Mutation in G6PD gene leads to loss of cellular control of protein glutathionylation: Mechanism and implication
Abstract
More than 400 million people are susceptible to oxidative stress due to glucose-6-phosphate dehydrogenase (G6PD) deficiency. Protein glutathionylation is believed to be responsible for loss of protein function and/or cellular signaling during oxidative stress. To elucidate the implications of G6PD deficiency specifically in cellular control of protein glutathionylation, we used hydroxyethyldisulfide (HEDS), an oxidant which undergoes disulfide exchange with existing thiols. G6PD deficient (E89) cells treated with HEDS showed a significant increase in protein glutathionylation compared to wild-type (K1) cells. In order to determine whether increase in global protein glutathionylation by HEDS leads to loss of function of an important protein, we compared the effect of HEDS on global protein glutathionylation with that of Ku protein function, a multifunctional DNA repair protein, using a novel ELISA. E89 cells treated with HEDS showed a significant loss of Ku protein binding to DNA. Cellular protein thiol and GSH, whose disulfide is involved in protein glutathionylation, were decreased by HEDS in E89 cells with no significant effect in K1 cells. E89 cells showed lower detoxification of HEDS, that is, conversion of disulfide HEDS to free sulfhydryl mercaptoethanol (ME), compared to K1 cells. K1 cells maintained their NADH level in the presence of HEDS but that of E89 cells decreased by tenfold following a similar exposure. NADPH, a cofactor required to maintain reduced form of the thiols, was decreased more in E89 than K1 cells. The specific role of G6PD in the control of such global protein glutathionylation and Ku function was further demonstrated by reintroducing the G6PD gene into E89 (A1A) cells, which showed a normal phenotype. J. Cell. Biochem. 103: 123–135, 2008. © 2007 Wiley-Liss, Inc.
More than 400 million people throughout the world are prone to oxidative stress due to genetic defects in glucose-6-phosphate dehydrogenase (G6PD), an X-linked gene [Baker et al., 1984; Vulliamy et al., 1992; Ruwende et al., 1995; Devi et al., 1997; Wells et al., 1997; Cocco et al., 1998; Ruwende and Hill, 1998]. We have previously demonstrated that the Chinese hamster ovary G6PD-null mutant (CHO; E89) has enhanced radiation sensitivity, loss of Ku protein function and an inability to repair DNA double-strand breaks (DSBs) after oxidative stress induced by hydroxyethyldisulfide (HEDS), a thiol specific oxidant [Ayene et al., 2000, 2002]. DNA repair is believed to be important since any one of the unrepaired DNA lesions, single-strand break (SSB), DSB, base alterations, DNA protein cross links, inter-strand and intra-strand cross links, will lead to mutations and cell death. Several DNA repair proteins are present in mammalian cells to repair these lesions [Aravind et al., 1999; Eisen and Hanawalt, 1999; Ronen and Glickman, 2001; Wood et al., 2001; Adachi and Lieber, 2002]. However, proteins are known to be susceptible to carbonyl formation, protein glutathionylation, and ubiquitin-dependent protein degradation during oxidative stress [Davies et al., 1987; Fisher et al., 1994; Jones, 2006; Townsend et al., 2006].
Protein glutathionylation is believed to be responsible for loss of function and/or cellular signaling during oxidative stress [Garner et al., 1992; Finkel and Holbrook, 2000; Klatt and Lamas, 2000; Eaton et al., 2003; Giustarini et al., 2003; Fratelli et al., 2005]. However, adverse effects of G6PD deficiency in cellular control of protein glutathionylation have never been demonstrated. To elucidate the implications of G6PD deficiency in cellular control of protein glutathionylation, we compared wild-type K1 with G6PD deficient E89 cells after exposure to a relatively non-toxic HEDS, which oxidizes thiols in G6PD deficient cells [Ayene et al., 2000]. In contrast, other oxidants such as hydrogen peroxide and oxidative stress are not specific for thiol oxidation since these are known to induce other types of protein damage such as protein carbonyl, which may cause protein degradation leading to irreversible loss of protein function [Ayene et al., 1992; Beal, 2002].
To understand the effect of cellular protein glutathionylation on a specific protein, we have determined the function of a multifunctional DNA repair protein Ku in cells undergoing protein glutathionylation. The Ku (auto) antigen was named after the first patient in which this protein was determined to cause an autoimmune dysfunction [Tuteja and Tuteja, 2000]. Subsequent work has shown that Ku is actually a heterodimer, consisting of Ku70 and Ku80, with molecular mass 69.6 and 81.9 kDa, respectively. This heterodimer (Ku70/80) is thought to bind to damaged DNA in association with other proteins of the non-homologous end joining (NHEJ) repair pathway [Getts and Stamato, 1994; Mizuta et al., 1996]. These additional proteins include DNA-PKcs, XRCC4 and ligase IV [Kemp et al., 1984; Jeggo et al., 1989; Taccioli et al., 1994; Jeggo, 1998; Featherstone and Jackson, 1999]. Mutations in any of these individual proteins have been shown to cause radiation sensitivity. DNA-PK (comprising Ku and the catalytic subunit DNA-PKcs) is also essential for V(D)J recombination and its mutation is responsible for the SCID phenotype [Blunt et al., 1995]. In 1993, Yaneva and colleagues showed that reduced cysteine residues are required for Ku binding to DNA [Zhang and Yaneva, 1993]. New functions of Ku are rapidly emerging. It is known to bind to multiple forms of DNA, though most strongly to DSBs. Ku protein has been associated with proper functioning of DNA synthesis in yeast (homologous proteins in yeast are Hdf1p and Hdf2p). It has been suggested that Ku is anti-apoptotic and necessary for maintenance of telomere length [Barnes and Rio, 1997; Polotnianka et al., 1998].
G6PD, the rate-limiting enzyme of oxidative pentose phosphate cycle (OPPC) in cells, is required for regeneration of NADPH from NADP+. NADPH, the major reducing equivalent produced by OPPC in cells, is required for redox regulation of glutathione [Ayene et al., 2000, 2002; Tuttle et al., 2000; Biaglow et al., 2003]. Glutathione is believed to be involved in the maintenance of intracellular redox status either by scavenging reactive oxygen species (ROS) directly or as a substrate for other enzymes that reduce oxidatively modified proteins including glutathionylated proteins. In this report, the role of G6PD in the control of such global protein glutathionylation and Ku function was demonstrated by comparing wild-type (K1) cells with G6PD deficient E89 cells. The E89 cells were also used after reintroduction of G6PD (A1A cells) in order to determine the specific effect of G6PD. In order to determine the biochemical mechanisms of protein glutathionylation in G6PD deficient (E89) cells, we measured the effect of HEDS on bioreduction (conversion of HEDS to ME), NADPH, NADH, GSH, and PSH. Using these genetic and biochemical approaches, we have demonstrated that the lack of OPPC/G6PD leads to loss of cellular control of global protein glutathionylation, which in turn was associated with loss of protein function during oxidative stress.
MATERIALS AND METHODS
G6PD Mutants and Cell Transfections
Cells deficient in G6PD were derived from K1 cells after mild mutagenesis with ethyl methane sulfonate. The clones were selected by histochemical stain for G6PD enzyme activity [Stamato et al., 1982]. The E89 clones were found to have no functional enzyme activity. The E89 mutant cells were used to transfect the G6PD gene by electroporation with hamster G6PD-cDNA (AF 044676) inserted into pcDNA3.1 constructs and selected in G418 medium. The clones with the maximum G6PD activity were selected by histochemical staining for G6PD activity and Northern analysis using G6PD-cDNA [Tuttle et al., 2000]. A1A clone has G6PD activity similar to wild-type K1 cells suggesting a 100% stably transfected cells.
Western Blot Analysis
Total cellular protein glutathionylation was quantified using Western blot analysis. Cellular extract preparations and electrophoresis were carried out under non-reducing conditions. Cells treated with HEDS were cooled in ice, rinsed with cell rinse three times and mixed with 150 µl of cold lysis buffer containing 20 mM Tris, pH 7.6, 420 mM NaCl, 2 mM EDTA, 10% glycerol, 1% Triton X-100, 1 mM orthovanadate, 2 mM phenyl methyl sulfonyl fluoride, and 6 µl of protease inhibitor cocktail (Sigma). The cells in lysis buffer in the dish were removed using a teflon scraper and transferred to an eppendoff tube using a micropipette. DNA was sheared using a 1 ml syringe (25 gauge needle), centrifuged at 6,000g for 2 min in a microfuge (Fisher 59A), and the supernatant stored at −80°C. Protein concentration was measured by using Bio-Rad protein reagent. The protein extracts (10 µg) were incubated in NUPAGE sample buffer at 70°C for 10 min. These samples were then electrophoresed on a 10% BIS/TRIS precast gel from NUPAGE at 200 V for 60 min in MOPS SDS running buffer (NUPAGE). The proteins were transferred to nitrocellulose by electrophoresis in NUPAGE transfer buffer at 30 V for 60 min. The nitrocellulose blot was incubated with 10 ml blocking buffer (5 g dry milk in 100 ml PBS) for 1.5 h at RT on a rocker and stored in the cold room overnight. The nitrocellulose paper was washed five times with PBS containing 0.1% Tween-20 (PBST) and then incubated with 20 µl of primary anti-glutathione antibody (D8 mouse monoclonal, Virogen, MA) in 10 ml of blocking buffer for 2 h at RT. The nitrocellulose paper was washed five times with PBST before incubation with secondary peroxidase-labeled anti-mouse Ab for 1 h, per the manufacturer's instruction, and the bands were detected using a standard ECL kit (Amersham, NJ).
Enzyme-Linked Immunosorbent Ku70-Binding Assay
For Ku-binding assay, nuclear and cytoplasmic extracts were prepared by using nuclear extract kit from Active Motif (CA). Cells grown in tissue culture dishes were rinsed with 5 ml ice-cold PBS (0.01 M phosphate buffer, pH 7.5, 0.15 M NaCl, 2.7 mM KCl) buffer. Three milliliters of ice-cold PBS was added to the dish and the cells were detached gently by teflon cell scraper. The cells were centrifuged for 5 min at 500 rpm and the cell pellet was used for nuclear and cytoplasmic extracts. Cytoplasmic fractions were collected in 500 µl 1X hypotonic buffer (20 mM HEPES, pH 7.5, 5 mM NaF, 10 µM Na2MoO4, 0.1 mM EDTA) and gently mixed by pipetting up and down several times. After 15 min incubation on ice, 25 µl detergent was added and vortexed for 10 s at highest speed. The cellular mixture was centrifuged for 30 s at 14,000g and the supernatant (cytoplasmic extract) was transferred into a prechilled microcentrifuge tube. The cytoplasmic extract was stored at −80° C.
The nuclear pellet was resuspended in 50 µl complete lysis buffer from Active Motif by pipetting up and down. The suspension was vortexed for 10 s at highest setting, and incubated for 30 min on ice on a rocking platform at 150 rpm. After incubation, the suspension was again vortexed for 30 s at highest speed, and centrifuged for 10 min at 14,000g in a microcentrifuge. The supernatant (nuclear fraction) was transferred into a prechilled microcentrifuge, and nuclear extract was stored at −80°C.
These extracts were used to quantify the Ku70 and Ku80 binding using Ku DNA repair kits from Active Motif as per the manufacturer's instructions. Forty microliters of AM6-binding buffer was added to each well of an eight-well strip. Nuclear or cellular extract in a total volume of 10 µl complete lysis buffer were then added to each well, and incubated for 1 h at room temperature on a orbital shaker at 100 rpm. After incubation, the wells were washed three times with 200 µl of 1X washing buffer. Ku70 antibody at a 1:1,000 dilution in 1X antibody-binding buffer was added to all wells, and incubated for 1 h at room temperature on an orbital shaker (100 rpm). After incubation, the wells were washed four times with 200 µl 1X washing buffer. For colorimetric reaction, 100 µl developing solution was added to each well, and incubated 4 min at room temperature in dark. The reaction was stopped by mixing it with 100 µl of stop solution. The OD was read in a microplate reader at 450 nm with a reference wavelength of 655 nm.
Estimation of Intracellular Protein Thiols
Cells treated with and without HEDS were cooled in ice, washed three times with ice cooled PBS and then washed three times with 5 ml SSA. Under these experimental conditions, the cells are lysed and precipitated in situ while eliminating all NPSH. The resultant samples were incubated with 1 ml of 100 mM phosphate buffer, pH 7.4, containing 1.5 mM DTNB for 15 min at 37°C. Protein thiols were estimated using an absorption coefficient for reduced DTNB of 13,600 at 412 nm [Tietze, 1969].
Quantification of Intracellular and Extracellular Thiols
The concentrations of the intracellular and extracellular thiols were estimated either by HPLC or 5,5-dithiobis-2-nitrobenzoic acid (DTNB) assays [Ayene et al., 2000, 2002; Biaglow et al., 2000, 2003, 2006]. Reverse phase C-18 column (Alltech) was used to separate the non-protein thiols using a mobile phase containing 100 mM phosphate, pH 2 with 15% methanol. Cells treated with and without HEDS after the desired time were cooled in ice. To quantify the extracellular thiols, half a milliliter of the extracellular medium was mixed with 0.5 ml of sulfosalicyclic acid (SSA) lysis buffer (100 mM SSA, 0.1 mM diethylenetriaminepenta acetic acid (DTPA), 0.1 mM diethyldithiocarbamic acid (DTC), 0.1 mM EDTA) in microfuge tube, and centrifuged at high speed in a Fisher 59A microfuge. To quantify the cellular thiols, the cells in the dish were rinsed three times with cell rinse, and the attached cells were mixed with 1 ml of ice-cold SSA buffer diluted (1 + 1) with water. After 15 min on ice, the cells were scraped with a teflon spatula, and centrifuged at high speed in a Fisher 59A microfuge. The supernatant of these samples were injected into the HPLC column and the thiols were quantified using an electrochemical detector (Waters 460).
Intracellular and extracellular extracts prepared as described above for HPLC measurements were also used to quantify non-protein thiols by DTNB assay. For extracellular thiol measurement, 150 µl of the extract was mixed with 1,200 µl of phosphate buffer (0.1 M NaH2PO4, 0.5 mM EDTA, pH 7.5) and 150 µl of 10 mM DTNB. For intracellular thiol measurement, 300 µl of sample was mixed with 1,050 µl of phosphate buffer, and 150 µl of 10 mM DTNB. The OD was measured at 412 nm and the concentration calculated as described above for protein thiols.
Estimation of Intracellular reductants NADPH and NADH by High-Performance Liquid Chromatography (HPLC)
Cells plated in 60 mm dishes were treated with desired concentrations of HEDS. The dishes were swirled every 5 min for 1 h incubation at 37°C in a humidified 5% CO2 incubator. NADPH and NADH were extracted by cooling the dishes on ice, then rinsing the cells with PBS and extracting by suspending into 70% methanol. NADPH and NADH were resolved using HPLC (Jasco, Japan) by eluting from C-18 reverse phase column (Altima) at 1 ml/min flow rate with 0.1 M KH2PO4, pH 6.1, and a step gradient of methanol: 5 min, 0% CH3OH; 6 min, 4% CH3OH; 5 min, 12% CH3OH; 14 min, 40% CH3OH as described previously [Ayene et al., 2000]. The NADPH and NADH were quantified by fluorescence detector (Waters 474) at an excitation and emission wavelength of 340 and 455 nm, respectively. The data are collected and analyzed using Jasco Borwin software.
RESULTS
The effect of G6PD mutation on protein glutathionylation was measured at 5 mM concentration of HEDS by Western blot analysis using a mouse monoclonal anti-glutathione antibody (Fig. 1). Under the experimental conditions, HEDS is relatively non-toxic (SF = 0.85 ± 0.05) to any of these cells. Several proteins were glutathionlyated in wild-type (K1), G6PD deficient (E89), and G6PD transfected E89 (A1A) cells even in the absence of HEDS. Further, G6PD deficient E89 cells have higher glutathionylation than wild-type K1 cells even in the absence of treatment. K1 cells treated with 5 mM HEDS did not show significant difference in protein glutathionylation compared to the untreated ones. In contrast, protein glutathionylation was increased in E89 cells both in terms of the number of bands and the intensity of bands compared to untreated ones. The protein glutathionylation level was decreased in G6PD transfected E89 (A1A) cells, and was similar to that of K1 cells. This indicates that protein glutathionylation caused by HEDS is controlled by G6PD. To determine if the effect of HEDS on protein glutathionylation in G6PD mutant cells was a direct consequence of protein thiol oxidation, we treated the E89 cellular extract with 0, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, and 1.0 mM of DTT. The glutathionylation was significantly reversed by DTT with complete deglutathionylation at 0.8 mM (Fig. 2).
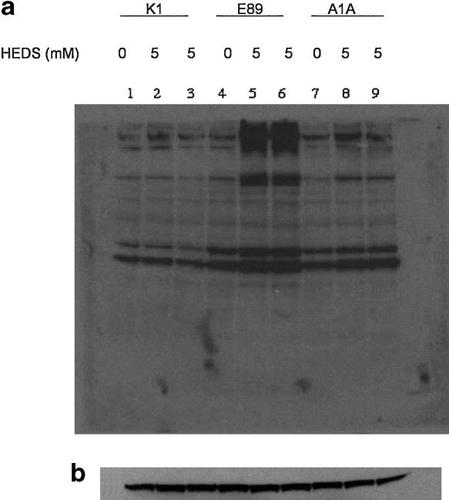
Effect of HEDS on protein glutathionylation in K1 (wild-type), E89 (G6PD-null mutant), and A1A (G6PD-transfected E89) cells. a: A representative Western blot of glutathionylated proteins. Several proteins were found to be glutathionlyated in K1 (lane 1), E89 (lanes 4), and A1A (lane 7) cells even in the absence of HEDS. HEDS (5 mM) treatment (lanes 5 and 6) increased protein glutathionylation in E89 cells both in terms of the number of bands and the intensity of bands compared to untreated ones. The protein glutathionylation level was not significantly changed by 5 mM HEDS exposure in K1 (lanes 2 and 3) and revertant A1A (lanes 8 and 9) cells. This indicates that protein glutathionylation caused by HEDS is controlled by OPPC. b: Loading control for the Western blot as determined by anti-β-actin antibody. The experiments were repeated at least three times.
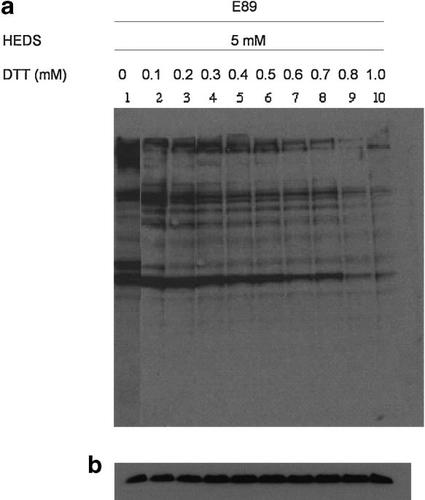
Effect of DTT on protein deglutathionylation in cellular extract prepared from HEDS treated E89 cells. a: A representative Western blot of glutathionylated proteins. Lane 1, cellular extract from cells treated with 5 mM HEDS; lanes 2–10, sample in lane 1 mixed with 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, and 1.0 mM of DTT, which showed a concentration-dependent decrease in protein glutathionylation. b: Loading control for the Western blot as determined by anti-β-actin antibody. Each experiment was repeated at least three times.
In order to determine whether global protein glutathionylation by HEDS leads to loss of function of a protein, we compared the effect of HEDS on global protein glutathionylation with that of Ku protein binding to DNA. We have modified the assay kit provided by Active Motif by adding an excess of circular plasmid DNA to adapt this assay kit to specifically measure Ku binding to DSB. In the absence of plasmid DNA, Ku protein will bind to both internal DNA sequences and DNA ends. As a result of the presence of circular plasmid DNA, proteins that bind to internal DNA sequences will be bound by the excess plasmid DNA, which does not contain DNA ends. Figure 3a,b shows 1 µg of puc18 reduced the overall binding by 36% for both 8 and 16 µg of K1 nuclear extract. The data suggest that 1/3 of the Ku binding to DNA is due to non-specific binding to internal sequences of DNA. All these measurements were made in the presence of DTT as suggested by the instructions to avoid spontaneous oxidation. However, our experimental conditions require the Ku binding to be measured in the absence of DTT since our goal was to measure oxidation of Ku by HEDS. Figure 3c shows a linear concentration-dependent increase in Ku binding for nuclear extract prepared from K1 cells even in the absence of DTT with absorption similar to that observed in the presence of DTT.
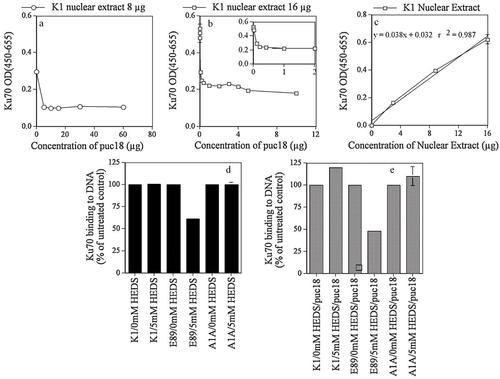
Effects of HEDS on Ku binding to DNA as measured by Active Motif ELISA technique. a,b: Ku binding in the presence of different concentrations of plasmid DNA for nuclear extracts (a, 8 µg; b, 16 µg) prepared from K1 with a maximum inhibition at 1 µg puc18. One microgram of puc18 reduced the overall binding by 36% for nuclear extract (see inset in Fig. 3b). The data suggests that 36% of the Ku binding to DNA is due to non-specific binding to internal sequences of DNA. c: Shows a linear concentration dependent increase in Ku binding for nuclear extract prepared from K1 cells even in the absence of DTT with OD similar to that observed in the presence of DTT. d,e: Effect of 5 mM concentrations of HEDS on Ku binding to DNA. Ku binding to DNA for nuclear proteins extracted from K1 cells was not affected by HEDS. In the absence of puc18, HEDS inhibited the binding of Ku to DNA only by 40% in E89 cells (d). However, the Ku binding to DNA is inhibited by 65% in the presence of puc18 (e). G6PD mutant cells transfected with G6PD (A1A cells) was also not affected by HEDS. Each experiment was repeated at least three times with SD as shown unless smaller than points plotted.
Using these conditions, we quantified the effect of 5 mM concentration of HEDS on these cells under conditions comparable to that used for protein glutathionylation (Fig. 3d,e). Ku binding to DNA for nuclear proteins extracted from K1 cells was not affected by HEDS. In the absence of puc18, HEDS inhibited the binding of Ku to DNA by 40% in E89 cells (Fig. 3d). However, the Ku binding to DNA was inhibited by 65% when the assay was carried out in the presence of puc18 (Fig. 3e). The 65% inhibition was consistent with our previous report that used electromobility shift assay to quantify Ku binding to DNA ends in the presence of puc18 (Ayene et al., 2002). G6PD mutant cells transfected with G6PD (A1A cells) were not affected by HEDS. These results demonstrated that the specific function of Ku in binding DNA ends was affected more than its binding to internal DNA sequences in G6PD deficient cells.
The effect of G6PD deficiency on protein thiol loss was measured at 5 mM concentration of HEDS in these cells. Under these conditions, HEDS decreased PSH by 30% in E89 cells with minimal effect on K1 and A1A cells (Fig. 4) indicating that the HEDS-mediated change in cellular glutathionylated proteins results in the loss of PSH.
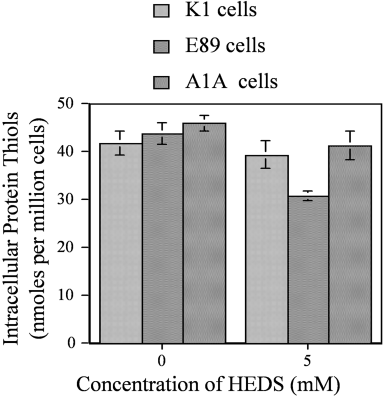
Loss of protein thiol (PSH) in K1 (wild-type), E89 (G6PD-null mutant), and A1A (G6PD-transfected E89) cells as measured by DTNB reactive intracellular protein. Under the conditions used in Figures 1-4, 5 mM HEDS decreased PSH by 30% in E89 cells with minimal effect on K1 and A1A cells. Each experiment was repeated at least three times with SD as shown unless smaller than points plotted.
We have also estimated the intracellular total non-protein thiols (NPSH) in K1, E89, and A1A cells using DTNB assay (Fig. 5). The total DTNB reactive intracellular non-protein thiols continued to increase with HEDS concentration in K1 and A1A cells. A maximum 15–20% increase in NPSH was observed at 5 mM HEDS. In contrast, the total intracellular NPSH showed a slight increase at lower concentrations of HEDS in E89 cells but a further increase in HEDS led to a concentration-dependent decrease in NPSH with a maximum 71% decrease at 5 mM HEDS.
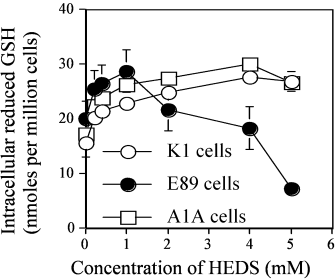
The effect of HEDS on the intracellular non-protein thiols (NPSH) in K1 (wild-type), E89 (G6PD-null mutant), and A1A (G6PD-transfected E89) cells. NPSH was measured by DTNB reactive intracellular non-protein thiols. HEDS at concentrations higher than 1 mM showed a concentration-dependent decrease in total NPSH level in G6PD deficient cells with no significant decrease in K1 and A1A cells. Each experiment was repeated at least three times with SD as shown unless smaller than points plotted.
Figure 6 shows the bioreduction of different concentrations of HEDS for K1, E89, and A1A cells as measured by the DTNB reactive extracellular non-protein thiols. At concentrations up to 0.6 mM concentrations of HEDS, these cells converted HEDS into mercaptoethanol (ME) at the same rate. These results suggested that G6PD-independent pathways are capable of reducing low levels of disulfides since G6PD activity was zero in the mutants. At concentrations higher than 0.6 mM HEDS, G6PD containing K1 and A1A cells produced more ME from HEDS compared to G6PD deficient E89 cells.
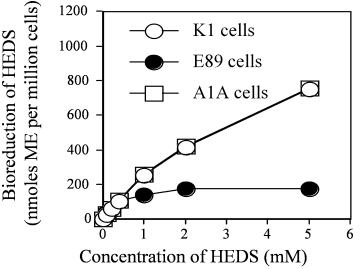
Effect of G6PD on cellular bioreductive capacity. The conversion of different concentrations of HEDS into mercaptoethanol by bioreductive pathways in K1 (wild-type), E89 (G6PD-null mutant), and A1A (G6PD transfected E89) cells were measured by DTNB. These cells were all incubated for 1 h at 37°C in the presence of different concentrations of HEDS. At concentrations greater than 1 mM of HEDS, G6PD deficient E89 cells had lower bioreductive capacity than the G6PD containing wild-type K1 and G6PD transfected A1A cells. Each experiment was repeated at least three times with SD as shown unless smaller than points plotted.
In order to identify the specific effect of HEDS on GSH and ME in these cells, we have resolved the various non-protein thiols using HPLC (Figs. 7 and 8). The HPLC data showed that ME, cysteine, and GSH standards had retention times of 5, 6, and 10 min, respectively (data not shown). The results indicated a HEDS concentration-dependent increase in ME production similar to that observed for DTNB assays (Fig. 7). Our HPLC measurements demonstrated that the extracellular thiols are mainly due to the presence of ME produced by bioreduction of HEDS. We did not see any difference in ME levels either measured by HPLC or DTNB assays. In the presence of HEDS, cells have GSH, ME, and cysteine. However, the intracellular GSH level was much higher than the ME level in K1 and A1A cells (results not shown). In E89 cells, the GSH level was decreased by sixfold with no significant effect on K1 and A1A cells (Fig. 8).
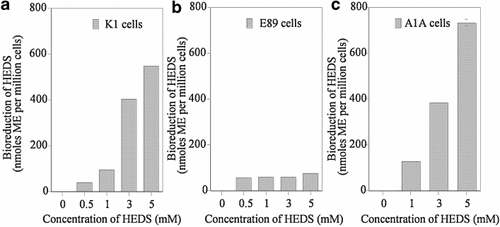
HPLC analysis of extracellular non-protein thiols. In order to identify the effect of G6PD deficiency on HEDS bioreduction, we have quantified the effect of HEDS by resolving the various non-protein thiols using HPLC and electrochemical detection. The three major NPSH ME, cysteine, and GSH standards are eluted at 5, 6, and 10 min, respectively (not shown). ME is the only NPSH detected in extracellular medium after cells were treated with HEDS (results not shown). a–c: HEDS bioreduction quantified from HPLC tracings. The data showed a HEDS concentration-dependent increase in the extracellular ME in K1 and A1A. G6PD deficient E89 cells did not show such a profile. The overall extracellular ME levels quantified by HPLC are almost similar to that measured by DTNB for all three cell lines. Each experiment was repeated at least three times with SD as shown unless smaller than points plotted.
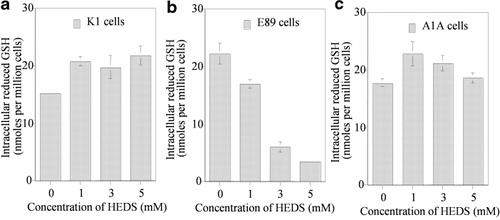
HPLC analysis of intracellular non-protein thiols. In order to identify the effect of G6PD deficiency on HEDS-mediated GSH oxidation, we have quantified the effect of HEDS by resolving the various non-protein thiols using HPLC and electrochemical detection. a–c: GSH quantified from HPLC tracings. At 5 mM concentrations of HEDS, GSH level is decreased by sixfold in E89 cells with no significant effect on wild-type K1 and A1A (G6PD-transfected E89) cells. The overall intracellular GSH levels quantified by HPLC indicated a maximum decrease in GSH at 5 mM HEDS in E89 cells. Each experiment was repeated at least three times with SD as shown unless smaller than points plotted.
Consistent with the above results, the NADH level was not altered in K1 and A1A cells either at low (280, 560 µM) or high (5 mM) concentrations of HEDS (Fig. 9). Although NADH level was not changed at 280 and 560 µM concentration in E89 cells, a significant decrease in NADH was observed at 5 mM concentration of HEDS. In contrast to NADH levels, NADPH was decreased at all three concentrations of HEDS in E89 cells but a significant decrease was observed only at 5 mM concentrations in K1 and A1A cells (Fig. 10). These results suggest that NADPH is likely the major cofactor required in maintaining the intracellular thiol redox homeostasis when the cells are under oxidative stress.
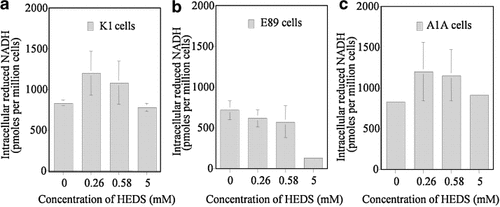
Quantification of NADH by HPLC and fluorescence detection. In order to identify the effect of G6PD deficiency on HEDS-mediated NADH and NADPH consumption, we have quantified these pyridine nucleotides by using HPLC and fluorescence detection. The two major intracellular pyridine nucleotides NADPH and NADH standards are eluted at 18 and 21 min, respectively (not shown). The data showed that NADH level was not altered in (a) K1 (wild-type) and (c) A1A (G6PD-transfected E89) cells at 260, 580 µM and 5 mM HEDS. NADH level was not affected at 260 and 580 µM concentration in (b) E89 (G6PD-null mutant) cells but a significant decrease in NADH was observed at 5 mM HEDS. Each experiment was repeated at least three times with SD as shown unless smaller than points plotted.
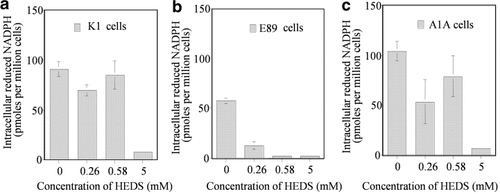
Quantification of NADPH by HPLC. Graph of the effect of HEDS on NADPH quantified from HPLC tracing. In contrast to NADH levels shown in Figure 9, NADPH levels were decreased at all three concentrations of HEDS in (b) E89 cells with significant effect on (a) K1 and (c) A1A cells only at 5 mM HEDS. Each experiment was repeated at least three times with SD as shown unless smaller than points plotted.
DISCUSSION
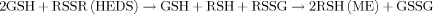 (1)
(1)The loss of GSH in E89 cells and not in K1 and A1A cells suggested that G6PD was an important factor in converting GSSG back to GSH.
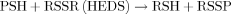 (2)
(2)Cellular extracts of HEDS-treated E89 cells showed dramatically increased glutathionylation of several proteins (Fig. 1) suggesting that GSSG is involved in the loss of PSH. The lack of protein glutathionylation when nuclear extracts were treated with DTT ex vivo has suggested that the protein glutathionylation caused by HEDS under these conditions in E89 cells is a reversible phenomenon (Fig. 2). DTT and ME are thiol reductants commonly used to reduce oxidized protein thiols. However, studies are underway to understand the role of glutaredoxin and thioredoxin systems in deglutathionylation of Ku protein using glutredoxin, thioredoxin, NADPH, etc. None of these HEDS-mediated effects occurred in wild-type CHO-K1 cells, or in E89 cells transfected with the G6PD gene (CHO-A1A). Comparison of our results with other studies that have used diamide suggested that several bands have appeared in untreated controls compared to a significant increase in diamide-treated normal cells [Eaton et al., 2002; Brennan et al., 2004]. Unlike diamide, HEDS did not increase the intensity of the bands in G6PD containing normal cells. However, HEDS did increase the number and the intensity of bands significantly in G6PD deficient cells. Although untreated controls looked similar between our data and theirs, HEDS is effective only in G6PD deficient cells. Our results have also shown that Ku binding to double-strand DNA ends was decreased in cells with enhanced glutathionylation (Fig. 3d,e). Shen et al. [1999] have shown that glutathione conjugates induced a destabilization of the DNA-PK complex (DNA-PKcs, Ku70, and Ku80) by thiolation of DNA-PK. Townsend et al. [2002] have also shown recently that cisplatin resistant ovarian cancer cells C70 and C200 cell lines have higher protein levels of DNA-PKcs, Ku70, and Ku80 compared to parental cells. Treatment of these cells to a novel π-activated prodrug {γ-glutamyl-α-amino-β[2-ethyl-N,N,N′,N′-tetrakis (2-chloroethyl) phosphorodiamidate]-sulfonyl-propionyl-(R) phenylglycine; TLK286} showed similar toxicity for both parental and cisplatin resistant cells. They have suggested that a drug-induced destabilization of the protein–protein interaction presumably due to thiolation of DNA-PK may be responsible for TLK286-induced cytotoxicity. Although the data from their reports have suggested that the DNA repair proteins are susceptible to thiolation induced by certain drugs in G6PD containing cells, our results demonstrated that the effectiveness of thiolation by thiol oxidants is determined by OPPC.
These results suggest that G6PD plays an important role in cellular defense against oxidative damage to proteins by inhibiting protein glutathionylation. This is an interesting effect since it implies multiple levels of interaction of oxidative damage with cellular processes. It is also possible that oxidative damage can interfere with signaling pathways in G6PD mutants since many signaling proteins are cysteine rich and their receptors often contain multiple extracellular disulfides. Our data on global protein glutathionylation and loss of function of individual Ku protein in mutant cells elucidate a new layer of control by G6PD/OPPC. The data specifically showed that minimally toxic oxidative modification of protein thiols can inactivate various cellular pathways and specific molecular interactions. Indeed, our laboratory has previously demonstrated that HEDS compromised DSB repair and survival in G6PD deficient cells after exposure to γ radiation (Ayene et al., 2000, 2002). In addition, the observation that oxidative damage can inactivate a DNA repair pathway suggests a new level of repair by G6PD. Our data addressed the question of “how might repair molecules be susceptible to oxidative damage and how are they protected against such damage?” The data suggests that critical cysteine residues may be susceptible to oxidation and that reducing equivalents provided by the pentose cycle can protect against or reverse such oxidation.
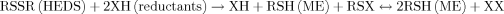 (3)
(3)However, K1 cells converted high levels of HEDS into ME more effectively than G6PD mutant cells suggesting that additional reductants required to convert HEDS are not produced in G6PD deficient cells.
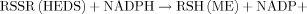 (4)
(4)The data on NADPH also indicated differences between G6PD containing and G6PD deficient cells at lowest concentrations of HEDS. NADPH, a cofactor required to maintain the thiols, showed a HEDS concentration-dependent decrease only in G6PD deficient cells with almost complete depletion at concentrations as low as 0.6 mM. However, depletion of NADPH at lower concentrations of HEDS did not affect NADH or GSH level suggesting that NADPH is a better indicator of low-level oxidative stress than NADH and GSH. NADPH produced by G6PD/OPPC is probably the primary reductant to counter the oxidative challenge posed by HEDS that includes maintaining GSH, PSH, NADH, control of protein glutathionylation and protein function.
In conclusion, mammalian cells are able to prevent thiol-disulfide exchange of their protein and non-protein thiols with disulfides via reducing equivalents produced by the OPPC, whose key regulatory enzyme is G6PD. The lack of G6PD will make the mammalian cells more susceptible to loss of protein function via glutathionylation during oxidative stress.
Acknowledgements
The authors thank Daphne Conde for excellent technical assistance. This study was supported by the National Institutes of Health grants (CA109604 to ISA), (CA92108 to CJK), (CA44982 to JEB), and (CA45277 to TDS).




