Inhibition of DNA topoisomerase II may trigger illegitimate recombination in living cells: Experiments with a model system
Abstract
We have developed a plasmid test system to study recombination in vitro and in mammalian cells in vivo, and to analyze the possible role of DNA topoisomerase II. The system is based on a plasmid construct containing an inducible marker gene ccdB (“killer” (KIL) gene) whose product is lethal for bacterial cells, flanked by two different potentially recombinogenic elements. The plasmids were subjected to recombinogenic conditions in vitro or in vivo after transient transfection into COS-1 cells, and subsequently transformed into E. coli which was then grown in the presence of the ccdB gene inducer. Hence, all viable colonies contained recombinant plasmids since only recombination between the flanking regions could remove the KIL gene. Thus, it was possible to detect recombination events and to estimate their frequency. We found that the frequency of topoisomerase II-mediated recombination in vivo is significantly higher than in a minimal in vitro system. The presence of VM-26, an inhibitor of the religation step of the topoisomerase II reaction, increased the recombination frequency by 60%. We propose that cleavable complexes of topoisomerase II are either not religated, triggering error-prone repair of the DNA breaks, or are incorrectly religated resulting in strand exchange. We also studied the influence of sequences known to contain preferential breakpoints for recombination in vivo after chemotherapy with topoisomerase II-targeting drugs, but no preferential stimulation of recombination by these sequences was detected in this non-chromosomal context. J. Cell. Biochem. 99: 598–608, 2006. © 2006 Wiley-Liss, Inc.
Recent investigations have shown that in many cases anti-tumor chemotherapy with topoisomerase II-specific agents results in secondary leukemias [Rowley, 1993; Rowley et al., 1997; Felix, 1998, Mao et al., 2000; Zhang et al., 2002]. These leukemias are often correlated with various chromosomal rearrangements. However, it has not yet been proven that chromosomal rearrangements are the direct cause of tumors (see [Mitelman, 2000] for review).
Topoisomerase II, a member of the DNA topoisomerase family, acts as a regulator of DNA topology [Champoux James, 2001; Wang, 2002]. It can relax positive and negative supercoils in circular DNA (or closed loop domains of eukaryotic DNA) and unknot or uncouple DNA molecules by introducing transient double-strand breaks (DSBs). Such breaks may lead to chromosomal translocations which, in turn, are believed to be common causes of tumor development [Elliott and Jasin, 2002]. A DSB may be repaired in three different ways—homologous recombination (HR), single-strand annealing (SSA) or non-homologous end joining (NHEJ). HR requires long regions of homology between the DNA fragments and may lead to exact recovery of the affected fragment. Direct repeats flanking the DBS are necessary for SSA to work (this type of DSB repair is sometimes considered as a particular case of HR); the complementary single-strand repeats are annealed and all non-homologous regions are deleted. No homology at all is needed for the protein complex which catalyzes NHEJ; this complex performs processing of DSB ends in which endo- and exonucleases, DNA polymerases, helicases, polynucleotide kinases, and ligases can take part and often results in duplication, inversion or deletion of DNA fragments [Elliott and Jasin, 2002]. Some translocations have been suggested to result from errors in V(D)J recombination [Jaeger et al., 1994, Welzel et al., 2001 and references therein], but this mechanism is likely to operate in lymphoid cells only.
The eukaryotic genome is known to consist of chromatin loops attached to the nuclear matrix in interphase nuclei and to the chromosomal scaffold in mitotic chromosomes [Cook et al., 1976; Razin and Gromova, 1995]. Regions where these loops are attached to the nuclear matrix are called matrix association regions or MARs [Cockerill and Garrard, 1986]. These elements are not identical but have a number of common properties: they are usually about 1,000 bp or slightly more, are AT-rich, they often form non-canonical DNA structures (palindromes or different distortions of the double helix), are often situated near cis-regulatory sequences (enhancers, promotors, insulators, and transcription factor binding sequences) and contain sites or regions hypersensitive to DNAse I. Moreover, MARs are enriched in topoisomerase II cleavage sites [Gasser and Laemmli, 1986; Boulikas, 1993, 1994; Bode et al., 1995; Yan and Qian, 1998; Porter et al., 1999; Razin, 2001; Holmes-Davis and Comai, 2002; Felsenfeld et al., 2004; Yusufzai and Felsenfeld, 2004].
Topoisomerase II is one of the main components of the nuclear matrix [Berrios et al., 1985; Adachi et al., 1989]. This enzyme seems to be able to induce illegitimate recombination in vitro, especially in the presence of some of its inhibitors [Bae et al., 1988; Charron and Hancock, 1991; Gale and Osheroff, 1992; Ikeda, 1994; Asami et al., 2002]. Two alternative mechanisms have been proposed for topoisomerase II-mediated illegitimate recombination. Historically, the first was the model of subunit exchange [Ikeda et al., 1981]. During the reaction each subunit of homodimeric topoisomerase II is covalently bound to one end of the introduced transient DSB, thus forming the cleavable complex [Liu, 1989; Nitiss and Wang, 1996]. Two cleavable complexes located close to each other are proposed to be able to exchange reciprocally their subunits with the attached DNA ends. If the resulting chimeric cleavable complexes completes the enzymatic cycle, illegitimate recombination will occur. In other words, topoisomerase II both introduces DSBs and repairs them. According to the second, “induction” model [Adachi et al., 2003, 2004; Bystritskiy and Razin, 2004], topoisomerase II only introduces DSBs in DNA but their repair is performed by some other enzyme(s), most probably components of the standard cellular repair systems. It is noteworthy that many topoisomerase II inhibitors such as doxorubicin, amsacrine, teniposide or etoposide block the catalytic reaction at the stage of the cleavable complex [Liu, 1989] and thus introduce potential DSBs into DNA. These DSBs are likely to be repared by NHEJ [Kantidze et al., 2006].
In order to analyze the role of topoisomerase II in illegitimate recombination and to clarify its mechanism, we have developed a test system. This is based on a plasmid construct containing an inducible “killer” (KIL) gene flanked by two different recombinogenic elements. The plasmid is placed under recombinogenic conditions and subsequently transformed into bacteria which are grown in medium containing the inducer of the KIL gene. Hence, only bacteria lacking the KIL gene survive. The KIL gene can be removed only by recombination between the flanking regions, and therefore this system allows to detect recombination events and to estimate their frequency. This test system may be used to study any aspect of illegitimate recombination; here we use it to find out if topoisomerase II takes part in illegitimate recombination and to analyze the influence of topoisomerase II inhibitors on the frequency of this process both in vitro and in vivo.
MATERIALS AND METHODS
Plasmid Construction
Our first step was to produce a plasmid able to replicate autonomously in simian COS-1 cells. The SV40 origin of replication obtained by PCR from the pSI vector (Promega®) using the primers actcatatgTCAGCAACCATAGTCC and actcatatgGTCAGAA-GAAT-CCAGC (restriction sites are shown with small bold letters and extra bases providing a dock for the restriction enzyme are underlined) was digested with NdeI and inserted into the vector pSP73 (Promega®) to yield plasmid pSP-ori. A fragment containing the breakpoint cluster region (bcr) from the human AML1 gene (corresponding to positions 4,6301–4,9309 of the GenBank accession AP000057.1) was amplified from total human genomic DNA with the primers actctcgagATGTTACTTACAGCCACAGATC and actctcgagCAAGCATTCCTTCT-TCCCT, and the 2,836 bp PCR-product was cut with XhoI and ligated into SalI-XhoI–digested pSPori yielding pSP-AML-ori. Then a fragment containing the bcr of the human ETO gene (positions 11,5939–11,8673 of GenBank AF181450.5) was amplified using the primers ctggatccAATTACTCTGGAAAGTGAAGGG and ctggatccCATGCAACTAGTCAGACA-AAGC and the 2,484-bp PCR product was digested with BamHI and inserted into BamHI- and BglII-digested pSP-ori-AML, yielding p2BCR-ori. To produce the final construct, pKIL2BCR-ori, the KIL gene was excised from plasmid pKIL18 [Bernard et al., 1994; Bernard, 1995] by digestion with PvuII and the 660 bp fragment was inserted into the filled-in BamHI site of p2BCR-ori.
To obtain a plasmid containing the ψTTC pseudogene, a 6,516 bp fragment (positions 34,501–28,052 of GenBank AL391495.16) was excised from bacmid clone RPCI11-324B6 [Osoegawa et al., 2001] (Children's Hospital Oakland Research Institute) by BseX3I. Because pSP-ori contained no BseX3I site, two oligonucleotides (GATCGGCCG and TCGACGGCC) were annealed to give an adaptor containing a BseX3I site, which was cloned into pSP-ori digested with BglII and XhoI. This construct was cut with BseX3I and the excised ψTTC pseudogene was inserted to produce plasmid pTTC-ori. To introduce the KIL gene into this construct, the 660 bp fragment excised from PvuII-digested pKIL18 was cloned into pTTC-ori digested with BstPAI yielding plasmid pKIL2TTC-ori.
Bacterial Strain and Transformation
The respective plasmids were transformed into E. coli XL-1 cells (XL-1 blue: recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZ.M15 Tn10 (TetR)]). Ampicillin-resistant (AmpR) colonies were selected by plating bacteria on plates containing ampicillin (75 mg/ml). Colonies lacking the KIL gene were selected by growth on plates containing ampicillin (75 mg/ml) and IPTG (5 mM).
Decatenation of Plasmids
Plasmid DNA (1 µg) was incubated with one unit of human topoisomerase II alpha (Topogen) in topoisomerase II assay buffer (50 mM Tris-HCl, pH 8.0, 120 mM KCl, 100 mM MgCl2, 0.5 mM ATP, 0.5 mM DTT, and 30 mg/ml BSA) in a total volume of 30 µl for 30 min at 37°C. The reaction was terminated by adding 1.2 µl 0.5 M EDTA and 3 µl 5 M NaCl followed by incubation for 10 min at 30°C. The mixtures were extracted with an equal volume of phenol and DNA was ethanol-precipitated. Bacterial cells were transformed by this DNA and grown on LB agar containing ampicillin (75 mg/ml) or ampicillin (75 mg/ml) and IPTG (5 mM). Some AmpR colonies were picked and grown in LB medium with ampicillin (75 mg/ml). Plasmid DNA was extracted and the full procedure repeated. Absence of IPTG-resistant clones was interpreted as proof of successful decatenation.
Induction of Recombination by Topoisomerase II In Vitro
Plasmid DNA (0.1 µg of each) were incubated with five units of topoisomerase II (Topogen) and 0.3 µg of VM-26 (Bristol-Myers Squibb) in cleavage buffer (30 mM Tris-HCl, pH 7.6, 60 mM NaCl, 8 mM MgCl2, 3 mM ATP, and 15 mM 2-mercaptoethanol) in a total volume of 30 µl for 30 min at 37°C, and DNA was isolated as described above. Bacteria were transformed with these reaction mixtures and grown on LB agar containing ampicillin (75 mg/ml) or ampicillin (75 mg/ml) and IPTG (5 mM).
Cell Culture, Transfection, and Isolation of Plasmid DNA From Transfected Cells
COS-1 cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Pan-Eco) with 10% fetal bovine serum (FBS; Invitrogen) in 100 mm dishes with 10 ml of medium and incubated for 24 h. Then cells were trypsinized and replated at a density of ∼2 × 105 cells/well in standard 6-well plates with 2 ml of medium/well. Transfection of plasmid DNAs into COS-1 cells was performed using Unifectin-56 (Unifect Group, Moscow) according to the manufacturer's protocol. When confluence reached 50%, 3 µl of Unifectin-56 in culture medium containing 0.5 µg of plasmid DNA was added to each well. The topoisomerase II inhibitor VM-26 was added to the medium 18 h later to a final concentration of 40 µM and the medium was replaced after 1 h with fresh medium without inhibitor. The cells were harvested after two more hours and plasmid DNAs were isolated using the Hirt extraction protocol [Hirt, 1967].
Radioactive Labeling of Oligonucleotides and Hybridization With DNA
Oligonucleotide probes spaced evenly on the recombinogenic target sequences were designed using Array Designer 3.0 software (PremierBiosoft) (Table I). The purified DNAs (0.1 µg each) from IPTG-resistant clones were dotted on Nytran SuPerCharge membrane (Schleicher & Schuell) using a MiniFold I vacuum slot-blotter (Schleicher & Schuell). The membrane was subsequently hybridized with an end-labeled oligo probe prepared by labeling 50 pmol of the corresponding oligo with [γ-32P]ATP (6,000 Ci/mmol; Izotop, Moscow) using T4 polynucleotide kinase (SibEnzyme, Novosibirsk) according to the manufacturer's recommendations. Hybridization with 50 µCi of probe was performed in 0.69 M NaH2PO4, 0.28 M Na2HPO4, 0.5% SDS, 0.7 mM EDTA at 50°C for 2 h using an HB-1000 Hybridizer (UVP). The membrane was washed for 15 min in 2 × SSC, 0.1% SDS and for an additional 15 min in 0.2 × SSC, 1% SDS, both at room temperature. The results of hybridization were visualized with a Cyclone Storage Phosphor System (Packard).
| Oligo name | Distance of the oligo from the vector part of the plasmid, bp | Oligo sequence |
|---|---|---|
| A0 | 43 | GGAAGCTCACCAGATAGGCTGTAGCTA |
| A1 | 372 | TATTGGAACCATAAGGCTGAGGAAGCA |
| A2 | 693 | TTTGAGACAATGTCAACTGTGCCAAGG |
| A3 | 978 | CTTTCATCCACAAAGCCGGTGCTTAAA |
| A4 | 1270 | CAGGCATTAGACTGTGGTAGGAGGTTAA |
| A5 | 1559 | CTAAGAGAGACACAACTGCCAAGAGCT |
| A6 | 1837 | TGAGAGTTATGCCTGAACACTTCTGCTA |
| A7 | 2151 | ATTTGGCACATGAGGTCTGATGGTTTC |
| A8 | 2578 | TGAGAAAATGGTACTAGAAGAATTTAGGACCTTTT |
| A9 | 2895 | TACAGCCACAGATCACAATGGTGTCTT |
| E0 | 32 | AAACTCATGGTCTCAGGTCACTTTGATTT |
| E1 | 324 | ATATTTAAGTTGTGTGTGCATTTATGTGTATGTGT |
| E2 | 618 | AAGGTAGATTCCTACATAGGCACTAGCAAG |
| E3 | 946 | CCTCCTCATCTTGACTAATGGCGAGTTA |
| E4 | 1253 | TCCAAATCCCTTGCATATGGAGACACT |
| E5 | 1533 | GAACTTGATTATATTCATTATTTTGGAGCCTGGG |
| E6 | 1838 | GCATTGAAAGAACTGAACAAATGACACATAAATG |
| E7 | 2137 | CTATGCACACATACTAATCTGCCACGTTTA |
| E8 | 2442 | TCTTACTTTCATTTATTAAAATTCTCCTTAACTGCA |
Treatment of Nuclei With Micrococcal Nuclease (MNase) and Southern Hybridization
We used digestion with MNase to study if the episomal DNA had a nucleosome organization. Cells transfected with pKIL2BCR-ori were harvested 24 h after transfection as described above and permeabilized with buffer A (50 mM Tris, pH 7.5, 5 mM MgCl2, 5 mM CaCl2) supplemented with 0.5% Nonidet P-40, 0.5 mM PMSF, and 100 mM NaCl for 30 min on ice. The nuclei were then collected by centrifugation, resuspended in buffer A, and incubated with different amounts of MNase (Fermentas) (see Fig. 2) for 30 min at 37°C. The reaction was stopped by adding 3 volumes of proteinase K solution (0.1 M EDTA, 1% SDS, and 0.5 mg/ml proteinase K). The mixture was left at 50°C overnight and the DNA was phenol extracted and ethanol precipitated.
The resulting material was separated by electrophoresis and capillary transferred as described above. The membrane was hybridized with linearized pKIL2BCR-ori DNA labeled with [α-32P]dATP (6000 Ci/mmol; Izotop) using a Megaprime labeling kit (Amersham Biosciences) as recommended by the supplier. Hybridization was performed as described above but at 66°C overnight. The membrane was washed once for 15 min in 2 × SSC, 0.1% SDS at room temperature and for additional 15 min in 0.2 × SSC, 1% SDS at 66°C. Hybridization was visualized as described above.
RESULTS
The Test System
We designed a shuttle vector (Fig. 1) which contains the ccdB gene, encoding an inhibitor of bacterial gyrase, under the control of the lac promotor, forming a KIL gene whose expression in bacteria leads to cell death. Deletion of ccdB as a result of illegitimate recombination between flanking elements allows bacterial cells to grow and form colonies on medium containing IPTG, an inducer of the lac promotor. The construct also contains a bacterial origin of replication (the common ColE1 plasmid origin), a eukaryotic origin of replication (from simian virus 40) and a selection marker for cultivation in bacteria (ampicillin resistance). The KIL gene was placed between sequences putatively involved in rearrangements of human chromosome in vivo; we chose the third bcr of the AML1 gene and the second bcr of the ETO gene, which participate in the translocation t(8;21)(q22; q22) which is strongly associated with acute myeloid leukemia [Zhang et al., 2002]. The resulting plasmid was named pKIL2BCR-ori. We also designed a control construct containing non-recombinogenic elements; the long processed ψTTC3 human pseudogene (also called LOC286495) was cloned into pSP-ori and the ccdB gene was inserted into the center of the fragment. This plasmid was named pKIL2TTC-ori.
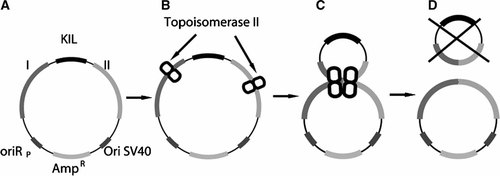
Principle of the test system for studying illegitimate recombination. A: The test plasmid construct containing an inducible “killer” gene (KIL) flanked by two different target sequences (I and II). The plasmid also contains a bacterial selection marker (AmpR) and two replication origins, prokaryotic (OriRP) and eukaryotic (SV-40 ori). B: Topoisomerase II binds to the regions flanking the KIL gene and forms cleavable complexes in the DNA. C: Two molecules of topoisomerase II interact with each other (according to the subunit exchange model). D: Exchange of topoisomerase II subunits results in illegitimate recombination giving rise to two circular molecules. One of these does not contain the plasmid origin of replication and therefore cannot survive in bacterial cells, while the other lacks the killer marker and is thus resistant to IPTG. Note that recombination events that do no eliminate the killer gene will not be detected by our assay.
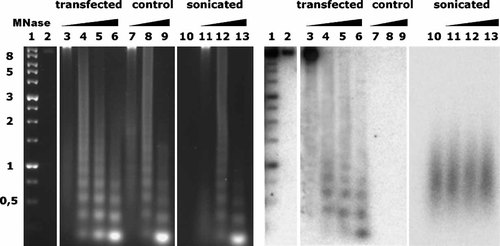
Plasmids are in a nucleosomal conformation in the nuclei of COS-1 cells. Ethidium bromide-stained agarose gel (left) and results of Southern hybridization (right) of the same gel with total pKIL2BCR-ori DNA. 1, markers (lengths in kb shown at left); 2, purified linearized pKIL2BCR-ori DNA; 3, total DNA from nuclei of COS-1 cells transfected with pKIL2BCR-ori; 4–6, same digested with increasing amounts of micrococcal nuclease (MNase); 7–9, MNase-digested COS-1 DNA (no transfection); 10, sonicated pKIL2BCR-ori DNA; 11–13, sonicated pKIL2BCR-ori DNA mixed with MNase-digested COS-1 DNA.
Catenanes containing two or more interlocked molecules could have been formed in the course of construct preparation, and deletion of the ccdB gene in one of the molecules would not be revealed in our test system because it would be masked by the normal ccdB gene in the second molecule. To assess the possible presence of catenanes, the DNA constructs were incubated with purified topoisomerase II under catalytic conditions and transformed into bacterial cells which were grown in medium containing IPTG. No IPTG-resistant colonies we observed in ∼5,000 total colonies for pKIL2BCR-ori and ∼7,000 colonies for pKIL2TTC-ori, confirming that the frequency of catenated molecules was negligible.
Recombination events and their frequency were then assessed by exposing the test constructs to potentially recombinogenic conditions and then transforming bacterial cells with the resulting material. The cells were grown in the presence of the inducer of the KIL gene, IPTG. The frequency of recombination was calculated as the ratio of the number of IPTG-resistant colonies to the total number of colonies on medium without IPTG.
Induction of Illegitimate Recombination by Topoisomerase II In Vitro
To find out whether topoisomerase II can directly mediate recombination when its religation activity is suppressed, we incubated the test constructs with purified topoisomerase II in the presence of the inhibitor of the enzyme's religation reaction VM-26 (teniposide). The test plasmids were then transformed into bacterial cells and the percentage of colonies with a deleted ccdB gene was calculated as described above. We did not observe any recombination events in the case of pKIL2BCR-ori and the presence of the inhibitor showed no effect. The recombination frequency for pKIL2TTC-ori was 0.06% in the absence of teniposide and 0.07% in its presence. These data do not allow us to exclude completely the possibility that topoisomerase II can mediate recombination, but the frequency of such an event is beyond the sensitivity of our system.
Replication and Nucleosomal Status of Transfected Plasmid DNA
To verify that our constructs replicated in an episomal fashion in COS-1 cells, the change in methylation pattern of plasmid DNA during replication [McWhinney and Leffak, 1990] was examined. The prokaryotic cells used to grow our constructs have the dam-methylation system, which is absent in eukaryotic cells, and moreover eukaryotic cells do not maintain preexisting dam-methylation in the course of replication so that each round of replication of an initially dam-methylated plasmid DNA will increase the amount of non-dam-methylated DNA while keeping the amount of dam-methylated DNA constant. The DpnI restriction endonuclease cleaves only dam-methylated DNA while MboI does not cleave dam-methylated DNA, so that by restricting the constructs isolated from transfected eukaryotic cells with these endonucleases one may calculate the percentage of replicated plasmid [McWhinney and Leffak, 1990]. Test constructs purified from E. coli were transfected into COS-1 cells, the cells were harvested after 18 h, and plasmid DNAs were isolated [Hirt, 1967] and digested with either MboI or DpnI. We found that the percentage of DpnI-resistant DNA increased linearly with time (data not shown), confirming that our test constructs replicated in COS-1 cells.
It was also important to know if autonomously replicating test constructs were arranged into nucleosomes. Thus nuclei of cells bearing the autonomously replicating construct pKIL2BCR-ori were treated with MNase and DNA fragments were separated by agarose gel electrophoresis and transferred onto a nylon filter. Hybridization with pKIL2BCR-ori DNA revealed a typical nucleosomal pattern (Fig. 2). To eliminate the possibility that fragmented plasmid DNA was adsorbed randomly on cellular nucleosomal bands, pKIL2BCR-ori DNA sonicated to 200–1,000 bp fragments was mixed with a nucleosomal ladder from non-transfected cells, and no nucleosomal ladder was observed (Fig. 2).
Induction of Illegitimate Recombination In Vivo and the Influence of Inhibitors
The frequency of recombination in the constructs propagating in COS-1 cells was then studied. The frequency of recombination between AML1 and ETO bcrs increased with time (Fig. 3), probably reflecting the fact that replication of plasmids in COS-1 cells stimulates recombination. In further experiments plasmid DNA was isolated 16–18 h after transfection to obtain sufficient quantities and to keep the background (spontaneous) recombination frequency at a relatively low level (about 8%). To test whether inhibition of DNA topoisomerase II affects the frequency of illegitimate recombination in vivo in autonomously replicating plasmids, COS-1 cells transfected with different test constructs were incubated with VM-26 for 1 h. The cells were then cultivated in drug-free medium for 2 h to allow the rejoining of the DNA which had been trapped in cleavable complexes with topoisomerase II. The plasmid DNA was extracted and the percentage of recombinants with a deleted ccdB gene was calculated. Treatment of cells with 40 µM VM-26 increased the frequency of illegitimate recombination 1.6-fold compared to untreated cells (Table II). Moreover, the frequency of recombination without VM-26 was higher in the control plasmid containing presumably non-recombinogenic elements than in the construct containing bcrs (Fig. 3).
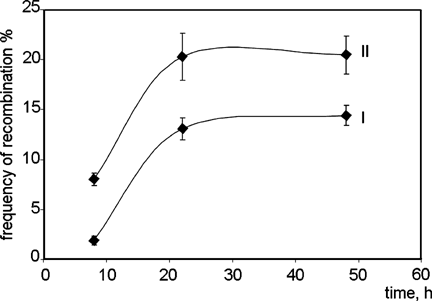
Influence of time on recombination frequencies during propagation of plasmids in COS-1 cells. Recombination frequency of pKIL2BCR-ori (I) and pKIL2TTC-ori (II) was determined after the specified intervals of time. Error bars show standard deviation (n = 4).
| Plasmid | Without inhibitor, % | With inhibitor, % |
|---|---|---|
| pKIL2BCR-ori | 8.21 ± 1.98 | 12.89 ± 1.73 |
| pKIL2TTC-ori | 15.61 ± 2.25 | 25.97 ± 3.11 |
Mapping of Breakpoint Regions in Recombinant Plasmids
We used a modified array-hybridization to map approximately the positions of the breakpoints in the recombinants obtained. A set of oligonucleotide (oligo) probes was prepared spaced evenly along the recombinogenic element studied (Table I; see also Fig. 4). These probes were labeled and hybridized with DNA of recombinant plasmids lacking KIL gene, and the absence of hybridization of one or several probes was considered as showing that the corresponding region had been deleted from this particular construct. The data obtained suggest that several breakpoint clusters existed both in the AML1 bcr and in the ETO bcr (Fig. 4). It was noteworthy that in some regions deletions occurred with an equal frequency in the absence and presence of VM-26, while in others a stimulation of recombination events by VM-26 was quite obvious. The regions A5, A7, A9, and possibly A8 of the AML bcr and regions E1, E4, E6, and E8 of the ETO bcr were particularly sensitive to VM-26.
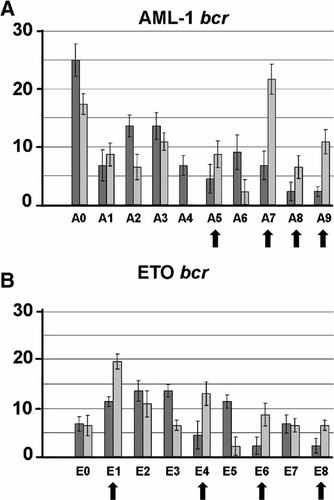
Breakpoints are distributed non-randomly in bcrs. The frequencies (in percent) of recombination in different regions of the bcrs are shown by dark and light columns representing respectively recombination in the absence and in the presence of the topoisomerase II inhibitor VM-26. Bold arrows indicate the regions where recombination was stimulated by VM-26. The vector region of the construct is at the left (A0/E0 probes) and the killer gene is at the right (A9/E8). Error bars show standard deviation (n = 5).
DISCUSSION
Topoisomerase II Is Responsible Only for the First Step of the Illegitimate Recombination Process
A test system based on positive selection of recombinant clones offers a simple and convenient approach to study illegitimate recombination. Our model containing a SV-40 replication origin allows the plasmids to replicate autonomously in eukaryotic cells. In designing this system, we particularly wished to study potentially recombinogenic elements which are involved in rearrangements in vivo, namely the bcrs of the human AML1 and ETO genes that are frequently involved in mutual translocations [Rowley, 1999; Okuda et al., 2001; Hart and Foroni, 2002; Zhang et al., 2002; Mitelman et al., 2003] which are often associated with lymphomas. These bcrs were inserted flanking a positive selection marker (the ccdB KIL gene) in our test construct, and the plasmid was exposed to potentially recombinogenic condition and subsequently transformed into bacteria. The bacteria were grown in medium containing IPTG so that only cells in which the KIL gene had been deleted from the plasmid survive, and deletion could occur only by recombination between the two bcrs. This system therefore allows detection of recombination events and estimation of their frequency, and we note that it can be used to test the recombinogenic effect of any nature on any DNA sequence element; one has only to substitute the bcrs tested in our experiments with other sequences of interest. The system has the limitations that recombination events that do no eliminate the KIL gene, as well as intermolecular recombination, will not be detected. However, it is hardly feasible that mechanisms of intramolecular and intermolecular recombination are principally different. The assay system based on detection of intramolecular recombination has one obvious advantage. In this system recombination evens occur with zero order kinetics and thus the results of quantitative analysis do not depend on concentration of the construct. That is why we have preferred this system over the test system based on recombination between two plasmids that can, for example, bring together two different markers.
At the present time a large number of topoisomerase II inhibitors are used as anticancer drugs [Fortune and Osheroff, 2000; Li and Liu, 2001]. Chemotherapy utilizing these substances often provokes secondary leukemias [Ratain and Rowley, 1992; Felix, 1998; Leone et al., 2001] which may have chromosomal rearrangements as their direct cause. These rearrangements usually result from illegitimate recombination. Recent investigations have shown that topoisomerase II may be involved in induction of illegitimate recombination, either directly via reciprocal exchange of enzyme subunits or indirectly via introduction of DSBs in DNA and subsequent activation of the NHEJ repair system [Adachi et al., 2003, 2004]. We failed to observe any recombination events directly mediated by topoisomerase II in vitro. This result is in contrast to previously published data reporting that topoisomerase II can induce illegitimate recombination in vitro [Bae et al., 1988], a difference which is possibly due to a lower sensitivity of our plasmid-based assay system compared to the bacteriophage system used by Bae et al. The frequency of illegitimate recombination observed in our experiments in living cells increased under conditions of topoisomerase II inhibition, consistent with the possibility that topoisomerase II may trigger illegitimate recombination in vivo.
bcrs Are Not Regions of Increased Recombinogenicity
The reasons for the non-random distribution of recombination frequencies along genes are not yet understood. To understand if recombinogenicity is determined by the sequence of the DNA fragment alone, one should compare the recombination frequency of test constructs carrying different types of targets. To examine if bcrs are really regions with increased recombinogenicity, we compared the construct pKIL2BCR-ori with another having non-recombinogenic sequences flanking the KIL gene. It is not easy to deduce how to find such a non-recombinogenic fragment in the genome, but we reasoned that open reading frames of functional genes are subject to rearrangements to a much lesser extent than non-coding regions because recombinogenicity in a coding region would probably be wiped out in the course of evolution as a source of instability. On the other hand, to have a really appropriate control the target sequences in both constructs should be of comparable length. The two bcrs used in pKIL2BCR-ori total about 6 kbp, but unfortunately such long continuous ORFs are not frequent in the genome. However, the human genome contains plenty of processed pseudogenes having long open reading frames which in most cases do not form functional transcripts and so are protected from selection pressure. Although this allows different mutations (including those increasing recombinogenicity) to accumulate, those in pseudogenes which are evolutionally recent should not differ significantly from those in the prototype gene. Hence, they should still be less recombinogenic than a random genomic fragment. To make a control test construct we therefore used the long processed pseudogene LOC286495 which is about 6.5 kbp long. It is a pseudo for the TTC3 human gene (whose function is still enigmatic [Tsukahara et al., 1996; Ohira et al., 1996]). The ψTTC3 is almost a nascent pseudogene, as it is absent in the closest H. sapiens relatives gorilla and chimpanzee. The sequence identity of ψTTC3 and TTC3 is 82% at the DNA level and 92% at the protein level. We assumed this to be enough to be sure that ψTTC3 has not yet acquire any recombinogenicity and was hence suitable for the control construct. Analysis of the frequencies of recombination observed with different test constructs led us to conclude that bcrs do not show any increased recombinogenicity compared to this presumably neutral pseudogene (Table II). If bcrs possess intrinsic features making them preferential targets for topoisomerase II-mediated cleavage, one would expect significant clustering of deletion breakpoints in the bcrs tested in our experiments, but although some clustering was observed it was not very impressive (Fig. 4). This unexpected result could be explained in two ways. First, the clustering of breakpoints within bcrs observed in previous studies could be influenced by clonal selection [Bystritskiy and Razin, 2004], and bcrs might not be the regions where primary recombination events are clustered. The second possibility is that primary recombination events are concentrated in bcrs but that their recombinogenic properties can be reproduced only in a chromosomal context.
In COS-1 cells the transfected constructs were arranged into nucleosomes, but higher levels of DNA packaging as well as proper spatial organization in the nuclei cannot be reproduced in our model system. Thus, we cannot exclude the possibility that in a chromosomal context bcrs are formed only at higher levels of chromatin packaging, including interaction of the ends of DNA loops with the nuclear matrix.
Acknowledgements
The authors are very grateful to Ronald Hancock for critical reading of the manuscript and valuable comments and suggestions. This work was supported by a grant of the Program on Molecular and Cellular Biology (MCB) of the Presidium of Russian Academy of Sciences, RFBR grant 05-04-22002-CNRS-a and Russian-French collaborative grant PICS 3207.