Identification of regions of leukotriene C4 synthase which direct the enzyme to its nuclear envelope localization
Abstract
Leukotrienes (LTs) are fatty acid derivatives formed by oxygenation of arachidonic acid via the 5-lipoxygenase (5-LO) pathway. Upon activation of inflammatory cells 5-LO is translocated to the nuclear envelope (NE) where it converts arachidonic acid to the unstable epoxide LTA4. LTA4 is further converted to LTC4 by conjugation with glutathione, a reaction catalyzed by the integral membrane protein LTC4 synthase (LTC4S), which is localized on the NE and endoplasmic reticulum (ER). We now report the mapping of regions of LTC4S that are important for its subcellular localization. Multiple constructs encoding fusion proteins of green fluorescent protein (GFP) as the N-terminal part and various truncated variants of human LTC4S as C-terminal part were prepared and transfected into HEK 293/T or COS-7 cells. Constructs encoding hydrophobic region 1 of LTC4S (amino acids 6–27) did not give distinct membrane localized fluorescence. In contrast hydrophobic region 2 (amino acids 60–89) gave a localization pattern similar to that of full length LTC4S. Hydrophobic region 3 (amino acids 114–135) directed GFP to a localization indistinguishable from that of full length LTC4S. A minimal directing sequence, amino acids 117–132, was identified by further truncation. The involvement of the hydrophobic regions in the homo-oligomerization of LTC4S was investigated using bioluminescence resonance energy transfer (BRET) analysis in living cells. BRET data showed that hydrophobic regions 1 and 3 each allowed oligomerization to occur. These regions most likely form transmembrane helices, suggesting that homo-oligomerization of LTC4S is due to helix–helix interactions in the membrane. J. Cell. Biochem. 98: 1517–1527, 2006. © 2006 Wiley-Liss, Inc.
Leukotrienes (LTs) are arachidonic acid derived molecules formed via the 5-lipoxygenase (5-LO) pathway in inflammatory and other cells [Samuelsson et al., 1980; Hammarström, 1983; Samuelsson, 1983]. Upon cell activation 5-LO is translocated to the nuclear envelope (NE) and peripheral endoplasmic reticulum (ER) from cytoplasmic and/or nuclear pools [Woods et al., 1993]. Arachidonic acid is hydrolytically released from membrane phospholipids by cytosolic phospholipase A2 and then oxygenated and dehydrated by 5-LO to form the unstable epoxide LTA4. Two 17 kDa proteins important for LT biosynthesis are found on the NE and peripheral ER membranes, namely five-lipoxygenase activating protein (FLAP) [Dixon et al., 1990], which may have a role in presenting arachidonic acid to 5-LO for the conversion to 5-HPETE [Mancini et al., 1993] and leukotriene C4 synthase (LTC4S) [Söderström et al., 1988; Nicholson et al., 1993; Lam et al., 1994; Welch et al., 1994; Surapureddi et al., 1996], which specifically conjugates LTA4 to form LTC4. Alternatively LTA4 is hydrolyzed to the chemotactic agent LTB4, by LTA4 hydrolase. LTC4 is transported out of cells by multidrug-resistance associated protein 1 [Loe et al., 1996] and then metabolized to LTD4 by removal of the γ-glutamic acid residue and further to LTE4 by elimination of a glycine residue. LTC4, D4, and E4, also known as slow reacting substance of anaphylaxis (SRS-A) have important functions in asthma and inflammation [Samuelsson et al., 1980; Hammarström, 1983; Samuelsson, 1983]. These LTs exert their action via two G-protein coupled receptors, cysLT1 and cysLT2 [Lynch et al., 1999; Heise et al., 2000].
LTC4S belongs to a protein family called “membrane associated proteins in eicosanoid and glutathione metabolism” (MAPEG) [Jakobsson et al., 1999]. MAPEG proteins are small integral membrane proteins of 150–160 amino acids with similar hydropathy plots showing three hydrophobic regions [Bresell et al., 2005]. The middle region seems to consist of two short transmembrane helices as judged by electron crystallography projection maps for microsomal glutathione S-transferase 1 (MGST1) [Holm et al., 2002] and LTC4S [Schmidt-Krey et al., 2004], which both revealed four transmembrane helices. An investigation of the membrane topology of LTC4S predicted that the two hydrophilic loops point to the luminal side of the ER or nuclear membrane [Christmas et al., 2002]. These regions contain residues involved in substrate binding and catalysis [Lam et al., 1997]. The arachidonic acid binding residues residing in the first hydrophilic loop of FLAP are directed towards the luminal side [Mancini et al., 1993; Vickers et al., 1993; Woods et al., 1993]. Interaction between LTC4S and FLAP may play a role in regulation of LTC4S catalytic activity [Mandal et al., 2004] and we have previously shown that interaction with MGST1, another MAPEG protein, alters the enzymatic activity of both enzymes [Surapureddi et al., 1996; Surapureddi et al., 2000]. Based on gel exclusion chromatography [Penrose et al., 1992; Nicholson et al., 1993; Lam et al., 1994; Welsch et al., 1994] it was postulated that LTC4S is a homodimeric protein. We showed, using bioluminescence resonance energy transfer (BRET), that LTC4S forms homooligomers in vivo and that the C-terminal part of LTC4S (amino acids 114–150), is important for oligomer formation [Svartz et al., 2003]. A recently published study on the 2D structure of LTC4S showed that it is a trimeric protein [Schmidt-Krey et al., 2004].
Molecular mechanisms for the targeting of LTC4S (and other MAPEG proteins) to the NE or ER are not known. We have now investigated the importance of the three hydrophobic regions in LTC4S for subcellular targeting and report that the third hydrophobic region, containing a putative transmembrane helix, acts as a NE localization signal. This region together with the first hydrophobic region is also involved in the homo-oligomerization of LTC4S.
MATERIALS AND METHODS
Materials
Dulbecco's modified Eagle's medium, fetal calf serum, antibiotics, restriction enzymes, T4 DNA polymerase, T4 DNA ligase and competent E. coli DH5α were obtained from Invitrogen (Paisley, Scotland). Monoclonal green fluorescent protein (GFP) antibody (B-2) was from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal 5-LO antibody was from Research Diagnostics (Flanders, NJ). Renilla luciferase fusion protein plasmid vector (pRlucC3), GFP fusion protein plasmid vector (pGFP2C2) and DeepBlueC™ (the coelenterazine substrate for luciferase used in BRET2) were from BioSignalPackard (Montreal, Canada). Red fluorescent ER marker protein vector (pDsRed2/ER) was from Clontech (Palo Alto, CA). Plasmid purification kit was purchased from Saveen (Malmö, Sweden). Mounting medium (SlowFade Antifade Kit) and the nuclear stain ToPro3 were from Molecular Probes (Eugene, OR). Western blot chemiluminescence reagent (Renaissance) was from NEN Life Science Products (Boston, MA). SDS–PAGE molecular weight standard (broad range) was purchased from BioRad (Hercules, CA). All other chemicals used were from Sigma-Aldrich (St Louis, MO).
Cell Culture
Human embryonic kidney (HEK) 293/T cells and COS-7 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal calf serum, penicillin 100 units/ml and streptomycin 100 µg/ml. Cell cultures were split 1:5 at confluency.
Prediction of Transmembrane Regions
The hydrophobicity profile of the amino acid sequence of human LTC4S (Swissprot accession number Q16873) was determined using freely available web applications predicting transmembrane regions and creating hydropathy plots (c.f. Supplementary information). The consensus of the predictions is shown as hatched boxes in the LTC4S sequence in Figure 1.
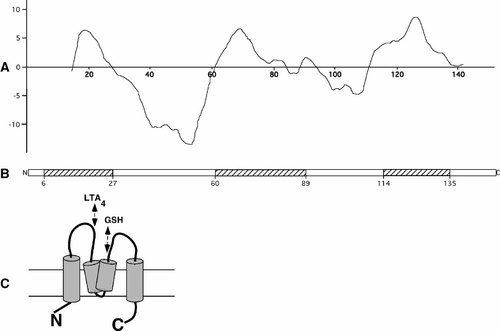
Hydrophobic regions in LTC4S and illustration of membrane topology. A: An illustrative hydropathy plot of the LTC4S sequence using the Wimley and White hydrophobicity scale [Wimley and White, 1996] is shown. Similar results are obtained with other methods used (c.f. Supplementary information). B: Boxes mark the three hydrophobic regions investigated: amino acids 6–27 (ALLAAVTLLGVLLQAYFSLQVI); amino acids 60–89 (FPLFLATLWVAGIFFHEGAAALCGLVYLFA); amino acids 114–135 (ALWLLVALAALGLLAHFLPAAL). C: The membrane topology illustrated is based on the putative membrane topology by Christmas et al. [2002] and modified according to new structural information from Schmidt-Krey et al. [2004]. The cylinders are illustrating transmembrane helices and double arrows indicate the positions, on the luminal side of the membrane, of residues important for LTA4 and glutathione binding, respectively [Lam et al., 1997].
Recombinant Plasmids
Preparation of GFP plasmids encoding the following truncated variants of human LTC4S (1–24, 1–58, 1–88, 1–115, 23–150, 57–150, 87–150, 114–150, 23–58, 23–88, 23–115, 57–88, 57–115, and 87–115) has been described [Svartz et al., 2003]. Constructs encoding short truncated variants of human LTC4S (amino acids 6–27, 9–24, 60–74, 77–89, 114–135, 114–127, 127–135, 117–132, 120–129, and 136–150) as GFP fusions were prepared as follows: Sense and antisense oligonucleotides (c.f. Supplementary information) encoding LTC4S amino acids were designed to create a 5′ Eco RI overhang and a 3′ Hind III compatible overhang after annealing. Annealed oligonucleotides were ligated into pGFP2C2 cleaved with Eco RI/Hind III. cDNA encoding amino acids 6–27 was subcloned from pGFP2C2 into the luciferase fusion protein plasmid pRlucC3. Two additional pairs of oligonucleotides without stop codons were designed to encode amino acids 114–135. One was ligated into the Eco RI site of a pEGFP/5-LO construct resulting in a plasmid expressing GFP/LTC4S114–135/5-LO fusion protein. The other was ligated into pGFP2N1 creating a construct encoding an LTC4S114–135/GFP fusion protein. All plasmids were purified using column chromatography and were analyzed by DNA sequencing. To exclude the possibility that GFP influences enzyme functionality of GFP/LTC4S this protein was expressed in HEK 293/T cells and assayed for enzymatic activity by incubation with LTA4 and glutathione as described [Söderström et al., 1990]. The formation of LTC4 was determined by rpHPLC separation and the product was identified by UV spectra and by co-elution with synthetic standard. The results showed that the enzymatic activity of GFP/LTC4S did not differ significantly from that of LTC4S.
Transient Transfections
One day prior to transfection, 3 × 105 per well HEK 293/T or COS-7 cells were seeded in 6-well dishes on cover slips. On the next day cells were washed, serum free medium was added prior to incubation for 1 h with a mixture containing: 1 µg of each plasmid in 20 µL of 10 mM polyethyleneimide and 5% glucose. After approximately 6 h, fetal calf serum was added to a final concentration of 10% and the cells were incubated for 16–20 h.
Western Blotting
Resuspended cells were washed in 25 mM Tris-HCl, collected by centrifugation, boiled in SDS sample buffer and analyzed by SDS–polyacrylamide gel electrophoresis and electrotransfer to nitrocellulose membrane. The membrane was blocked with bovine serum albumin, incubated over night at +4°C with anti-GFP or anti-5-LO. It was then washed, and incubated with HRP conjugated goat-anti-mouse IgG, washed again and incubated with chemiluminescence reagent prior to recording emission using a Fuji 1000CH LAS camera.
Fluorescence Microscopy
Transfected cells were washed with PBS, fixed in paraformaldehyde and incubated with the nuclear stain ToPro3. GFP (excitation with 488 nm Ar laser), DsRed ER marker protein (excitation with 543 nm He/Ne laser) and ToPro3 (excitation with 633 nm He/Ne laser) fluorescence, respectively, were imaged using a Nikon C1 confocal unit. Average images from five laser scans performed in channel series were obtained. For each construct, at least three pictures of different representative cells were recorded and analyzed by visual inspection.
BRET2 Interaction Assay
Protein-protein interaction studies were performed using bioluminescence energy transfer (BRET) analysis as described [Svartz et al., 2003]. BRET readings were recorded in dual luminescence mode using a Victor2 1420 Multilabel Counter (Wallac, Perkin Elmer). Transmission filters (Packard instruments) 410 nm (bandwidth 80 nm) and 515 nm (bandwidth 30 nm) were used to detect Rluc and GFP emission, respectively. Energy transfer is calculated as BRET ratio (Emission515–Emission410 × Cf)/Emission410 (Cf = Emission515/Emission410 for the Rluc construct expressed alone in the same set of experiments).
RESULTS
Based on prediction of transmembrane regions (c.f. Supplementary information), GFP fusion proteins were constructed with truncated LTC4S sequences starting and/or ending near the hydrophobic regions: Amino acids 6–27 (ALLAAVTLLGVLLQAYFSLQVI) constituting the first region; amino acids 60–89 (FPLFLATLWVAGIFFHEGAAALCGLVYLFA) constituting the middle region; and amino acids 114–135 (ALWLLVALAALGLLAHFLPAAL) constituting the third region (Fig. 1).
The Third Hydrophobic Region in LTC4S Directs GFP to a NE Localization
Confocal fluorescence microscopy of HEK 293/T cells and COS-7 cells transiently transfected with constructs encoding GFP/LTC4S fusion protein and DsRed2/ER showed complete colocalization of LTC4S and the red fluorescent ER marker protein at the NE and ER. GFP fluorescence was strongest around the nucleus (Fig. 2A) whereas the GFP control and was localized in the cytoplasm and nuclear matrix. COS-7 cells showed a more pronounced ER staining for both GFP/LTC4S and DsRed2/ER with complete colocalization also in this case (Fig. 2A).
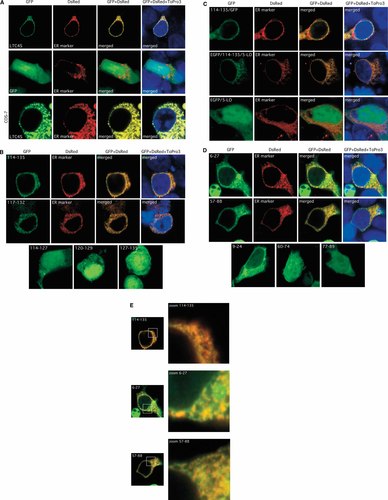
The third hydrophobic region in LTC4S (aa 114–135) is sufficient for correct subcellular localization. Confocal microscopy was performed on HEK 293/T cells (or COS-7 cells as indicated) cotransfected with a GFP construct vectors and a fluorescent ER marker protein vector DsRed2/ER. After fixation, the cells were stained with the fluorescent nuclear dye ToPro3. The left column shows green fluorescence after excitation of GFP constructs with a 488 nm Ar laser, the second column shows red fluorescence obtained after excitation of the DsRed2/ER protein with a 543 nm He/Ne laser. The third column shows pictures from the first and second columns merged. The right column shows in addition fluorescence obtained after excitation of the nuclear dye ToPro3 (with a 633 nm He/Ne laser). The resulting red emission of ToPro3 stained nuclei is pseudo colored as blue. Panels A–D: different constructs as indicated; Panel E: membrane structures are magnified for constructs containing amino acids 6–27, 57–88, and 114–135.
The localization of GFP fused to the third hydrophobic region (114–135) was undistinguishable from that of GFP fused to full length LTC4S. Amino acids 117–132 was the shortest sequence giving a clear NE/ER localization as a GFP fusion protein. GFP fusion proteins with LTC4S amino acids 114–127, 127–135, or 120–129, were located in the cytoplasm and nucleus (Fig. 2B).
Both GFP/LTC4S(114–135) and LTC4S(114–135)/GFP showed the same localization. To investigate if the LTC4S 114–135 sequence could also direct the localization of another protein we placed it between GFP and 5-LO. Also positioning of the LTC4S part in between these two proteins did not alter its subcellular localization. The fluorescence staining of this protein was located at the NE/ER, in the same way as GFP/LTC4S. In contrast GFP/5-LO was localized like GFP in cytosol and nuclear matrix (Fig. 2C).
Localization of Constructs Including the First and Middle Hydrophobic Regions of LTC4S
The first hydrophobic region in LTC4S (6–27), expressed as a GFP fusion protein gave less specific membrane localization including some cytoplasmic distribution, which was more pronounced for the shorter 9–24 variant which also stained the nuclear matrix. LTC4S amino acids 1–24 gave a subcellular distribution very similar to the 6–27 (data not shown). The second hydrophobic region, construct encoding amino acids 57–88, localized GFP mostly to the NE and ER. Two shorter variants (60–74 and 77–89) both showed up as cytoplasmic proteins (Fig. 2D). These two variants are lacking the charged residue positions His-75 and Glu-76.
Table I summarizes the results: Of all constructs investigated in general constructs including the third hydrophobic region showed absolute NE/ER localization as determined by co-localization with the ER marker Ds2Red/ER whereas those lacking this region did not exclusively localize to the NE or ER, but were in various degrees found in the cytoplasm and/or the nucleus. The difference in localization between 6–27, 57–88, and 114–135 is more obvious in Figure 2E where the membrane parts are magnified. The localization of the constructs was similar in COS-7 and Western blot analyses showed that all GFP fusion constructs expressed proteins of expected molecular weights in the transfected cells (data not shown).
| GFP/LTC4S constructs | Co-localization with the ER marker protein DsRed2/ER |
|---|---|
| 1–150 | +++ |
| 23–150 | +++ |
| 57–150 | +++ |
| 87–150 | +++ |
| 114–150 | +++ |
| 114–135 | +++ |
| N-term 114–135 | +++ |
| GFP/114–135/5-LO | +++ |
| 117–132 | +++ |
| 114–127 | − |
| 127–135 | − |
| 120–129 | − |
| 136–150 | − |
| 1–115 | ++ |
| 1–88 | ++ |
| 23–115 | ++ |
| 57–115 | +++ |
| 87–115 | − |
| 23–88 | + |
| 57–88 | ++ |
| 60–74 | − |
| 77–89 | − |
| 1–58 | − |
| 1–24 | + |
| 6–27 | + |
| 9–24 | − |
The First and the Third Hydrophobic Region of LTC4S are Sufficient for Oligomerization
We recently showed that a C-terminal LTC4S domain (amino acids 114–150) was sufficient for homo-oligomerization [Svartz et al., 2003]. The ability of the 114–135 sequence to oligomerize with full length LTC4S was tested. BRET assays showed that these 22 hydrophobic amino acids also resulted in oligomerization with full length LTC4S. A shorter 117–132 variant, which had the same subcellular localization, did not interact with full length LTC4S, indicating that increased BRET ratio is not simply an effect of proteins being in close proximity due to subcellular colocalization (Fig. 3A). In a previous study amino acids 1–24 and 57–88 of LTC4S did not interact with full-length protein [Svartz et al., 2003]. However, when the first hydrophobic region, amino acids 6–27 was tested for the ability to interact with full-length protein, high BRET ratios were obtained (Fig. 3A). Hydrophobic region 1 fused to Rluc also resulted in increased BRET ratios when tested against amino acids 114–135 and especially against the construct also including the C-terminal domain, amino acids 114–150 (Fig. 3B). It is highly probable that amino acids 6–27 and 114–135 form or are part of transmembrane helices, indicating that LTC4S trimers are formed by homo-oligomerization due to helix–helix interactions in the membrane in a head-to-tail manner (Fig. 4).
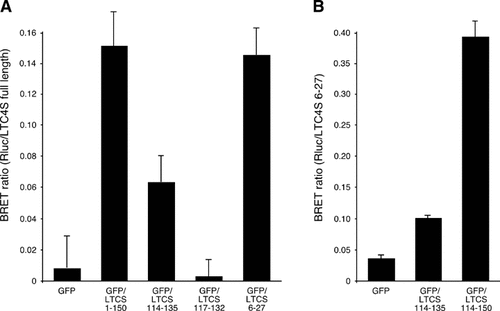
The first and the third hydrophobic regions of LTC4S are sufficient for oligomerization. Interactions between GFP/LTC4S constructs and Rluc fused to A: Full length LTC4S or B: LTC4S amino acids 6–27, were investigated using bioluminescence resonance energy transfer (BRET) assays as described [Svartz et al., 2003]. Briefly, HEK 293/T cells were seeded in 6-well dishes and transfected the next day with 0.25 µg pRluc/LTC4S and 1 µg pGFP construct. After 24 h cells were detached and BRET2 assays were performed in 96 well plates by light-emission acquisition with 410 and 515 nm transmission filters immediately after addition of the luciferase substrate DeepBlueC™. BRET ratio = (Emission515–Emission410 × Cf(/Emission410, (Cf = Emission515/Emission410 for Rluc/LTC4S construct expressed alone in the same experiments). Data expressed as mean values ± standard deviation (n = 3). For further details, see Materials and Methods.
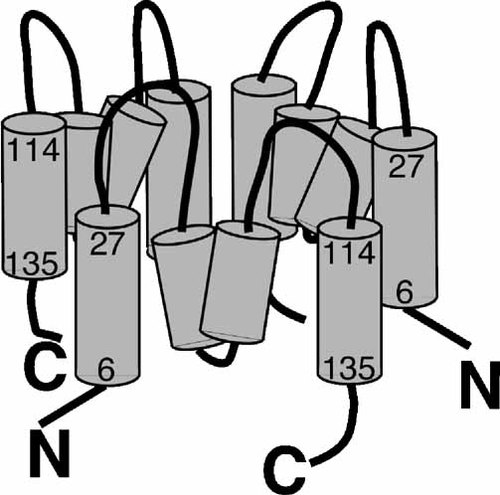
Model of the orientation and topology of a LTC4S homotrimer. The figure shows a model of the orientation of three LTC4S monomers oligomerized as a homotrimer. As indicated the interaction is in a head-to-tail manner where the first N-terminal hydrophobic region (amino acids 6–27) interacts with the third C-terminal region (amino acids 114–135) of a neighboring monomer.
DISCUSSION
The proteins necessary for LTC4 production, 5-LO, FLAP, and LTC4S are all located on or translocated to the NE or ER membranes. LTC4S has 150 amino acids organized into two hydrophilic loops surrounded by three hydrophobic regions. According to the membrane topology suggested by Christmas et al. [2002] the short hydrophilic N- and C-terminal parts point to the cytosolic side of the membrane, while the hydrophilic loops, containing the catalytically active residues Arg-51 and Tyr-93 [Lam et al., 1997], are on the luminal side. Thus, the first and third hydrophobic domains are transmembrane and, probably in α-helical conformation. The second hydrophobic region does not pass the membrane as a single spanning segment with this topology. There are two charged residues, His-75 and Glu-76 in the middle of hydrophobic domain 2, supporting the notion that it is made up of two short transmembrane segments divided by charged residues. Electron crystallography confirmed the presence of four helices per LTC4S monomer [Schmidt-Krey et al., 2004].
LTC4S forms homooligomers and amino acids 114–150, are sufficient for this interaction to occur [Svartz et al., 2003]. In the present study the ability of various LTC4S regions to direct the enzyme to ER and NE was determined. Membrane topology prediction (c.f. Supplementary information) identified three hydrophobic regions: 6–27, 60–89, and 114–135. Localization was determined using confocal microscopy of GFP fusion proteins coexpressed with the ER marker protein DsRed2/ER (Figs. 2A–E). Proteins containing the first hydrophobic region (amino acids 6–27) of LTC4S were partly membrane bound; partly present in the nuclear matrix, (Figs. 2D–E; Table I). The membrane location decreased further for amino acids 9–24. Constructs containing the second hydrophobic region gave localization similar but less pronounced compared to that of full-length protein. Both the segments ahead of (60–74) and that after (77–89) the charged residues His-75 and Glu-76 in hydrophobic region 2 gave cytosolic GFP fusions (Fig. 2D; Table I). The sequence with amino acids 57–88 was the shortest for region 2 giving nearly complete colocalization with the ER marker protein. However, all constructs containing the third LTC4S hydrophobic region were localized identically to full length LTC4S. Because the GFP fusion constructs had the LTC4S sequence C-terminally of GFP, membrane uptake of GFP-LTC4S does not require an N-terminal hydrophobic region (c.f. [White and von Heijne, 2004]). It was recently suggested that membrane insertion is based on a thermodynamic reasoning and that the hydrophobicity of the emerging polypeptide is scanned cotranslationally [Hessa et al., 2005]. The results of the present study suggested that the third hydrophobic region suffices to effect membrane insertion. To prove this we investigated if the 114–135 sequence placed N-terminally or as a linker between GFP and cytosolic 5-LO protein. These fusion proteins were localized to NE and ER, identically to full-length LTC4S, whereas GFP/5-LO without the LTC4S sequence was cytosolic. Further truncation of LTC4S identified amino acids 117–132 as the shortest sequence giving specific NE/ER localization. Many transmembrane proteins are synthesized with leading signal peptides (13–36 residues) to which the signal recognition particle complex binds and thereby prevents the protein from being released into the cytosol [Blobel and Potter, 1967; Gilmore et al., 1982]. Transmembrane proteins contain a 20-residue membrane-anchor or stop-transfer sequence that arrests the passage of the growing polypeptide chain through the membrane [Hegde et al., 1998]. It is interesting that the third transmembrane region provides information for ER retention and that no individual hydrophobic domain is expressed on other membranes, for example, the plasma membrane.
Sequence comparisons of LTC4S with other MAPEG proteins did not show much conservation in the third hydrophobic regions. In contrast, amino acids in the hydrophilic loops are those most highly conserved during evolution of these proteins [Jakobsson et al., 1999; Bresell et al., 2005]. These loops contain both catalytic and substrate binding residues [Lam et al., 1997]. The localization of FLAP, another MAPEG family member, on the inner nuclear membrane [Christmas et al., 2002], differs from that of LTC4S. Sequence differences in the third hydrophobic region might explain this difference [Rolls et al., 1999]. Proteins unique to the inner NE typically bind specifically to lamin or chromatin [Holaska et al., 2002]. The total hydrophobic moment [Wimley and White, 1996] of this region of LTC4S (amino acids 114–135), is greater than that of the corresponding FLAP region (amino acids 118–139). Electron crystallographic analyses have demonstrated that LTC4S [Schmidt-Krey et al., 2004], MGST1 [Schmidt-Krey et al., 1999; Holm et al., 2002], and microsomal prostaglandin E synthase-1 [Thorén et al., 2003] each are homotrimers. BRET analyses showed that the first and third hydrophobic regions in LTC4S are sufficient for oligomerization with full length LTC4S as well as with each other suggesting that the formation of LTC4S oligomers is due to helix–helix interactions between membrane helices 1 and 3 in a head-to-tail manner (Fig. 4). In summary, we have shown that the third hydrophobic region of LTC4S is important for both membrane insertion and localization as well as for oligomerization through interaction with hydrophobic region 1 of a neighboring LTC4S monomer.
Acknowledgements
Human 5-LO cDNA (pEGFP/5-LO) was a kind gift from Dr. T. Izumi, Gunma University, Japan. Human embryonic kidney (HEK) 293/T cells and COS-7 cells were kindly provided by Drs. J. Löfling, Karolinska Institutet, Sweden, and J. Paulsson, Linköping University, Sweden, respectively.