Regulation of dHAND protein expression by all-trans retinoic acid through ET-1/ETAR signaling in H9c2 cells
Abstract
dHAND is thought to be a cardiac-restricted transcription factor during embryonic development. Vertebrate heart development involves many transcription factors such as Nkx2.5, GATA, and tbx5. All-trans retinoic acid (AtRA), the oxidative metabolite of vitamin A, can regulate the expression of these factors to affect embryonic heart development. However, the action of atRA on the expression of dHAND is rarely reported. To clarify whether atRA regulate the dHAND expression, we exposed cultured H9c2 cells (rat embryonic cardiomyocytes) to atRA and detected the protein expression of dHAND by Western blot analysis. We observed atRA can regulate the dHAND expression in a dose- and time-dependent manner. AtRA also inhibited endothelin-1 (ET-1) expression in a time-dependent manner. Further studies revealed that pretreatment with 10 µM BQ-123, a selective endothelin-1 receptor (ETAR) antagonist, for 2 h can significantly counteract the inhibition of 5 µM atRA treatment for 2 h of dHAND mRNA and protein expression. Taken together, these results suggest that atRA regulates dHAND expression by ET-1/ETAR signal transduction pathway in H9c2 cells. The mechanism of ET-1/ETAR signaling in controlling the level of dHAND protein is to reduce the levels of dHAND mRNA. It is possible for atRA to exert its cardiac teratogenesis during vertebrate embryonic development in this way. J. Cell. Biochem. 99: 478–484, 2006. © 2006 Wiley-Liss, Inc.
Abbreviations used:
ET-1/ETAR, endothelin-1/endothelin-1 receptor; atRA, all-trans retinoic acid; bHLH, basic helix-loop-helix; ZAP, zone of polarizing activity.
Vertebrate heart development involves many transcription factors such as dHAND, Nkx2.5, and GATA [Lyons, 1996. The proper temporal and spatial expression of these factors governs the normal development of the vertebrate heart. Any situation affecting correct expression of these heart specific transcription factors may influence cardiac development. It has been reported that a deficiency of vitamin A and its active derivatives such as retinoic acid (RA) results in congenital heart defects [Smith et al., 1998. RA is necessary for early fetal cardiogenesis. Supplementing with excess vitamin A or RA during the gestational period leads to neonatal cardiac malformations [Tembe et al., 1996; Collins and Mao, 1999.
Retinoic acid may regulate vertebrate cardiogenesis by affecting some transcription factors such as Nkx2.5 [Pellizzer et al., 2004, GATA-4, and tbx5 [Liberatore et al., 2000. dHAND is a basic helix-loop-helix (bHLH) transcription factor. It is expressed in mesodermal and neural crest-derived structures during heart development. dHAND plays an essential role in the formation of the right ventricle, trabeculae, and neural crest-derived aortic arches. It is also involved in the regulation of chamber specification, cardiac looping, and the cardiac neural crest [Srivastava et al., 1997. During the zone of polarizing activity (ZPA) formation in vertebrate limb development, RA acts early to activate expression of dHAND as an upstream signal [Mic et al., 2004. All-trans retinoic acid (AtRA) suppresses endothelin-1 (ET-1) mRNA expression in cultured endothelial cells [Yokota et al., 2001. ET-1 is a 21-amino acid peptide. It has been found to be produced and secreted by diverse types of cells and to elicit a wide spectrum of biological effects. ETA receptors are preferentially activated by ET-1 and mediate biological actions of ET-1 [Chandan et al., 2003. ET-1 mainly acts as a locally-active autocrine/paracrine factor rather than as a circulating hormone [Giannessi et al., 2001. Many studies [Charite et al., 2001; Mina, 2001; Ivey et al., 2003; Yanagisawa et al., 2003; Fukuhara et al., 2004 have indicated that ET-1/endothelin-1 receptor (ETAR) signaling is crucial for the expression of dHAND in embryogenesis. dHAND serves as a downstream target for ET-1/ETAR signaling.
All the facts and studies mentioned above prompted us to think that it is possible for RA to regulate the expression of dHAND by ET-1/ETAR signaling during vertebrate heart development. AtRA may exert its cardiac teratogenesis in this way. Most previous studies used were practiced by vertebrate whole-embryo culture and explant cultures but rarely done on cultured cells in vitro. In this article, we aimed to explore whether atRA regulates the expression of dHAND in H9c2 cells (rat embryonic cardiomyocytes) and if so, its underlying mechanism(s). We present data showing that atRA inhibits the expression of dHAND in H9c2 cells in a time- and dose-dependent manner. AtRA may suppress the dHAND expression by ET-1/ETAR signaling. The mechanism of ET-1/ETAR signaling in controlling the level of dHAND protein is to reduce the levels of dHAND mRNA. These results may be helpful for interpreting the mechanisms of cardiac teratogenesis by atRA and vitamin A.
MATERIALS AND METHODS
Materials
All-trans retinoic acid, Dulbecco's modified Eagle' medium (DMEM), bovine serum albumin (BSA), BQ-123, Western blotting luminol reagent were purchased from Sigma (St.Louis, MO). Fetal calf serum (FCS) was from HyClone. The polyclonal antibodies to dHAND and β-actin were from Santa Cruz Biotechnology (Santa Cruz, CA). The anti-Propre-endothelin-1 IgG was obtained from Phoenix Pharmaceuticals, Inc.
H9c2 Cell Line and Culture
H9c2 cells from the American Type Culture Collection (ATCC, CRL-1446) were maintained by our laboratory. Cells were cultured in DMEM supplemented with 10% FCS, 100 U/ml of penicillin and 100 µg/ml streptomycin, in an atmosphere of 5% CO2/95% humidified air. H9c2 cells were grown to approximately 80% confluence, at which point the medium was changed to serum-free DMEM. Twenty-four hours later, different doses of atRA [dissolved in dimethyl sulfoxide (DMSO)] were added and cells were incubated in the dark for various times and different doses for different purposes.
Western Blot
After treatment with or without atRA, H9c2 cells were rapidly chilled on ice, washed twice with ice-cold washing buffer (10 mM Tris-HCl, 150 mM NaCl, 1 mM Na3VO4 [pH7.5]), and then lysed in 300 µl (per 106 cells) lysis buffer [10 nM Tris (pH7.4), 300 nM NaCl, 1 nM EDTA, 10 mM MgCl2, 2 mM DTT, 5 mM phenylmethysulfonyl fluoride, 10 µg/ml aprotinin, 10 µg/ml leupeptin, and 0.5% Nonidet P-40]. The lysis products were centrifuged at 12,000g for 15 min, the supernatent was collected, and protein concentrations were measured by Bradford method. Samples of H9c2 cell extract were subjected to SDS/PAGE and then electrophoretically transferred to a PVDF membrane (Millipore Co.). Non-specific binding was blocked by incubating the membranes with phosphate buffered saline containing 1% BSA and Tween-20 (PBST). Western blotting was performed using 1:200 dilution of dHAND or propre-endothelin-1 antibody as the primary, a 1:10,000 dilution of horseradish peroxidase-conjugated anti-antibody as the secondary and an ECL detection system. The membranes were exposed to Kodak XAR film. Quantification of the relative abundance of dHAND or endothelin-1 protein was done by volume densitometric analysis of the films using SmartView software (Shanghai FURI Science & Technology Co., Ltd). All the values were normalized to the values obtained with β-actin to correct for differences in loading and transfer efficiency.
RNA Isolation and Northern Blot Analysis
After H9c2 cells were treated with 5 µM atRA for 2 h in the presence or absence of 10 µM BQ-123, total cellular RNA was isolated with Trizol reagent (Invitrogen) following the recommended protocol. Twenty micrograms of total RNA was size-fractioned on formaldehyde-agarose gel (1%), transferred onto a nylon membrane (Hybond N+; Amersham Biosciences) by capillary blotting in the presence of 20 × SSC (300 mM sodium chloride and 300 mM sodium citrate), and then immobilized by incubating the membrane for 2 h at 80°C. Blots were hybridized with random prime-labeled rat cDNA probes for dHAND or glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The probes were prepared as previously described [Thomas et al., 1998 and were labeled with [32P] dCTP by a Prime-a-Gene labeling system (Promega). The membrane was washed with 0.1 × SSC containing 1% sodium dodecyl sulfate (SDS) at 42°C for 30 min and then exposed to X-ray film at −70°C. Blots of specific mRNA bands were detected by autoradiography and analyzed with a densitometer (Shanghai FURI Science & Technology Co., Ltd). The level of expression of dHAND mRNA was quantified and was normalized to the GAPDH signal.
Statistical Analysis
Data are presented as mean ± SD (standard deviation). The data were evaluated by unpaired Student's t-test or one-way ANOVA using SPSS 13.0 (SPSS, Inc., Chicago, IL) program. Following the one-way ANOVA, the least significant difference (LSD) test or the Student-Newman-Keuls (S-N-K) q test was performed as a post hoc multiple comparision. P < 0.05 was considered statistically significant.
RESULTS
The Regulation by AtRA of dHAND Protein Expression in H9c2 Cells
To investigate the effects of atRA on the protein expression of dHAND, we employed the Western blot assay. AtRA (5 µM) significantly inhibited the protein expression of dHAND after atRA treatment for 2, 6, 12, and 24 h (P < 0.05; Fig. 1). When H9c2 cells were exposed to atRA for 2 h, the dHAND protein expression is the lowest among all the treatment groups compared with control cells that were treated only with vehicle (indicated as 0 µM). AtRA also inhibited the dHAND protein expression in a dose-dependent manner (Fig. 2). AtRA (5 µM and 10 µM) inhibited the dHAND protein expression by 88.51% and 89.53%, respectively compared with 0 µM atRA as the control (P < 0.05). The level of dHAND following 24 h exposure to RA increased compared with the 2 h treatment (P < 0.05).

The time course of effects of atRA on dHAND protein expression. (A) H9c2 cells were exposed to atRA (5 µM) for the times indicated. The protein expression of dHAND was determined by Western blotting. (B) The protein levels of dHAND were quantified by densitometric scanning and normalized to the levels of β-actin. Data represent the average fold of controls from six independent experiments (mean ± SD). *P < 0.05 versus control.
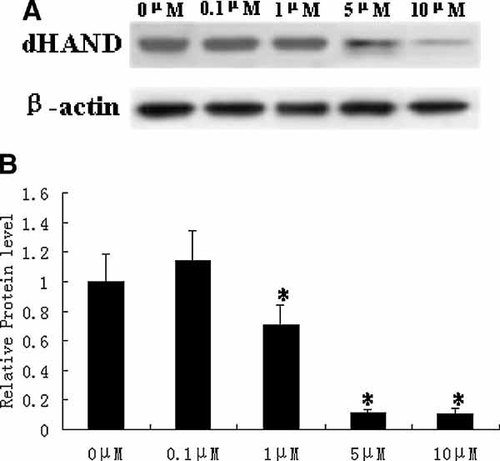
AtRA inhibits the protein expression of dHAND in a dose-dependent manner. (A) H9c2 cells were treated with different doses of atRA for 2 h. Cell extracts were subjected to 10% SDS–PAGE and immunoblotted with an antibody specific for dHAND. (B) The protein levels of dHAND were quantified by densitometric scanning and normalized to the levels of β-actin. Data represent the average fold of controls from six independent experiments (mean ± SD). *P < 0.05 versus control.
The Regulation by AtRA of ET-1 Protein Expression in H9c2 Cells
Based on the effects of atRA on dHAND protein expression, we treated H9c2 cells by 5 µM atRA for different times and found the ET-1 protein level was lowered versus control after atRA treatment for 2, 6, 12, and 24 h (P < 0.05; Fig. 3). When H9c2 cells were exposed to atRA for 2 h, the ET-1 protein expression is the lowest among all the treatment groups compared with control cells that were treated only with vehicle (indicated as 0 µM). ET-1 protein level also increased after 24 h of RA treatment compared with the 2 h treatment (P < 0.05).
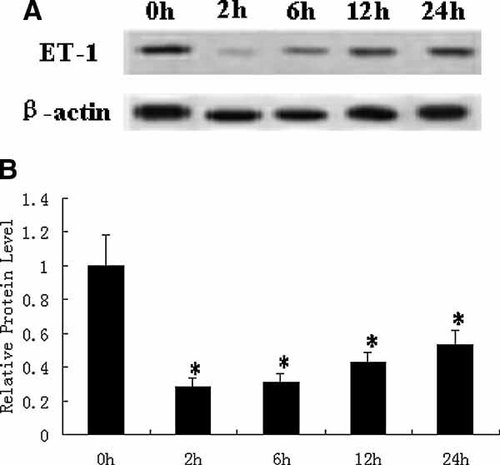
The time course of effects of atRA on ET-1 protein expression. (A) H9c2 cells were exposed to atRA (5 µM) for the times indicated. The protein expression of ET-1 was determined by Western blotting. (B) The protein levels of ET-1 were quantified by densitometric scanning and normalized to the levels of β-actin. Data represent the average fold of controls from six independent experiments (mean ± SD). *P < 0.05 versus control.
The Role of ET-1/ETA Signaling in the Regulation of AtRA to dHAND Protein Expression
To test whether ET-1/ETA signal pathway plays a role in the regulation of atRA to dHAND protein expression, H9c2 cells were pretreated with 10 µM BQ-123 for 2 h and then exposed to 5 µM atRA for 2 h. BQ-123 is a kind of selective blocking agent for ETA receptor and can block ET-1/ETA signal transduction pathway effectively. We found 5 µM atRA treatment for 2 h decreased the dHAND protein level while 10 µM BQ-123 pretreatment for 2 h before 5 µM atRA treatment can significantly attenuate the inhibition by atRA of dHAND protein level (P < 0.05; Fig. 4). This indicates atRA exert its inhibition of dHAND protein expression by ET-1/ETA signal pathway.
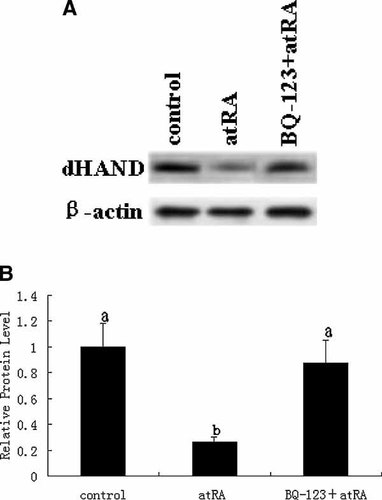
Role of ET-1/ETA signal pathway in the atRA-induced decrease in dHAND expression. (A) H9c2 cells were exposed to atRA (5 µM) for 2 h, in the presence or absence of BQ-123 (10 µM). Cell extracts were subjected to 10% SDS–PAGE and immunoblotted with an antibody specific for dHAND. (B) The protein levels of dHAND were quantified by densitometric scanning and normalized to the levels of β-actin. Data represent the average fold of controls from six independent experiments (mean ± SD). a, b, different letters represent P ± 0.05 versus each other.
The Role of ET-1/ETA Signaling in the Regulation of AtRA to dHAND mRNA Expression
Based on the results mentioned above, we pretreated H9c2 cells with 10 µM BQ-123 for 2 h and then exposed the cells to 5 µM atRA for 2 h. We found 5 µM atRA treatment for 2 h decreased the dHAND mRNA level by 71.57% while 10 µM BQ-123 pretreatment for 2 h before 5 µM atRA treatment can significantly attenuate the inhibition of atRA to dHAND mRNA level (P < 0.05; Fig. 5). This indicates atRA exerts its inhibition of dHAND mRNA expression by ET-1/ETA signal pathway.
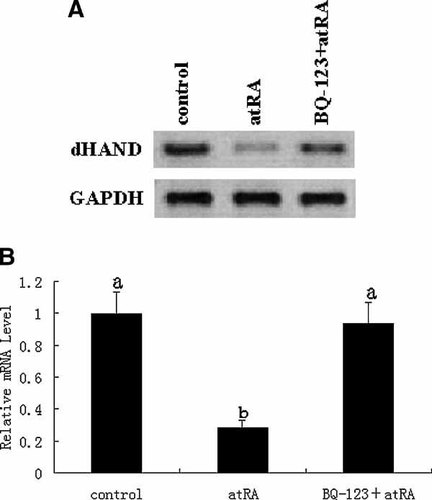
Northern blot analysis of dHAND mRNA in H9c2 cells. (A) H9c2 cells were exposed to atRA (5 µM) for 2 h, in the presence or absence of BQ-123 (10 µM). Total RNA was extracted from cells and Northern blot analyses were performed. (B) The mRNA levels of dHAND were quantified by densitometric scanning and normalized to the mRNA levels of GAPDH. Data represent the average fold of controls from six independent experiments (mean ± SD). a, b, different letters represent P ± 0.05 versus each other.
DISCUSSION
The present study demonstrated that atRA suppressed dHAND protein expression in a time- and dose-dependent manner in cultured H9c2 cells. AtRA (5 µM) inhibited the expression of ET-1 protein time-dependently. AtRA (5 µM) treatment for 2 h inhibited the expression of dHAND protein and ET-1 protein to the greatest extent. If blocked ETA receptors using their specific repressor BQ-123 beforehand, we observed the dHAND protein expression increased compared with that of only 5 µM atRA treatment for 2 h. BQ-123 (10 µM) also can significantly attenuate the inhibition of atRA to dHAND mRNA level. AtRA appears to regulate the protein expression of dHAND through the ET-1/ETAR signaling pathway. The mechanism of ET-1/ETAR signaling in controlling the level of dHAND protein is to reduce the level of dHAND mRNA.
dHAND is a bHLH transcription factor cloned in 1995. It was given different names such as HAND2, Th2, and exd [Srivastava et al., 1997. In the developing heart, dHAND is initially expressed in the precardiac mesoderm. It is also expressed throughout the linear heart tube, but becomes restricted predominantly to the future right ventricular compartment during cardiac looping [Srivastava et al., 1995, 1997. dHAND mutants exhibit hypoplasia of the right ventricle, branchial arches, and aortic arch arteries [Srivastava, 1999. RA, derived from the nutrient vitamin A, has potent cardiac teratogenicity. RA receptors are expressed throughout heart. A major function of RA is transcriptional regulation through its nuclear receptors. RA-responsive genes are found in all cardiac compartments (myocardium, endocardium, cushion mesenchyme). Various cardiac malformations can be observed in RA receptor null mutant mice [Smith and Dickman, 1997. It has been reported that RA affects heart development by regulating transcription factors such as Nkx2.5, GATA, and tbx5. But whether RA regulates dHAND to affect heart development still remains to be elucidated. Mic et al. 2004 proved that RA acts early to activate the expression of dHAND during ZPA in vertebrate limb development. This caused us to assume that atRA may regulate dHAND expression in embryonic cardiomyocytes. Our results also indicate that atRA regulates the expression of dHAND in a time- and dose-dependent manner in cultured H9c2 cells. AtRA (5 µM) treatment for 2 h inhibited the expression of dHAND to the maximal degree. High levels of atRA may exert their cardiac teratogenesis by suppressing dHAND expression. There is an apparent increase in the level of dHAND following 24 h exposure to RA compared with the 2 h treatment. This is caused by the time course effects of atRA on the protein levels of ET-1.
We next explored the possible mechanism(s) of the inhibitory effect of atRA on dHAND protein expression. Endothelins (ETs, including ET-1, ET-2, and ET-3) is a peptide family of 21 amino acids that were originally isolated from the culture medium of porcine aortic endothelial cells [Yanagisawa et al., 1988. ETs exerts a variety of biological effects including vasoconstriction and cell proliferation via G protein-coupled endothelin receptors type A and B [Kurihara et al., 1999; Kedzierski and Yanagisawa, 2001. ETA receptors exist mainly on vascular smooth muscle cells and respond to ET-1. Many types of cells can produce and secrete ET-1 which mainly acts as a locally-active autocrine/paracrine factor. Signalings through ET receptors are essential for normal embryonic development of subsets of neural creast cell derivatives. ET-1, as well as ETA receptor, also has an important role in cardiovascular development, as observed by the variety of abnormalities related to neural crest-derived tissues in mouse embryos deficient in a member of the ET-1/ETAR pathway [Giannessi et al., 2001. RA, leptin, prostaglandins, hypoxia, etc. are modulators of the ET system. Yokota et al. 2001 have proved that atRA suppressed ET-1 mRNA expression in cultured endothelial cells. Consistent with this, we further observed that atRA inhibited the expression of ET-1 in a time-dependent manner. AtRA (5 µM) treatment for 2 h can significantly inhibited the expression of ET-1 in cultured H9c2 cells. While the level of ET-1 following 24 h exposure to RA apparently increased in compared with that of the 2 h treatment. This may be the adaptive response of cells to extraneous stimulating factors. It is possible for cells to initiate some potential protective mechanism(s) unknown to us. Taking cues from the consistency of the time course effects of atRA on dHAND and ET-1 protein expressions, we assumed that there should be some linkage(s) between the expression of dHAND and ET-1 in H9c2 cells.
It has been proved that ET-1 can regulate dHAND expression during embryonic development. dHAND is a ET-1-dependent transcription factor. In ET-1 mutant embryos, dHAND expression in the branchial arches is downregulated, implicating it as a transcriptional effector of ET-1 action [Charite et al., 2001. ET-1/ETAR and subsequent epithelial signals are sequentially involved in branchial arch development by maintaining dHAND and Dlx6 expression [Fukuhara et al., 2004. Yet the role of ET-1/ETAR signaling in the regulation of atRA to dHAND expression in heart development still remains to be determined. We tentatively used BQ-123, a selective inhibiter of ETAR, to block the ET-1/ETAR signaling pathway before atRA treatment in cultured H9c2 cells and observed the dHAND protein expression was significantly increased compared with that of only atRA treatment. This caused us to hypothesize that atRA first inhibited ET-1/ETAR signaling, while cells produced and secreted ET-1 to act on themselves and cells in the peripheral region adjacent to them. With the inhibition of ET-1/ETAR signal, dHAND protein expression reduced correspondingly. AtRA may exert its teratogenic action and lead to cardiac malformations in this way eventually. Therefore, we came to the conclusion that atRA regulates the expression of dHAND through ET-1/ETAR signaling pathway in cultured H9c2 cells. Further exploration indicated that the mechanism of ET-1/ETAR signaling in controlling the level of dHAND protein is to reduce the levels of dHAND mRNA.
These results may have significance in illuminating the signal transduction pathways of atRA in embryonic developing heart. They may also be helpful to understand the cardiac teratogenesis of atRA and its underlying mechanism(s). Vertebrate heart development is a very complicated course involving many cellular and molecular events. Various related protein-signaling molecules interact with each other and form a complex network fine-tuning cardiac development temporally and spatially. Factors in vivo and in vitro affecting these molecules will cause a delicate imbalance of this network and influence heart development. We prove that atRA suppresses dHAND expression through ET-1/ETAR signal transduction pathway in cultured H9c2 cells. AtRA may induce cardiac malformations in vertebrate developing embryos. Further studies are needed to explore whether there are other signaling molecules such as Dlx6, etc. functioning as segments of the signaling pathway from ET-1 to dHAND. AtRA may also regulate dHAND expression through other pathways. The regulation by atRA of dHAND may be different in different types of cells and tissues. All these prospective questions may generate future research projects.