sartori et al., 2009].
Our results indicate that deletion of myostatin in vitro increased expression of Akt, mTOR, and rpS6 at the protein but not at the mRNA level. This has some logic with our data showing that genetic deletion of myostatin improved the global translation rates. This suggests that myostatin controls the expression of a specific subset of proteins that function as a size checkpoint in muscle cell. Deciphering how the translation machinery preferentially translates these mRNAs needs further investigations. In this line it would be interesting to analyze whether the translation of these specific transcripts (Akt, mTOR, rpS6) is improved in myostatin KO cells. It is also possible that myostatin acts directly on some regulators of translational efficiency which in turn could play a role in the translation of specific mRNAs. This is supported by the report that several genes encoding translation initiation and elongation factors are expressed at higher levels in muscle of young mice with constitutive myostatin deficiency [Steelman et al., 2006; Chelh et al., 2009]. More recently, it has been shown that overexpression of the translational factors eIF3f or eIF2Bε induced the same effect as myostatin inactivation, that is, cap-dependent translation and skeletal muscle hypertrophy suggesting that myostatin action on protein synthesis could be an indirect effect [Lagirand-Cantaloube et al., 2008; Csibi et al., 2010; Mayhew et al., 2011]. In addition to protein translation initiation, the rate of cell mass accumulation is dependent on the cellular level of functional ribosomes (translational capacity). Ribosomal gene transcription (rDNA transcription) is a major rate-limiting step in the biogenesis of ribosomes. In this line, our preliminary data have shown that myostatin KO cells present a significant increase of total RNA levels which could represent an increase in ribosomal RNA (Inset Fig. 4D). This suggests that myostatin effects on protein synthesis may impact on the activities required for ribosomal biogenesis although we cannot exclude the regulation of specific mRNA sub-classes.
In summary, using satellite cells isolated from myostatin knock-out mice we have shown that myostatin genetic deletion induces myotube hypertrophy by increasing translational efficiency in vitro. Specifically, we have demonstrated that myostatin regulates both cap-dependent translation and accumulation of rRNA. Future studies will be to determine how myostatin controls ribosomal biogenesis.
Acknowledgements
The authors would like to thank Drs G. Carnac, J. Mercier, and B. Picard for their useful discussions. We thank Drs V. Ollendorff and A. Csibi for the gift of the phRLuc-C2 and the bicistronic reporter plasmid pRL-5'-IRES-FL, respectively. We would like to thank the Montpellier RIO imaging facility (RIO, Campus La Gaillarde, INRA, Montpellier). We thank Miss B. Bonafos for taking care of animals used in the present study. J.R. is the recipient of a doctoral fellowship cofinanced by INRA and Région Languedoc-Roussillon. M. T. is supported by the ANR Myotrophy program.




