Ribonucleoprotein immunoprecipitation (RNP-IP): A direct in vivo analysis of microRNA-targets†
The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Abstract
We have developed a novel Ribonucleoprotein Immunoprecipitation (RNP-IP) method to isolate miR-RISC complexes, associated microRNAs and target mRNAs. Our method characterizes mRNAs present in immunoprecipitates containing miR-RISC complexes that were obtained using GW182 and AGO2 antibodies. MicroRNA bound transcripts were reverse transcribed and amplified using seed sequence and 3′UTR derived primers. This flexible IP-based assay is a straightforward method to identify miRs participating in gene regulation and their cognate mRNAs in real time. J. Cell. Biochem. 110: 817–822, 2010. © 2010 Wiley-Liss, Inc.
Post-transcriptional control of gene silencing by microRNAs (miRs) is a ribonucleoprotein-driven process, which involves specific RNA binding proteins, microRNAs and their mRNA targets. This multi-component RNA-induced silencing complex (miR-RISC) regulates the stability and translation of mRNAs that are partially or fully complementary to specific miRs. Molecular and genetic studies have shown that Argonaute (AGO) proteins are essential components of the translational repression complex that blocks the translation initiation of the messenger RNA associated with microRNA [Liu et al., 2004; Meister et al., 2004]. The recent discovery of the association of AGO proteins with P-body associated molecules such as GW182 family proteins and co-interacting proteins (Dcp1, Lsm1 and Xmn1) provided important insight into our understanding of the mechanism of gene attenuation by microRNAs [Pillai et al., 2007; Pillai, 2005]. Given the complexity of subunit interactions and pathways involved in small RNA processing, it is necessary to develop innovative techniques to identify and characterize the dynamic composition of RNA-associated proteins in miR-RISC complexes in a physiological response [Zhang et al., 2007]. Here we report a tandem strategy to immunoprecipitate microRNA-associated silencing complexes from polysomal extracts of MC3T3-E1 cells. Using specific antibodies to GW182 and AGO2, we isolated miR-RISC associated mRNA that was reverse transcribed into cDNA using specific miR seed-sequence based primers. The resulting cDNAs were then analyzed using gene-specific 3′-UTR primers of previously identified miR targets to establish the specificity of this RNP-IP strategy. This method permits rapid empirical validation of specific mRNA targets of miRs in vivo that were predicted by bioinformatic analysis.
MATERIALS AND METHODS
Cell Culture
Mouse preosteoblasts MC3T3-E1 cells were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA) and maintained in αMEM (Invitrogen-GIBCO, Carlsbad, CA) containing penicillin (100 µg/ml)-streptomycin (100 units/ml) and 200 mM L-glutamine (complete αMEM) supplemented with 10% FBS (HyClone, Logan, UT, USA).
Antibodies
Affinity purified anti AGO2 (N-13, sc-32659), a goat polyclonal antibody raised against the N-terminus of the human protein, and affinity purified anti GW182 (M-17, sc-47036) a goat polyclonal antibody, were purchased from Santa Cruz Biotechnology, Inc (Santa Cruz, CA, USA). Normal goat IgG from Santa Cruz (sc-2028) was used as a negative control.
RNA Electrophoretic Mobility Shift Assay (REMSA)
Polysomal extracts were prepared from 1 × 106 freshly subcultured MC3T3-E1 cells according to Keene et al. [Keene et al., 2006]. The extracts were stored initially in liquid nitrogen and later at −80°C for further use. Probe: The single stranded RNA oligo (20 pmole) corresponding to mature mmu-miR-27a (22nt) from Ambion (Applied Biosystems/Ambion, Austin, TX, USA) was labeled with [γ-32P]ATP for 30 min at 37°C with T4 polynucleotide kinase (New England Biolabs, Beverly, MA) and purified using a quick-spin G-25 Sephadex column (Roche Molecular Biochemicals, Indianapolis, IN, USA). Substrate: In vitro transcripts from pMIR-LUC containing the complementary seed sequence for miR-27a at the 3′UTR of luciferase mRNA were generated by T7 RNA polymerase. The reaction was carried out in 20 µl of buffer containing 40 mM Tris-HCl (pH 7.9), 6 mM MgCl2, 2 mM spermidine, 10 mM NaCl, 2.5 mM rNTPS and 20 units of T7RNA Polymerase. The in vitro reactions were treated with DNase I (5 units/µg of template, Roche Applied Science, Indianapolis, IN, USA) and transcripts were purified by alcohol precipitation with 2.5 M ammonium acetate salts. The probe-substrate mix (5 µl) was prepared using 50 fmol of radiolabeled probe and 200–300 ng Luciferase mRNA transcript containing a miR-27 binding site (substrate) in 2X binding buffer (1X = 50 mM Tris-HCl pH 7.5, 0.1% NP-40, 10 mM DTT, 10% Glycerol, 10% Sucrose, 5 mM MgCl2, 1 mM EDTA and 800 units/ml RNase out from Invitrogen-GIBCO, Carlsbad, CA). The mixture was heated at 65°C for 5 min, followed by 2 min in ice, then annealed at room temp for another 10–15 min and kept in ice until further use. The REMSA reaction was performed with 5 µg of polysomal extract in a total volume of 10 µl in ice for 30 min to 1 h. Complexes were resolved by electrophoresis in 4.5% (40:0.5) native acrylamide gels. Anti GW182 or AGO2 antibody (200 ng) was incubated with polysomal extract in ice for 30 min prior to the probe addition. The samples were electrophoresed at 200 V for 2 h. After completion, the gels were dried and autoradiographed at −70°C or room temperature according to the signal intensity.
Polysomal extracts and Ribonucleoprotein Immunoprecipitation (RNP-IP)
The original materials and methods [Keene et al., 2006] have been modified for preosteoblasts MC3T3-E1 cell lines. For preparation of polysomal extracts MC3T3-E1 cells (2–5 × 106) were collected by centrifugation at 1000 g for 10 min at 4°C and washed several times with 10 ml of ice cold 1 × PBS/Phosphatase Inhibitors (Active Motif, Carlsbad, CA, USA). The cell pellet was resuspended with an equal volume of polysome lysis buffer supplemented with RNase inhibitor (100 mM KCl, 5 mM MgCl2, 10 mM HEPES (pH 7.0), 0.5% NP40, 1 mM DTT, 800 units per ml RNase inhibitor, 1 X Complete Protease inhibitor (Roche Biochemicals), 1 mM PMSF and 25 µM MG132. The polysomal lysate was allowed to incubate in ice for 5–10 min and stored at -100°C for several months. Protein-A/G Agarose beads (Santa Cruz Biotechnology) or Protein G Sepharose beads (Upstate Biotechnology) were equilibrated with NT2 buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM MgCl2, 0.05% NP40, 1 X Complete Protease inhibitor (Roche Biochemicals), 1 mM PMSF and 25 µM MG132) supplemented with 5% BSA to a final ratio of 1:5 (Protein A/G: NT2 buffer) in a rocking platform for at least 1 h. The pelleted bead volume was 60 µl per IP in 500 µl protein A/G-BSA slurry. GW182 or AGO2 (5 µg) antibody was incubated with 500 µl of NT2 buffer-protein A/G −BSA slurry overnight, in a rotating wheel at 4°C. Normal goat antibody was used as a non-specific control to check the background RNA contamination. The antibody coated beads were thoroughly washed with 1ml ice cold NT2 buffer 4-5 times at 4°C in a rotating wheel to remove the unbound antibodies. After the final wash, the beads were resuspended in 850 µl of ice-cold NT2 buffer supplemented with 800 units/ml of RNase inhibitor, 400 µM Vanadyl ribonucleoside complexes, 1 mM PMSF, 25 µM MG132, 1 mM DTT and 20 mM EDTA and kept in ice.
The polysomal lysate was thawed on ice and spun at 15,000 g for 15 min to clear. The lysate was then precleared with Protein A/G beads (60 µl beads/500 µl lysate) for 1 h to reduce the non-specific background. Cleared lysate (100 µl containing 200 µg, total proteins) was added to antibody coated Protein A/G mixture, mixed several times and incubated overnight at 4°C in a rotating wheel. Ten percent (10%) of the lysate was kept as input at −70°C for further analysis for input protein or total RNA; supernatant may be stored at −70°C for several months. The beads were washed 4–5 times with 1 ml of ice-cold NT2 buffer supplemented with 400 µM Vanadyl ribonucleoside complexes, 1 mM PMSF, 25 µM MG132, 1 mM DTT and 20 mM EDTA and finally resuspended in 100 µl of NT2 buffer supplemented with 100 µg per ml Proteinase K to release the RNP components. The reactions were incubated for 30 min at 55°C, and the RNA from the immunoprecipitated pellet was isolated by adding Trizol reagent (Invitrogen, Carlsbad, CA) directly to the beads. The RNA was then precipitated with glycogen (10 µg/reaction) and the pellet was resuspended in a volume appropriate for DNaseI digestion according to the manufacturer's protocol (Roche Molecular Biochemicals, Indianapolis, IN, USA). The DNase I treated RNA was dissolved in 10 µl of RNase free water and concentrations were estimated by NanoDrop spectrophotometer (ND1000 Spectrometer, NanoDrop Technologies, Wilmington, DE, USA). Examples of this type of analysis have been demonstrated in yeast [Gerber et al., 2004], Drosophila [Gerber et al., 2006] and mammalian cells [Tenenbaum et al., 2000; Yang et al., 2005; Brown et al., 2001].
Western Blot Analysis
The immunoprecipitation efficiencies of GW182 and AGO2 antibodies were confirmed by western blot analysis probed with anti GW182 and AGO2 antibody. After immunoprecipitation of the GW182 or AGO2 RISC complex (as above), protein A/G beads were suspended in 2X SDS sample buffer. The samples were boiled for 10 min and loaded in 8% SDS-PAGE. After completion of the run the proteins were transferred to PVDF (Immobilon-P) membrane and probed with anti GW182 or AGO2 antibody. The protein signals were visualized by Western Lightning® Plus–ECL, Enhanced Chemiluminescence substrate kit (PerkinElmer, Waltham, MA).
cDNA synthesis and Hot PCR Analysis
The RNA isolated from RNP-IP was subjected to cDNA synthesis using primers derived from specific seed sequence for miR-27a and miR-9 or group-specific seed for group miR-Let-7/98 (Table I) using a SuperScript first-strand cDNA synthesis kit (Invitrogen) according to the manufacturer's protocol. The first strand cDNA was further amplified with forward and reverse primers derived from the immediate upstream sequence of the 3′UTR target of the mRNA (Table II). The radioactive α 32P dCTP was used in the regular PCR mix to amplify the 3′UTR of the target genes. The amplified products were resolved in 6% PAGE, dried and autoradiographed at −70°C.
| Target Gene | Gene Name | Conserved Sites | Poorly Conserved Sites | miRs | Context Score |
|---|---|---|---|---|---|
| IGF2BP1 | Insulin-like Growth factor2 mRNA Binding protein 1 | 5 | 0 | mmu-let-7f/98 | −0.73 |
| COL1A1 | Collagen, type 1, alpha 1 | 1 | 0 | mmu-let-7d/98 | −0.19 |
| TGFBR1 | Transforming Growth Factor Beta Receptor 1 | 2 | 0 | mmu-let-7g/98 | −0.53 |
| MEIS3 | Meis homeobox 3 | 1 | 0 | mmu-let-7a/98 | −0.10 |
| HIC2 | Hypermethylated in Cancer | 4 | 1 | mmu-let-7d/98 | −0.61 |
| SMARCAD1 | SWI/SNF-related, Matrix-associated, Actin-dependent Regulator of Chromatin | 2 | 0 | mmu-let-7f/98 | −0.51 |
- Context score depends on the degree of seed match, the miR-target complementarity outside the seed region and the AU content 30 nt upstream and downstream of the predicted site. With all four features, a more negative score is associated with a more favorable site.
- Conserved site indicates the number of sites conserved across human (H), mouse (M), rat (R), and dog (D).
- Poorly conserved site indicates if the site is conserved across any other combination of species.
| Gene | Forward | Reverse |
|---|---|---|
| mHLX1 3′UTR | 5′TAGACTGGACCGAGGGCATA 3′ | 5′GGGAGGGGGAACCTTAAAAC 3′ |
| mIGF2BP1 3′UTR | 5′CTCCCTCCCGATCCTGTACT 3′ | 5′CACACGTCACGGTAGCTTTG 3′ |
| mSMARCAD1 3′UTR | 5′GCGGAATTCCTGATTGATGA 3′ | 5′GGAGAATCCAGTTAGTGGCAAA 3′ |
| mTGFBR1 3′UTR | 5′ CACAACTCAGCCAACAGGAA 3′ | 5′ GGGAAGCTTTCAGTTGACCA 3′ |
| mCOL1A1 3′UTR | 5′GGGGTTCTTGGACTGTTGAA 3′ | 5′GTCCGATGTTTCCAGTCTGC 3′ |
| mMEIS3 3′UTR | 5′ ACCAGCCTTGCACCTAATTG 3′ | 5′ GCTTTGTCTTTGCCCTTGAG 3′ |
| mHIC2 3′UTR | 5′CCCCTTCCTAGAAGCCAAAG 3′ | 5′GATTAAAGCTCTGGGCGTGA 3′ |
| mGAPDH 3′UTR | 5′TGGACAGCACTGACTTCCAG 3′ | 5′CAAAGCATCGACCAGTGCTA 3′ |
| miRs | Seed sequence (corresponding DNA oligonucleotides) | |
| mmu-miR-Let-7/98 | 5′ GAGGTAG 3′ | |
| mmu-miR-27a(S) | 5′TTCACAGTG 3′ | |
| mmu-miR-9 (S) | 5′TTGGTTTCT 3′ |
RESULT AND DISCUSSION
To develop this method we used polysome extracts from MC3T3-E1 osteoblast cells and two specific miRs (miR-27a and miR-Let7/98) that are known to be upregulated upon treatment of these cells with BMP-2 or during osteoblastic differentiation (Fig. 1a) [Li et al., 2009]. MC3T3-E1 polysomal extracts were immunoprecipitated with anti- GW182 and AGO2 antibodies. To exemplify the efficiency of immunoprecipitation, western blot analysis clearly revealed a several fold enrichment of endogenous GW182 by the anti-GW182 antibody compared to the nonspecific IgG control (Fig. 1b). We next established that the AGO2/GW182 complexes can associate with specific miRs in vitro using a RNA–electrophoretic mobility shift assay (REMSA). Polysomal extract (5 µl, 1 µg/µl concentration) was incubated with 5 µl probe-substrate mix (containing 50 femtomole 32P-labeled miR-27a and 200 ng of RNA spanning the LUC-3′UTR sequence obtained by in vitro transcription) at room temperature for 30 min. After resolution in a 4.5% native polyacrylamide gel, three major types of miR-27a– RNP complexes (C1, C2 and C3) were observed (Fig. 1c, indicated by arrows). All three of the observed RNP complexes compete with unlabeled pre-miR-27a oligo nucleotides (10 pmole) but not with non-specific miR (10 pmole) (Fig. 1c). RNP complex 1 (C1) is not detected in the presence of excess unlabeled pre-miR-27a or anti-miR-27a oligonucleotides but is not competed with non-specific miR (Fig. 1C), suggesting that the isolated miR-RISC complex C1 is a specific miR-27a-RNP complex. The identity of the miR-RISC complex was confirmed by immuno-reactivity (‘blockshift’) of the RISC-miR-mRNA ternary complex with GW182 antibody (data not shown). This result demonstrates that miR-RISC complexes present in the soluble polysomal fraction are functional and specific.
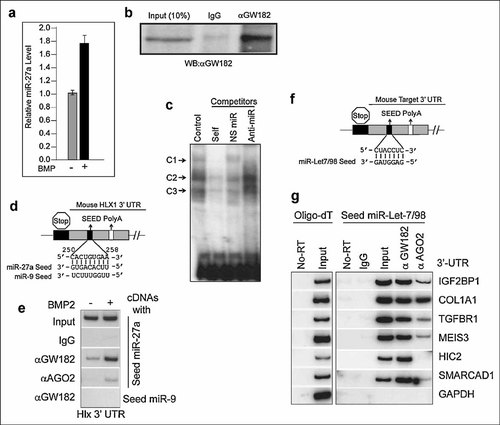
Analysis and Identification of microRNA Targets by RNP-IP (A) MC3T3-E1 cells were treated with BMP2 and miR-27a analyzed by RT-QPCR. (B) Immunoprecipitation of GW182 from polysomal extracts; Left, Input; Center, nonspecific (IgG) and Right, αGW182 pulldown. C) REMSA assay; Probe: Labeled miR-27a. Target: LUC 3′UTR with miR-27a seed sequence. Competitors: Self (pre-miR-27a), NS (non-specific miR), and anti-miR (anti miR-27a). (D) A conserved miR-27a binding site is present in Hlx1 mRNA 3′UTR. (E) RNP-IP followed by cDNA synthesis with miR-27a but not with miR-9 seed amplifies the 3′UTR of Hlx1. (F) Heptamer conserved seed sequence present in the miR-Let7/98 family. (G) RNP-IP with the indicated antibodies followed by cDNA synthesis with the miR-Let-7/ 98 specific heptamer seed (Fig. 1F) and 3′UTR amplification with primers for specific target genes. Several negative controls were included.
One of the most challenging aspects of understanding gene regulation by microRNAs is the multitude of potential gene targets within the cell for a given miR. For example, several target prediction programs yield hundreds of mRNA targets with varying degrees of seed complementarity for miR-27a. To experimentally validate these targets we isolated total RNA complexes by immunoprecipitation with anti-GW182 or anti-AGO2 antibodies from polysomal extracts of MC3T3-E1 osteoblasts. The RNA was purified and used for cDNA synthesis with reverse transcription primer based on the miR-27a or miR-9 seed sequences. We selected the gene for the homeobox transcription factor Hlx1 to validate our cDNA synthesis with the miR-27a-seed as this gene contains a highly conserved complementary seed sequence binding site for mmu-miR-27a in the 3′-UTR of the mRNA (Fig. 1d). We designed primers specific to the 3′ UTR of Hlx1 and performed PCR on miR-27a-generated cDNA. We observed that Hlx1 mRNA was associated with miR-27a-GW182 or miR-27a-AGO2 RNP silencing complex but not detected in control IgG immunoprecipitates (Fig. 1e). We also did not detect Hlx1 mRNA in immunocomplexes using miR-9 seed based primers. MiR-27a levels in MC3T3-E1 cells increase upon BMP2 stimulation (Fig. 1a); accordingly, we observed increased association of Hlx1 mRNA in both GW182 and AGO2 immuno-complexes in BMP2-treated cells (Fig. 1e). Thus, our RNP-IP method can identify specific miR targets from mRNA isolated from the miR-RISC complex in a biologically relevant context.
We also identified targets of the miR-Let-7/98 family to demonstrate that our RNP-IP method is broadly applicable to larger miR families. Large miR families or super-families (e.g., miR-Let-7/98) present a greater challenge for the identification of potential mRNA targets, as the smaller, shared conserved seed sequence of the family can potentially target thousands of mRNA sequences (Fig. 1f). Similar to the method used for identifying miR-27a targets, we used miR-Let-7/98 seed sequence-based primers to generate cDNA from miR-RISC-associated mRNA, followed by amplification of the 3′-UTR of several predicted miR-Let 7/98 mRNA targets (Table I). From our RNP-IP we confirmed that several of the predicted mRNA targets for the miR-Let-7/98 family (IGF2BP1, COL1A1, TGFBR1, MEIS3, HIC2 and SMARCAD1) were present in cDNA preparations primed using the Let-7/98 seed sequence (Fig. 1g). Absence of PCR products in non-specific IgG and no RT controls ruled out the possibility of non-specific immunoprecipitation or DNA contamination. Absence of amplification of the GAPDH 3′-UTR, which lacks the miR-Let-7/98 seed sequence, reflects seed specificity of the assay (Fig. 1g).
Many RNA complexes may associate with GW182 in P bodies; however, these silencing complexes are quite stable and their components are responsible for the final repression of mRNA targets [Meister et al., 2004; Pillai, 2005; Pillai et al., 2007]. It is striking that there is a difference in mRNAs associating with AGO2 compared with GW182. The Argonuate silencing complex may be transient, as it is initiated first as a repressive complex that then moves to the P body for final fate determination of mRNA stability [Pillai, 2005]. COL1A1 mRNA (the gene responsible for extracellular matrix formation in osteoblasts) associates with both GW182/P-bodies and AGO2 silencing complexes with equal affinity (Fig. 1g). However, Hypermethylated-In-Cancer 2 (HIC2) mRNA interacts only with the P-body associated silencing complex (Fig. 1g). Understanding the significance of this finding will require further experimentation. This level of analysis may provide significant insights into the mechanisms or kinetics of specific mRNA degradation in a cell-type specific manner.
The novel strategy validated in this study involves the immunoprecipitation of whole miR-silencing complexes containing miRs and associated target mRNAs and is capable of directly analyzing microRNA function in a specific cellular context (see Fig. 2 for an overall summary). The primary advantage of this technique is that it is a comprehensive, rapid, straightforward and simplified method for microRNA target identification in biological models. The target mRNAs identified in this co-immunoprecipitation study are associated with miR-27a and miR-Let-7/98 specific silencing complexes during osteoblastic differentiation. The method can be expanded by combining with high throughput sequencing to generate unbiased targets. We have been able to correlate the biological function(s) of the targets (i.e., COL1A1) to narrow down the list of potential microRNAs to be validated experimentally. Furthermore, immunoprecipitation of the in vivo RISC complex under different physiological or pathological conditions may generate microRNA target interaction maps. Matching of miRs to mRNAs can deduce a microRNA network for post transcriptional control of gene expression. This in vivo approach also provides an important balance to in silico methods of predicting microRNA targets, which, while growing in power, still fail to provide a comprehensive picture of microRNA regulatory networks.
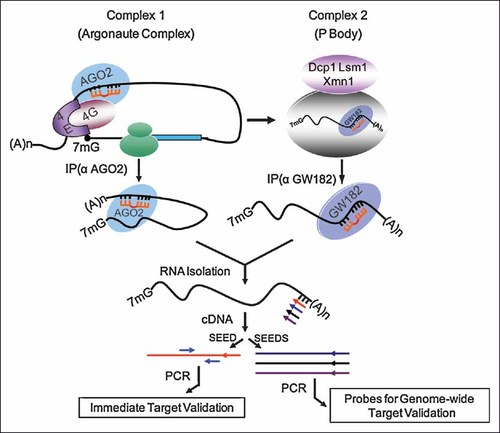
Overall Strategy for RNP-IP and Seed-Specific Amplification of miRNA Targets. Polysomal cytosolic fractions were immunoprecipitated with antibodies to GW182 (for P body complex) or AGO2 (for Argonaute complex). cDNAs were made from immunoprecipitated RNA using miR-specific or miR family-specific seed as primer. Targets were validated by PCR using primers derived from the mRNA 3′-UTR sequences of the respective miR targets. Alternatively, the labeled cDNA can be used as a probe for microarray analysis to screen genome-wide miR targets.