Stable formation of mutated p53 multimers in a Chinese hamster cell line causes defective p53 nuclear localization and abrogates its residual function
Abstract
We have previously described a methotrexate-resistant cell line (MTX M) characterized by amplified dihydrofolate reductase (DHFR) genes, cytoplasmic p53 localization, and p53 stable tetramers. To investigate the p53 functionality in MTX M, the effect of chemical/physical agents was studied. In MTX M cells, DNA damage did not induce p53 or mdm-2 protein, while in the parental V79 cells, a residual p53 activity was found. cDNA sequencing showed that V79 and MTX M cells share the same mutations, indicating that the complete loss of p53 function in MTX M cells was due to cytoplasmic sequestration of a mutated p53 with residual activity. In Chinese hamster, both p53 and DHFR genes map on short arm of chromosome 2 suggesting that p53 itself might be amplified. However, fluorescence in situ hybridization with a hamster p53 probe showed only a single signal. Thus, the presence of p53 stable tetramers in MTX M cells, although correlated with DNA amplification, could not be the consequence of either p53 or DHFR gene amplification. Expression of a C-terminal human p53 peptide does not induce p53 nuclear accumulation, indicating that the cytoplasmic localization is due to a mechanism different from that already described in cancer cell lines. Treatments with Sodium Butyrate induced β-tubulin polymerization, but did not apparently organize a normal microtubule network, which is shown to be important for the p53 localization. Our data indicated that in MTX M cells, p53 is sequestered in the cytoplasm by a novel mechanism that abrogates p53 residual function. J. Cell. Biochem. © 2006 Wiley-Liss, Inc.
Inactivation of p53 function by mutation of the p53 gene, often associated with nuclear accumulation of the mutated protein, is the most common alteration found in human cancer [Hollstein et al., 1991. However, different mechanisms of p53 inactivation have been described [Ko and Prives, 1996; Levine, 1997; Funk and Galloway, 1998. In a variety of tumors, lack of p53 function is linked to defective nuclear localization [Moll et al., 1992, 1995; Bosari et al., 1994. Defective p53 nuclear localization can be due to cytoplasmic sequestering, defective transport through the nuclear membrane, or active nuclear exclusion. The defect can reside in the p53 protein itself, in the architecture of the nucleus, or in other nuclear or cytoplasmic components. A number of proteins have been proposed to serve as cytoplasmic anchor proteins to impair nuclear localization [Gannon and Lane, 1991; Knippshild et al., 1996, including vimentin [Klotzsche et al., 1998 and β-tubulin [Giannakakou et al., 2000. Recently, the identification of a new factor, p53-associated parkin-like cytoplasmic protein (Parc), that binds to p53 in the cytoplasm and prevents its transport into the nucleus has been reported [Nikolaev et al., 2003. It has also been reported that p53 conformational changes may play a role in protein subcellular localization [Zerrahn et al., 1992. Moreover, the presence of a single mutation in a cis-acting sequence has been demonstrated to cooperate with C-terminal domain in the sequestration of p53 [Liang et al., 1998. More detailed explanations have also been proposed in the case of p53 cytoplasmic localization in neuroblastoma: in tumor-derived cell lines, cytoplasmic sequestration of wt p53, which impairs the G1 checkpoint after DNA damage [Moll et al., 1995, was found to be due to masking of the C-terminal domain, preventing the exposure of nuclear localization signals (NLSs) [Ostermeyer et al., 1996. It was also demonstrated that in such cell line, a hyperactive nuclear export contributed to cytoplasmic accumulation [Stommel et al., 1999. In these cells, Parc is highly expressed and its reduction mediated by RNAi relocalizes p53 in the nucleus [Nikolaev et al., 2003.
We reported a novel example of cytoplasmic p53 localization, which occurs in a methotrexate (MTX)-resistant Chinese hamster cell line (MTX M), carrying amplified dihydrofolate reductase (DHFR) genes. MTX M cells, which derived from a cell line (V79) with p53 nuclear accumulation, showed cytoplasmic p53 localization and, in immunoblot, several extra bands in the high molecular weight region, besides the expected 53-kDa band. In these cells, p53 cytoplasmic localization and the appearance of high molecular weight bands (HMWBs) appeared to be correlated. Furthermore, by comparing different MTX cell lines with increasing resistance to MTX, the appearance of HMWBs seemed to be correlated also with the degree of DNA amplification, although amplification of DHFR itself was not involved [Ottaggio et al., 2000. Cell fusion experiments demonstrated that p53 cytoplasmic localization in MTX M cells was a dominant phenotype and suggested that the defect causing lack of nuclear localization in this cell line did not reside in the nucleus. Experiments aimed at changing the p53 phosphorylation status or at alkylating p53, quantitatively influenced the formation of HMWBs, suggesting that in the cytoplasm of MTX M cells, post translational modifications of p53 occur that result in the formation of stable p53 multimers that are unable to re-enter the nucleus [Ottaggio et al., 2000.
In the present study, the functionality of p53 in response to DNA damage, the mutational status, and the gene copy number of p53, together with the integrity of microtubule network in MTX M, were investigated. Our data confirmed that in MTX M cells, p53 is sequestered in the cytoplasm by a novel mechanism that abrogates p53 residual function.
MATERIALS AND METHODS
Cell lines and Culture Conditions
The Chinese hamster embryo fibroblast (CHEF/18) cell line, kindly provided by Prof R. Sager (Dana-Farber Cancer Institute, Boston, MA) was grown in α-MEM supplemented with 10% FCS; the Chinese hamster ovary (CHO) cell line, strain AA8, obtained from Dr. M.Z. Zdziedmicka (University of Leiden, The Netherlands) was grown in F10 supplemented with 10% FCS; the Chinese hamster lung (V79) cell line, Thymidine Kinase-deficient derivative B7, obtained from Dr. C. Colella (IMD, CNR, Pisa, Italy) was grown in DMEM supplemented with 5% FCS; the MTX-resistant Chinese hamster cell line (MTX M), with DHFR gene amplification, was selected from B7 in our laboratory using a multistep protocol [Miele et al., 1989 and was grown in DMEM supplemented with 7.5% dialyzed FCS.
Exposure to DNA Damaging Agents
The medium was removed from sub confluent cells grown on Petri dishes, then about 6–7 × 106 cells were UV irradiated using a Multiband UV lamp, model UVGL-15UV (maximum emission at λ = 254 nm) at 20–40 J/m2. Radiation flux was measured using a UV meter, model UVX-Digital radiometer (UVP, Inc.). After UV exposure, the medium was replaced, and 3–5 h after treatment, cells were harvested for extracts preparation. Sub confluent cultures (about 6–7 × 106 cells) were exposed to 5–10–20 µg/ml Mitomycin C (MMC) (Kyowa Italiana) in complete medium for 6–18 h, then rinsed with PBS and harvested for extracts preparation.
SDS–PAGE and Western Blotting
To normalize the gel loading, two Petri dishes for each experimental point were set up. Cells in one dish were trypsinized and counted, while cells in the second one were scraped in PBS and, after centrifugation, the pellet was resuspended in lysis buffer (20 mM Tris-HCl pH 8, 137 mM NaCl, 10% glycerol, 1% NP-40, 10 mM EDTA, 2 mM PMSF) at a density of 107 cells/100 µl of lysis buffer. After incubation on ice for 30 min, the lysate was centrifuged again, the supernatant was collected and the equivalent of 2 × 106 cells/lane was fractionated on a 7.5% SDS–PAGE and transferred to nitro-cellulose using a Protean or miniProtean Electophoresis apparatus (Bio Rad). For detection, the filters were blocked with 1% low fat milk in PBS 0.1%, Tween 20 (PBS-T) and then probed 1 h with anti-human p53 (DO-7) or anti-hmdm2 (clone smp14) antibody. After washing in PBS-T, filters were incubated 1 h with a 1:2,000 dilution of a goat anti-mouse peroxidase-conjugated immunoglobulin (Sigma). Filters were washed three times again with PBS-T and once with PBS and then detected using ECL system (Amersham). Blots probed with anti β-actin antibodies (ICN Biochemicals) confirmed the reproducibility of the normalization procedures.
P53 Sequencing
One µg of total RNA was reverse transcribed using oligo(dT)16 and SuperscriptII (Gibco). The whole p53 coding sequence was PCR amplified using the following pair of primers: HAp53-5.1: 5′-AGC CAC AGT CAG ACC TCA GCA-3′ and HAp53-3.1: 5′-TCA GTC CGA GTC AGG CCC CT-3′. PCR products were purified from agarose gel and sequenced with Thermo Sequenase dye terminator cycle sequencing pre-mix kit, version 2.0 (Amersham) on an Applied Biosystems 377 Automated Sequencer (Perkin Elmer). The primers used for sequencing were HAp53-5.1, HAp53-3.1, and the following internal primers: HAp53-5.2: 5′-CGT ACT CCC CTT CCC TAA AT-3′; HAp53-3.2: 5′-AGC AGG TTA CCA CTG GGG T-3′; HAp53-5.3: 5′-AGA CCC CAG TGG TAA CCT GC-3′; HAp53-3.3: 5′-AAG GGG AGT TGC ACG TG-3′.
In Situ Hybridization and Fluorescence In Situ Hybridization (FISH)
pGEM-hap53 probe was prepared by PCR amplification of V79 cells cDNA using HAp53-5.1 and HAp53-3.1 primers. The PCR product was directly cloned in pGEM-T vector. pDHFR11 was a generous gift from Dr. RT Schimke (Stanford University, Stanford, CA). For in situ hybridization, pDHFR11 was nick translated with 3HdUTP. For FISH, both probes were biotin labeled by nick translation. Hybridization and cy3-conjugated avidine (Amersham) detection were performed according standard procedures. Metaphases were counterstained with Giemsa stain or diaminophenylindole (DAPI).
Transfections
V79 and its derivative MTX M were transfected by electroporation with the NeoRpcDNA3HAp53Ct obtained from G. Del Sal (LNCIB, Trieste, I) [Gostissa et al., 1999, or with the NeoRpC53-SN3 obtained from B. Vogelstein, (John Hopkins Oncology Center, Baltimore, MD) [Baker et al., 1990 encoding human C-terminus HA-tagged p53 (AA294–393) or murine full length p53, respectively. Cells (4 × 106) and 20 µg of plasmid DNA were used for each transfection. Electroporation was performed in a ElectroporatorII System (Invitrogen) with 0.4 cm cuvettes with the following parameters: 330 V, 25 mA, 25 W capacitance was set at 1,000 mF and resistance to infinite. Transfected cells were plated on two coverslips and p53 immunodetection was performed 48 h later. Alternatively, cells were plated on Petri dishes and selection with 400 µg/ml G418 (Gibco) was started 24 h later. Stably transfected clones were pooled and used for immunofluorescence experiments as described below.
Immunocytochemistry
Cells (2 × 105) were plated on two coverslips and grown for 24 h. Slides were washed three times with PBS and fixed in 1:1 methanol:acetone or with 3.7% paraformaldehyde followed by methanol permeabilization. They were subsequently incubated with the appropriate primary antibody. p53 was detected using mouse monoclonal antibodies DO-7, pAb241, pAb248, or pAb421 followed alternatively by: (1) an immunoperoxidase technique with a biotinylated anti-mouse antibody followed by ABC complex (SPA-Bio Division) and diaminobenzidine (Sigma) as chromogen; the slides were counterstained with hematoxylin and eosin according to the Papanicolau technique; (2) an immunofluorescence technique with an anti-mouse FITC conjugated antibody (Sigma); the slides were counterstained with DAPI. The ectopic p53 C-terminal peptide fused to HA epitope was detected with PAb421 followed by anti-mouse FITC-conjugated antibody (Sigma) or with biotinylated anti-HA antibody (Boehringer Mannheim), followed by cy3-conjugated avidine (Amersham). β-tubulin was detected using a mouse monoclonal antibody anti β-tubulin (ICN Biochemicals) followed by anti-mouse FITC-conjugated antibody (Sigma).
Treatment With Sodium Butyrate
Cultures at 50% confluence were incubated in Sodium Butyrate (NaB)-supplemented medium (5–10 mM) for 24 h. Cells were then harvested for extracts preparation, immunocytochemistry, and cytological preparations, or medium was replaced with chemical-free medium for 48 h and harvested as above.
RESULTS
P53 Is Induced by Chemical/Physical Agents in the Parental V79 cells but not in the Derivative MTX M Cells
To ascertain whether DNA damage triggers p53 induction in cells with cytoplasmic p53, we studied the effects of MMC and UV treatments on the parental V79 cells, with nuclear p53, and on the derivative MTX M cells, with cytoplasmic p53. CHEF/18, a non-transformed Chinese hamster fibroblast cell line at early passage, was used for normal control cells [Moro et al., 1995; Lee et al., 1997. CHO, a transformed Chinese hamster cell line, was used for p53-mutated control cells [Lee et al., 1997. Cells were treated with 5–20 µg/ml MMC for 6 or 18 h. In CHEF/18 cells, p53 increases 6 h after a 10 µg/ml treatment with MMC, and this increase was time-dependent (Fig. 1a) and dose-dependent (not shown), while the transformed CHO cells did not show any p53 increase, as expected (Fig. 1a). In V79 cells, a marginal reproducible increase in p53 was detected in three independent experiments, exclusively 18 h after 20 µg/ml MMC treatment (the highest dose used) (Fig. 1a), while no significant p53 induction was discernible in MTX M cells, even at the highest dose (Fig. 1a). Similar results were obtained after UV treatments: in CHEF/18 cells, the increase in p53 level was observed already 3 h after a 40 J/m2 UV treatment, and it was time-dependent (Fig. 1b) and dose-dependent (not shown). In V79 cells, an increase of p53 was observed only 5 h after treatment at the highest dose (Fig. 1b), while no change in protein level was seen in MTX M or CHO cells (Fig. 1b). Since the mdm-2 gene is regulated by p53, we determined the level of mdm-2 protein in the same samples. In CHEF/18 cells, the level of mdm-2 increased 3 h after UV treatment, while in V79 cells, a slight increase was observed only 5 h after treatment (Fig. 1b). No change in protein level was seen in MTX M or CHO cells (Fig. 1b). These data indicate that in MTX M cells, p53 is not inducible by genotoxic stresses and cannot regulate mdm-2.
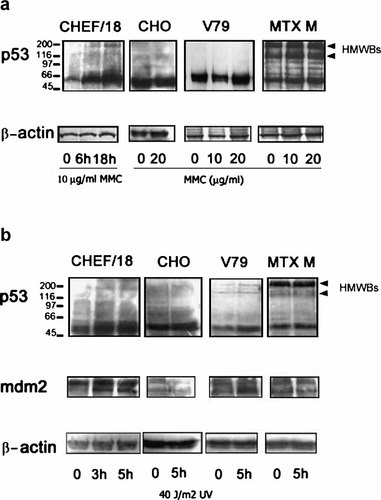
p53 expression after genotoxic stresses. a: p53 wild type control cells (CHEF/18) treated with 10 µg/ml MMC for 6 and 18 h; p53 mutated control cells (CHO), parental V79, and derivative MTX M cells treated at the indicated MMC doses for 18 h. Proteins were separated using a miniProtean apparatus and Western blots were probed with DO-7 to detect p53. The same extracts were probed with anti β-actin antibody for loading normalization. b: p53 wild-type control cells (CHEF/18) 3 and 5 h after 40 J/m2 UV treatment; p53 mutated control cells (CHO), parental V79 cells, derivative MTX M cells 5 h after 40 J/m2 UV treatment. Proteins were separated using a miniProtean apparatus and Western blots were probed with DO-7 to detect p53. The same extracts were probed with spm14 antibody to detect mdm2 protein or with anti β-actin antibody for loading normalization.
No Further Mutations Are Responsible for p53 Inactivation or for Cytoplasmic Accumulation in MTX M Cells
The whole p53 coding sequence, including the NLSs and the tetramerization domain, was sequenced in all cell lines. The non-transformed CHEF/18 cell line showed only a silent polymorphism at codon 210, already described [Lee et al., 1997. The parental V79 Chinese hamster cell line showed two base pair substitutions, giving rise to aminoacid changes at codons 133 and 135 (leucine to glutamine and cysteine to tryptophan, respectively) already described in V79 [Chaung et al., 1997, together with the silent polymorphism at codon 210. In the derivative MTX M cell line, we found the same mutations observed in the parental cell line V79 (Fig. 2). This indicates that the p53 cytoplasmic localization in MTX M was not due to further p53 mutations in the C-terminal domain or in a cis-acting sequence. The CHO cell line showed a single base pair substitution at codon 211, originating a threonine to lysine substitution.
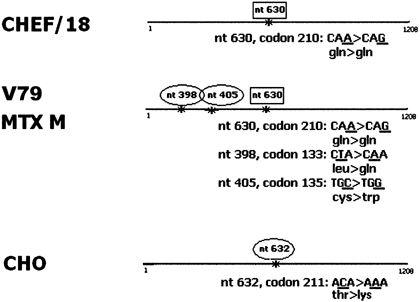
p53 sequencing of the CHEF/18, V79, MTX M, CHO cell lines.
P53 Gene does not Co-Amplify With DHFR Gene in MTX M Cells
A major phenotypic trait of MTX M cells is gene amplification and the question arises whether this trait could be causally related to the cytoplasmic localization of p53 in these cells. In Chinese hamster, both p53 and DHFR genes map on chromosome 2p. It could then be imagined that in MTX M cells, p53 co-amplifies with DHFR and p53 overexpression causes in some non-obvious way its cytoplasmic localization. This, however, does not seem to be the case, since MTX M cells do not contain more p53 protein than cells without DHFR amplification. As a confirmation, in situ hybridization in MTX M cells with pGEM-hap53 probe showed a single signal on a small submetacentric chromosome (Fig. 3c), while the pDHFR11 probe showed, as expected, a large amplified signal on the marker chromosome (Fig. 3b). In situ hybridization with pDHFR11 probe performed on cells early after MTX-stepwise selection, showed multiple amplification signals interspersed on long arm of a marker chromosome (Fig. 3a). These results indicated that in MTX M cells, the DHFR amplicon does not contain the p53 gene.
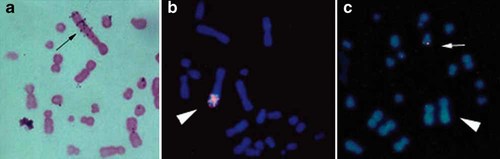
In situ hybridization with pDHFR11 and pGEM-hap53 probes. a: In situ hybridization with pDHFR11 probe on MTX M cells early after MTX-stepwise selection. Black arrow: marker chromosome. b: FISH with pDHFR11 probe on MTX M cells after prolonged subculturing with selective medium. White headarrow: marker chromosome. c: FISH with pGEM-hap53 probe on MTX M cells after prolonged subculturing with selective medium. White arrow: p53 gene signal on a small submetacentric chromosome. White headarrow: marker chromosome. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Reverse Transformation, β-Tubulin Polymerization, p53 Expression, and Localization Induced by Sodium Butyrate
To assess the relationship between the integrity of microtubule network and p53 localization, we treated V79, MTX M, and CHO cells with NaB, a compound known to induce reverse transformation (RT) and to directly affect the expression of p53 protein in colonic epithelial cells [Palmer et al., 1997. In CHO cells, a 20-h treatment with 5 mM NaB induced RT, in terms of change in cell shape (Fig. 4a,b), a decrease in the level of p53 (Fig. 4c) and, interestingly, a change of its cellular localization (Fig. 4b and Table I). β-tubulin, that is depolymerized in untreated, transformed cells, became polymerized (not shown). These effects were transient, as cell morphology and p53 localization partially reverted to transformed-like phenotype after 48 h from NaB removal. In V79, cells NaB treatment did not affect any of the parameters studied. In MTX M cells, no change in cell shape nor in the p53 localization was observed, while immunocytochemistry showed an increase in the p53 level (not shown) and immunoblot showed an increase in the p53 HMWBs, typical of this cell line [Ottaggio et al., 2000, while the 53 kDa bands slightly decreases (Fig. 5c). Moreover, β-tubulin became polymerized but does not apparently organize a normal microtubule network (Fig. 5a,b).
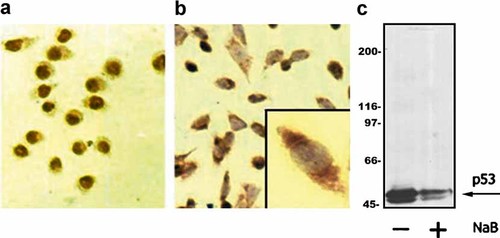
NaB treatment of CHO cells. a: p53 immunoperoxydase detection of untreated CHO cells with transformed phenotype (round cell shape and p53 nuclear accumulation) (40×). b: p53 detection of NaB-treated CHO cells with RT phenotype (fibroblast-like shape and p53 cytoplasmic accumulation) (40×). Box: The same cells at higher magnification (100×). c: Proteins were separated using a Protean apparatus and Western blots were probed with DO-7 to detect p53. Cells were treated with 5 mM NaB for 20 h. Gel loading was normalized by cell counting as described in Materials and Methods. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
| Cell line | Dose (mM)a | Cell shape | p53b |
|---|---|---|---|
| CHO | 0 | T | +++ (N) |
| 5 | RT | ++ (Cyt) | |
| 10 | RT | ++ (Cyt) | |
| 5 + 48 h recovery | RT/T | ++(N/Cyt) | |
| V79 | 0 | T | +++ (N) |
| 5 | T | +++ (N) | |
| 10 | T | +++ (N) | |
| MTX M | 0 | T | ++ (Cyt) |
| 5 | T | +++ (Cyt) | |
| 10 | T | +++ (Cyt) |
- T, transformed; RT, reverse transformed; N, nucleus; Cyt, cytoplasm.
- a Cells were treated for 24 h in complete medium.
- b Determined by immunocytochemistry.
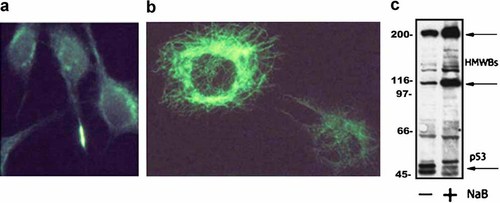
NaB treatment of MTX M cells. a: β-tubulin detection of untreated MTX M cells: β-tubulin is in depolymerized form and only the midbody is brightly stained. b: β-tubulin detection of NaB-treated MTX M cells: β-tubulin is polymerized even if a fully organized cytoskeleton is not visible. c: Proteins were separated using a Protean apparatus and Western blots were probed with DO-7 to detect p53. Cells were treated with 5 mM NaB for 20 h. Gel loading was normalized by cell counting as described in Materials and Methods. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Ectopic Expression of the p53 Tetramerization Domain does not Affect p53 Localization in MTX M Cells
To verify whether ectopic expression of the human p53 tetramerization domain could affect p53 subcellular localization, both the MTX M cells and the parental V79 were transiently or stably transfected with a neomycin-resistant expression plasmid encoding human p53 C-terminal peptide HA-conjugated (NeoRpcDNA3HAp53Ct). Forty-eight hours after transient transfection cells grown in duplicate slides were incubated with PAb421, which recognizes the human C-terminal epitope but not hamster p53 [Rotter et al., 1983, or DO-7, which recognizes hamster p53 [Vojtesek et al., 1992 but not the human C-terminal peptide. The results, are shown in Figure 6 and Table II, and can be summarized as follows. The human peptide was detected in a minority of cells (7%, which represents the transfection efficiency). Its localization was exclusively nuclear in V79 cells and both nuclear and cytoplasmic in MTX M cells. The results of the stable transfection experiments were more clear-cut. In this case, the endogenous p53 was detected with DO-7 and the ectopic p53 C-terminal peptide with anti-HA antibody in the same slide. In V79 cells, all nuclei showed localization of both hamster and human p53. These proteins were detected in the cytoplasm of most or all MTX M cells. These results indicate that in MTX M cells, the human peptide was unable to relocate the hamster p53 from the cytoplasm to the nucleus and was itself localized mostly in the cytoplasm.
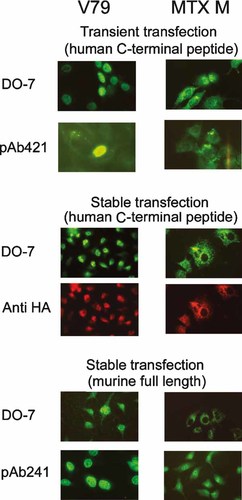
Subcellular localization of p53: representative pictures of transient and stable transfectant of parental V79 and derivative MTX M cells with human p53 C-terminal peptide of murine full length p53. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
| V79 | MTX M | |||
|---|---|---|---|---|
| % positive nuclei | % positive cytoplasm | % positive nuclei | % positive cytoplasm | |
| Transient transfection (human C-terminal peptide) | ||||
| DO-7 | 100 | 0 | 3 | 100 |
| pAb421 | 7 | 0 | 2.3 | 4.7 |
| Stable transfection (human C-terminal peptide) | ||||
| DO-7a | 100 | 0 | 0 | 100 |
| Anti-HAa | 100 | 0 | 10 | 100 |
| Stable transfection (murine full length) | ||||
| DO-7 | 100 | 0 | 0 | 100 |
| pAb241, pAb248 | 100 | 0 | 100 | 0 |
- Cells (1,000–1,500) were counted for each experimental point.
- a DO-7 and anti-HA detection were performed sequentially on the same slide.
A different result was found when cells were stably transfected with a murine p53 and probed with PAb241 and PAb248, which, at the used concentrations, recognize murine but not Chinese hamster p53 [Ottaggio et al., 2000. Unlike the human peptide, the murine p53 localized in the nucleus in both V79 and MTX M cells. However, like the human peptide, it was unable to relocate the endogenous p53 in the nucleus.
DISCUSSION
We have previously described a MTX-resistant cell line (MTX M) displaying an abnormal cytoplasmic p53 localization and the presence of HMWBs containing p53 epitope in immunoblot. The formation of stable p53 multimers is favored by hypophosphorylation, and, in some way, prevents p53 to be transported into the nucleus [Ottaggio et al., 2000. To better investigate the p53 functionality in MTX M cells, the effect of chemical and physical agents was investigated and the results compared with those obtained on different hamster cell lines. It was demonstrated that despite p53 mutations, V79 cells exhibit G1 arrest, apoptosis or p53 increases after treatment with chemical or physical agents [Chatterjee and Berger, 2000; Karmakar et al., 2001; Mi et al., 2001. In our experimental conditions, the V79 cells showed a slight p53 increase after treatment with both chemical and physical agents. This increase was observed late after treatment and at higher doses with respect to normal CHEF/18 cells. In V79, cells p53 also retained the capability to induce mdm-2. On the contrary, in the V79-derivative MTX M cells, p53, and mdm-2 were no more inducible by DNA damage.
To investigate whether the lack of function in MTX M cell line was due to p53 mutations, the p53 cDNA of all cell lines was sequenced. We chose to sequence the whole cDNA to detect possible mutations in the NLSs and in the tetramerization domain or in a cis-acting sequence, which could be responsible for impaired p53 nucleus import and/or tetramerization. The parental V79 and the derivative MTX M cell lines shared the same two mutations already described in V79 cell line [Chaung et al., 1997. These mutations did not cause the complete lack of function of p53 protein, as V79 retained residual p53 activity. No further mutations were found in MTX M cells, which could justify their complete p53 lack of function or their lack of p53 nuclear localization, suggesting that the complete loss of p53 function in these cells was due to cytoplasmic sequestration of a mutated p53 with residual activity. CHO cells showed a different single mutation [our results, Lee et al., 1997, Hu et al., 1999 that alone caused the complete lack of function of p53. CHEF/18 cells were wild type for p53, showing only a silent polymorphism.
In Chinese hamster, p53 and DHFR genes both map on short arm of chromosome 2, although rather apart: the DHFR gene maps closer to the centromere in 2p23 [Funanage and Myoda, 1986, while the p53 gene is closer to the telomere in 2p31 [Zatteroni et al., 1997. We have previously described that in MTX M cell line, a pericentric inversion containing the p32 region had occurred during DHFR amplification process [Nuesse et al., 1992. A similar pericentric inversion was described in another Chinese hamster cell line, and 2p32 band was identified as a hot spot for γ ray-induced breaks in CHO cells [Nuesse et al., 1992. Such a region could then be involved in rearrangements. Moreover, in MTX M cell line early after MTX-stepwise selection, the amplicon size was very large, giving rise to a discontinuous signal pattern after hybridization with DHFR probe. It was then possible that p53 itself might be included in the amplicon, and therefore, amplified. However, FISH with pGEM-hap53 probe performed on MTX M cells late after selection, showed only one signal on a small chromosome different from the marker chromosome carrying the amplified DHFR genes. Indeed, in these cells, DHFR amplification signals localized in a compact zone near the telomere of marker chromosome long arm, indicating that at this time of selection, the amplicon size was reduced. Probably, the small chromosome carrying the p53 gene originated from chromosome 2 during the amplification process. These results clearly indicated that the presence of p53 stable tetramers in MTX M cells could not be the consequence of p53 gene amplification.
It has been demonstrated that p53 is associated with cellular microtubules and after DNA damage accumulates in the nucleus of cells with a functional microtubule network [Giannakakou et al., 2000. Disruption of the microtubule network by treating with paclitaxel or vinblastine prevents p53 translocation to the nucleus. All the transformed cell lines used in this study showed depolymerized β-tubulin (data not shown). Despite this, all cell lines but MTX M accumulated p53 in the nucleus. We have previously demonstrated that in CHO cells, compounds able to induce RT (in terms of reduced ability to form colonies in soft agar and changes in cell shape), strongly reduced the expression of p53 [Moro et al., 1995. To assess the relationship between the integrity of microtubule network and the localization of p53 in our cell lines, we treated cells with NaB, a potent biological modifier, that among other effects, can induce reverse transformation [Kruh, 1982 and reduce the expression of p53 [Larno et al., 1989. In NaB-treated CHO cells both the disappearance of hallmarks of transformed phenotype and p53 reduction were observed and, interestingly, p53 localization changed from nucleus to cytoplasm. However, no p53 epitope containing-HMWBs were observed in immunoblot, suggesting that in CHO cells, the p53 cytoplasmic localization induced by NaB is due to a different mechanism that does not imply the formation of stable multimers. The modulation of p53 expression by NaB was already documented [Mi et al., 2001, Madigan et al., 1999, as well as its effect on tubulin [Larno et al., 1989. On the contrary, the change in p53 localization, to our knowledge, has never been described before. In V79 cells, NaB treatment did not affect any of the studied parameters, whereas in MTX M cells, only the p53 epitope containing-HMWBs increased after treatment.
Recently, a cytoplasmic anchor protein (Parc) associated with p53 was identified [Nikolaev et al., 2003. This protein binds to p53 in the cytoplasm and prevent its transport into the nucleus. These authors found that Parc is highly expressed in neuroblastoma cells and that inactivation of endogenous Parc expression in these cells by RNAi leads to nuclear p53 localization and restores a p53-dependent stress response. This finding is consistent with the previous observation that C-terminal p53 peptide, which in theory could compete for binding to anchor protein, efficiently induces nuclear accumulation of endogenous p53 in neuroblastoma cells [Ostermeyer et al., 1996. In our case, transfection experiments with C-terminal p53 peptide, which contains p53 NLSs, showed that this peptide is efficiently localized to the nucleus in parental V79 cells, as previously demonstrated in other cells [Gostissa et al., 1999, but is localized only in 10% of MTX M nuclei. These data indicated that the defect of nuclear localization in these cells involved not only the endogenous p53 but also the exogenous C-ter domain. Moreover, transfection with this domain, which is known to relieve p53 cytoplasmic sequestration in other cell lines [Ostermeyer et al., 1996, does not induce endogenous p53 nuclear accumulation in MTX M cells, indicating that in these cells, the cytoplasmic localization is due to a different mechanism. On the other hand, when MTX M cells were transfected with murine full length p53, murine p53 but not hamster p53 accumulated in the nucleus, indicating that: (i) cellular nucleus/cytoplasm transport machinery is intact in these cells; (ii) if cytoplasmic accumulation in these cells is mediated by stable p53 tetramer formation, murine and Chinese hamster p53 do not tetramerize. These finding are in keeping with our previous observations based on microinjection experiments in which MTX M cells microinjected with full length murine p53 showed PAb248 positive nuclei but only DO-7 positive cytoplasms [Ottaggio et al., 2000. All these data are in agreement with the notion that in MTX M cells, p53 heterooligomerizes with human but not mouse p53. That the stable formation of oligomers is the cause of cytoplasmic sequestration of endogenous and ectopic C-ter domain remains to be established.
Although p53 can enter the nucleus in an oligomerized form, it was suggested that the p53 monomer can enter the nucleus more efficiently than the oligomer [Liang and Clarke, 1999. Moreover, ectopic expression of the tetramerization domain was able to restore the nuclear retention of p53 in cells with enhanced nuclear export [Stommel et al., 1999, suggesting that tetramerization can inhibit p53 nuclear export by masking the nuclear exporting sequences. Although all these observations were done using ectopic expression of C-terminal peptide, it was hypothesized that p53 oligomerization itself is regulated, switching p53 from a tetramer to a monomer at the time that p53 needs to be transported in or out of the nucleus. Moreover, Contegno et al. 2002 demonstrated that proteins that are able to self-associate naturally, such as p53, could be inactivated by inducing the formation of stable oligomers. These authors demonstrated that fusion of the self-associating coiled-coil (CC) domain of nuclear factor of promyelocytic leukemia to p53 caused the formation of stable high molecular weight complexes. CC-p53 chimeras caused the formation of stable heterooligomers, delocalized wild-type p53 to the cytoplasm and caused loss of p53 function. However, in this article, the oligomerization chain reaction is presented as a general strategy for targeting p53 inactivation with therapeutical potential, and it is not known whether the formation of p53 stable oligomers represents a naturally occurring mechanism of p53 inactivation.
CONCLUSIONS
We have described a cell line, MTX M, where p53 is inactive and is retained in the cytoplasm. Some obvious explanations for this phenotype, namely specific p53 mutations and defective cytoplasm/nucleus transport machinery, were excluded. Considering that MTX M cells derive from V79 cells and that the more conspicuous difference between the two cell lines resides in the amplification of a chromosomal region in MTX M cells, it could be speculated that a clue for stable oligomers formation and cytoplasmic localization of p53 should be searched for in gene amplification. We have excluded p53 amplification in MTX M cells, but other genes are likely to be included in the large DHFR amplicon. It can be further speculated that overexpression of some unknown product could be involved in the p53 oligomer stabilization and this might in turn cause cytoplasmic sequestration. The effect of the siRNA approach in our experimental system will be investigated.
Since gene amplification is a common feature of cancer cells, especially in cells which display drug resistance after chemotherapy, it would be of interest to investigate in human cancer cell lines whether such a mechanism might be involved in p53 inactivation.
Acknowledgements
We thank D.P. Lane for supplying antibodies (DO-7, pAb241, pAb248, pAb421, and smp 14) and helpful suggestions.