SP100B is a repressor of gene expression
Abstract
Mammalian cell nuclei exhibit discrete sites where specific proteins characteristically localize. PML nuclear bodies (PML NBs) (nuclear domain 10s (ND10s)) are the primary localization site for the promyelocytic leukemia (PML) protein and the SP100 autoantigen. The observations that some PML and SP100 isoforms can function as transcriptional regulators, that both the size and number of PML bodies increase in response to interferon treatment, and that many mammalian viruses encode proteins that mediate disruption of PML bodies suggest that these sites suppress viral infection, perhaps by repressing viral gene expression. We hypothesized that a component of PML NBs functions as a repressor of gene expression. To test this hypothesis, we characterized the effect of PML or SP100 isoforms on expression of transfected reporter genes. PML-I, PML-VI, and SP100A did not repress reporter gene expression. In contrast, SP100B repressed reporter gene expression, especially under conditions in which the reporter gene expression was elevated by a viral transactivator or addition of trichostatin A to the culture medium. The SP100B DNA binding domain was required for repression. SP100B had no detectable effect on the amount, methylation pattern, or topological form of plasmid DNA in the nuclei of transfected cells. The demonstrated repressive activity of SP100B supports the hypothesis that SP100B is a component of an innate immune response that represses expression of ectopic DNA. © 2005 Wiley-Liss, Inc.
The mammalian cell nucleus contains a complex set of components that are spatially arranged and specifically interlinked to facilitate processes such as chromosomal replication, gene expression, and RNA processing. Among these components are a group of nonchromosomal organelles that are localized to discrete subnuclear sites and are referred to as PML nuclear bodies (PML NBs), nuclear domain 10 bodies (ND10s), or PML oncogenic domains (PODs) because there are approximately ten sites in the nuclei of mammalian cells that stain with antibody against the PML (promyelocytic) protein [Ishov et al., 1999]. PML NBs are complex organelles that exhibit changes in size, composition, and number during the cell cycle and in response to physiologic stress such as heat shock or viral infection [Stadler et al., 1995; Grotzinger et al., 1996; Eskiw et al., 2003]. PML proteins, of which there are more than a dozen isoforms encoded by splice variants [Fagioli et al., 1992], are required for the structural integrity and specific functions of PML NBs [Ishov et al., 1999; Jensen et al., 2001]. In addition to PML, the SP100A autoantigen is a constitutive component of PML NBs [Szostecki et al., 1990; Sternsdorf et al., 1999]. There are numerous factors that associate with PML NBs transiently or under specific conditions, including transcription regulators (Daxx, CREB-binding protein, p53), viral proteins (arenavirus RING protein, adenovirus E4ORF3, herpes simplex virus ICP0, Epstein–Barr virus BZLF1), chromatin, and viral genomes (herpes simplex virus, SV40 virus) [Everett, 2001; Negorev and Maul, 2001; Borden, 2002; Sourvinos and Everett, 2002; Eskiw et al., 2004].
The size (0.1–1.0 μm), compositional complexity, and dynamics of PML NBs suggest that these subnuclear organelles have multiple functions. Based on experimental results, investigators have proposed that PML NBs serve as storage depots for transcription factors, play a direct role in transcriptional activation or repression, regulate mRNA processing or translation, contribute to the control of cellular senescence and apoptosis, and serve as sites that either promote or inhibit viral replication [Negorev and Maul, 2001; Borden, 2002; Strudwick and Borden, 2002; Eskiw et al., 2004]. However, the fact that PML−/− knockout mice exhibit a relatively normal phenotype [Wang et al., 1998], other than a lack of detectable PML NBs and an increased sensitivity to some virus infections [Bonilla et al., 2002], indicates that neither PML proteins nor PML NBs are essential for normal development and suggests that PML NBs may have an ancillary function that is not critical in the context of the laboratory-raised mouse. The observations that genes encoding some PML NB components are induced by interferon and that the number and size of PML NBs increase in response to interferon [Lavau et al., 1995; Grotzinger et al., 1996] suggest that PML NBs may contribute to the interferon-induced resistance to viral infection. If so, this may explain why some DNA and RNA viruses encode proteins that disrupt PML NBs [Everett, 2001].
In addition to the functions provided by specific PML isoforms [Li et al., 2000; Kahn et al., 2001; Wu et al., 2001], there is growing evidence that isoforms encoded by the SP100 gene contribute to the function of PML NBs [Szostecki et al., 1990; Dent et al., 1996; Guldner et al., 1999; Seeler et al., 2001]. Analyses of the structure and function of SP100 isoforms have provided compelling evidence for the hypothesis that some SP100 isoforms are transcriptional regulators. SP100A (480 aa) is constitutively expressed in human cells and associates exclusively with PML NBs. SP100A stimulates transcription activation by ETS-1 approximately fivefold [Wasylyk et al., 2002] and inhibits transcription activation by Bright (B cell regulator of immunoglobulin heavy chain transcription) approximately sixfold [Zong et al., 2000]. A GAL4-SP100A fusion protein functions as a repressor of promoters with multiple GAL4 binding sites [Seeler et al., 1998; Bloch et al., 1999]. These results suggest that SP100A may function as either a co-activator or co-repressor, depending on the protein with which it interacts.
Unlike SP100A, the SP100B isoform has been detected only in cells that have experienced stress or been exposed to interferon [Guldner et al., 1999]. SP100B is essentially equivalent to SP100A (the first 476 aa are identical) plus a C-terminal extension that contains an high mobility group (HMG) interaction domain (residues 476–528) [Lehming et al., 1998] and a SAND domain (residues 590–688). The SAND domain functions as a DNA binding domain in several nuclear proteins that regulate transcription from chromosomal genes, including AIRE-1 (autoimmune regulator 1), NUDR (nuclear DEAF-1 related), DEAF-1 (deformed epidermal autoregulatory factor), and GMEB/PIF (glucocorticoid modulatory element binding protein/parvovirus initiation factor) [Bottomley et al., 2001; Burnett et al., 2001].
Based on the observations that interferons stimulate expression of PML and SP100 isoforms, that viral DNA associates with PML NBs, and that most DNA viruses encode a protein that disrupts PML NBs, it has been hypothesized that one or more components of PML NBs function to repress viral gene expression [Maul et al., 1996; Taylor et al., 2000]. When we began this project, our working hypothesis was that one or more components of PML NBs repress expression of extracellular DNA that enters the nucleus by infection or transfection. In this report, we have characterized the effect of PML or SP100 isoforms on expression of reporter plasmids introduced into cells by transfection. Under the conditions of our assays, neither PML-1, PML-VI, nor SP100A exhibited repressive activity. In contrast, SP100B mediated potent repression of both viral and cellular promoters introduced into cells by transfection.
MATERIALS AND METHODS
Cells
Vero cells (ATCC CCL-81) are an African green monkey kidney cell line and U2OS cells (ATCC HTB-96) are a human osteosarcoma cell line that were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% bovine calf serum and gentamicin (25 μg/ml).
Plasmids
A plasmid containing the cDNA encoding the 561 aa form of the PML protein (PML-1a or PML-VI) [Jensen et al., 2001] was obtained from G. Hayward and J. Ahn (Johns Hopkins University, Baltimore, MD). To construct pJLT33, the PML-VI coding region was inserted into pCMV-HA (a derivative of pCMV-mcs [Xiao et al., 1997] that contains the CMV major immediate early promoter followed by a translation initiation codon and 11 codons that encode the influenza virus hemagglutinin (HA) epitope. A plasmid containing the cDNA encoding the 882 aa form of the PML protein (PML-I) [Jensen et al., 2001] was obtained from E. Solomon (Guy's Hospital, London, England) and used to construct plasmids that express PML-I from the CMV promoter as a fusion protein with either an N-terminal HA epitope (pXK1334) or 3XFLAG epitope (pXK1340). Plasmids expressing N-terminal HA-tagged SP100A (pXK1302) and SP100B (pXK1303) from the SV40 early promoter [Seeler et al., 1998] were obtained from A. DeJean (Institut Pasteur, Paris, France). Plasmids using the CMV promoter to express N-terminal 3XFLAG-tagged SP100A (pXK1385) or SP100B (pXK1383) were constructed by inserting the corresponding cDNAs into derivatives of p3XFLAG-CMV-7.1 (Sigma, St. Louis, MO). Plasmids expressing 3XFLAG-tagged SP100B variants lacking residues 295–332 (pXK1368), 335–466 (pXK1369), 335–587(pXK1470), 470–587 (pXK1392), 477–514 (pXK1465), 542–688 (pXK1370), 594–688 (pXK1472), or 631–659 (pXK1384) were derived from pXK1383 by conventional techniques using the appropriate restriction enzymes and oligonucleotide linkers. Plasmid pXK1316 contains the HSV-1 ICP4 coding region inserted between the ICP4 promoter (−331 to +27) and the ICP4 polyA signal. Plasmid pXK350 contains the ICP4 coding region inserted downstream from the CMV major immediate early promoter [Xiao et al., 1997].
Reporter plasmids pXK859, pXK552, pXK543, pXK554, pXK866, pXK877, pXK557, pXK548, and pXK865, contain the promoter from the HSV-1 UL26.5 gene (−463 to +99), ICP0 gene (−130 to +55), ICP4 gene (−331 to +27), ICP27 gene (−274 to +78), LAT gene (−138 to +65), VP5 gene (−360 to +249), gD gene (−391 to +11), gC gene (−82 to +110), and TK gene (−197 to +53), respectively, inserted upstream of the luciferase gene in pGL3-basic (Promega, Madison, WI) as previously described [Bruce and Wilcox, 2002]. Reporter plasmids pXK1244 and pXK1239 contain the HSV-1 LAT gene promoter and ICP27 gene promoter inserted upstream of the luciferase gene in pGL3-enhancer (Promega). Reporter plasmids pXK867 and pXK1232 are identical to plasmids pXK866 and pXK1244, except for a mutation that abolishes the ICP4 binding site in the LAT promoter [Batchelor et al., 1994]. Reporter plasmids with the cellular serum response factor (SRF) and c-fos gene promoters linked to luciferase gene in pGL2-basic (Promega) were kind gifts from Dr. Ravi Misra (Medical College of Wisconsin). Plasmid pXK1231 was constructed by insertion of an oligonucleotide (5′-acgcgtgctagcgcgcgtacgttcgaacgttgccggctcgag-3′ and its complement) immediately upstream of the LAT promoter in pXK866. This oligonucleotide contains ten CpG dinucleotides within cleavage sites for seven restriction enzymes (MluI, BssHII, BsiWI, BstBI, AclI, NgoM4, XhoI) that are blocked by CpG methylation. The pCMV-β-galactosidase plasmid was constructed by inserting the β-galactosidase gene from the pSV β-galactosidase control vector (Promega) into pCMV-mcs [Xiao et al., 1997].
Transient Transfections for Reporter Gene Assays
Vero cells (1 × 106 cells in 60 mm dishes) were transfected in duplicate by the calcium phosphate precipitation procedure [Bruce and Wilcox, 2002]. U2OS cells (1 × 106 cells in 60 mm dishes) were transfected in duplicate with NovaFECTOR (VennNova, LLC, Pompano Beach, FL) using 10 μl NovaFECTOR per 1 μg of DNA per dish in 2 ml of DMEM. At 6 h post transfection, 2 ml of DMEM supplemented with 20% bovine calf serum, 50 μg/ml gentamicin, and 200 nM trichostatin A (TSA) (where indicated) was added to each dish. The response of a reporter gene (100 ng/dish) encoding luciferase to effector proteins encoded by co-transfected plasmids was used to determine effector activity. To measure reporter gene activity, transfected cells were lysed in 0.5 ml Reporter Lysis Buffer (Promega) 24–48 h after transfection and assayed for luciferase using the Luciferase Assay System (Promega) or for β-galactosidase (β-gal) activity using the Galacto-Star assay system (Applied Biosystems, Foster City, CA). Luciferase values from duplicate samples were generally similar to within 10%; therefore, to avoid clutter, only the average values are shown on the plots. In some cases (see below), cells were co-transfected with pCMV-β-galactosidase, and luciferase activities were normalized to β-galactosidase activity to correct for variations in transfection efficiency. Because the CMV promoter in the pCMV-β-galactosidase plasmid was repressed approximately threefold by SP100B and activated approximately threefold by TSA, normalization of luciferase activity to β-galactosidase activity was not appropriate in many experiments. Absolute luciferase values and fold activation or repression differ among similar experiments performed on different days. Therefore, all data within a panel were derived from transfections performed on the same day and assayed as a set.
Analysis of Transfected DNA by Southern Blot
U2OS cells (1 × 106 cells in a 60 mm dish) were transfected with pXK1231 (500 ng) plus or minus pXK1383 (200 ng) using NovaFECTOR. TSA was added to 100 nM at 6 h post transfection. The cells were harvested at 24 h post transfection by scraping into ice-cold phosphate buffered saline supplemented with 0.5 mM EDTA and pelleted in a microcentrifuge (2,000 rpm, 5 min). The cells were resuspended in 1 ml of NB1 (10 mM Tris-Cl pH 7.5, 10 mM NaCl, 1% Triton X-100, 10 mM β-mercaptoethanol), a hypotonic solution that causes rupture of the cytoplasmic membrane without nuclear disruption. Sucrose (0.2 ml of a 50% solution) was added and the nuclei were pelleted in at 2,000 rpm for 5 min. The nuclei were resuspended in 1 ml TEC (50 mM Tris-Cl pH 8.1, 10 mM EDTA, 1% CHAPS) and lysed by passage 10 times through a 25 Ga needle. The nuclear lysate was subjected to centrifugation (10,000 rpm, 10 min in a microcentrifuge). Under these conditions, most of the cellular and plasmid DNAs are found in the pellet. The pellet was resuspended in 0.45 ml of TE (10 mM Tris-Cl pH 8.0, 1 mM EDTA) supplemented with 250 mM NaCl. DNA was purified by phenol/chloroform extraction and resuspended in 20 μl TE. The DNA was digested with restriction enzymes, subjected to electrophoresis in an agarose gel, and then transferred to a Nytran membrane (Schleicher & Schuell, Keene, NH). Bands containing the luciferase gene from pXK1231 were detected by hybridization to a luciferase gene probe obtained by digestion of pGL3-basic with XbaI plus HindIII and radiolabeled by the random primer method using Ready-to-Go DNA labeling Beads (Amersham Biosciences) supplemented with the DNA fragment (25 ng) and [32P]-dCTP (50 μCi).
RESULTS
SP100B Represses Expression From an Activated Promoter
To identify repressors associated with PML NBs, we chose to focus on isoforms of the PML and SP100 proteins because interferon induces expression of these proteins [Lavau et al., 1995; Grotzinger et al., 1996] and some viruses induce degradation of these proteins [Everett, 2001]. Our decision as to which isoform to test was based in part on availability of cDNA clones and in part on how extensively the tested isoforms represent the genomic exons. For example, PML-I and PML-VI are encoded by the most and next-to-least number of exons, respectively [Jensen et al., 2001]. Transient transfection assays in Vero cells were initially used to determine whether a PML or SP100 isoform can repress expression from a reporter plasmid that contains the luciferase gene driven by the HSV-1 UL26.5 gene promoter. A plasmid encoding HSV-1 ICP4, which is an immediate early viral protein that activates transcription from most herpes simplex virus (HSV) early and late gene promoters [Xiao et al., 1997], was included in the initial transfections to elevate luciferase expression above the low levels driven by the UL26.5 promoter alone. Vero cells were cotransfected with the UL26.5-luciferase reporter plasmid, an activator plasmid (ICP4 promoter linked to the ICP4 gene), and a plasmid encoding an isoform of PML or SP100 (Fig. 1A). PML-I or PML-VI increased luciferase expression approximately threefold, SP100A had no effect, and SP100B reduced luciferase expression up to 25-fold (Fig. 1B). A similar pattern was observed in U2OS cells (Fig. 1C), except that the repression of luciferase activity by SP100B was substantially greater than that observed in Vero cells. This increased repression by SP100B in U2OS cells may be a consequence of the fact that ICP4 activates the UL26.5 promoter approximately 150-fold in U2OS cells, compared to only 30-fold in Vero cells under the conditions of these assays [Bruce and Wilcox, 2002].
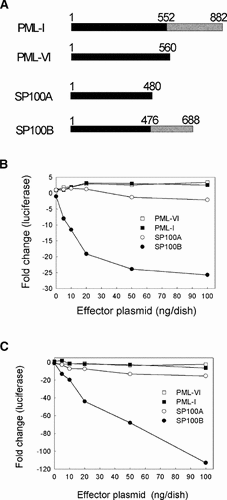
Effect of PML-nuclear-body proteins on gene expression. Panel A: Schematic diagram of splice variants of PML and SP100 proteins. The numbers correspond to amino acid positions. The break in the patterns for PML-I and SP100B indicate where these proteins differ from PML-VI and SP100A, respectively. Panel B: Effect of PML or SP100 proteins on ICP4 activation of the UL26.5 promoter. Vero cells were co-transfected with a reporter plasmid (pXK859) containing the UL26.5 promoter linked to luciferase, an activator plasmid (pXK1316) encoding ICP4 (10 ng/dish), and the indicated amount of an effector plasmid encoding PML-I (pXK1334), PML-VI (pJLT33), SP100A (pXK1302), or SP100B (pXK1303). Cells were harvested 46 h post transfection and assayed for luciferase. The fold change (negative values represent a decrease) in luciferase activity relative to the activity (approximately 140,000 U) observed in the absence of an effector plasmid is shown on the Y-axis. Panel C: U2OS cells were co-transfected with the same plasmids as in panel B and harvested 46 h post transfection. Luciferase activity in the absence of an effector plasmid was approximately 2,750,000 U.
SP100B Represses Basal Promoter Activity
The simplest interpretation of the results described above is that SP100B inhibits the transactivation activity of ICP4. An alternative interpretation is that SP100B directly represses both the ICP4 promoter (thus reducing the amount of ICP4 available to activate the UL26.5 promoter) and the UL26.5 promoter. To determine whether SP100B functions as a repressor in the absence of ICP4, U2OS cells were co-transfected with a plasmid encoding SP100B and reporter plasmids containing different viral gene promoters linked to the luciferase gene. The results (Fig. 2A) indicated that SP100B represses the basal activity of these promoters no more than two to threefold. A similar pattern was observed in Vero cells (data not shown), but the results were more subtle because expression of these transfected promoters is lower in Vero cells than in U2OS cells. These data appear to add support for the hypothesis that SP100B inhibits activation by ICP4. However, we favor an alternative interpretation of these data that is based on evidence that promoters associated with transfected plasmids are in a partially repressed state in the absence of activators, for example ICP4 (see below). In this model, partially repressed promoters associated with transfected DNA would be minimally susceptible to further repression by SP100B.
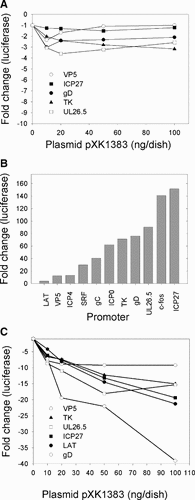
Effect of SP100B on promoter activity in the presence or absence of TSA. Panel A: U2OS cells were co-transfected with a reporter plasmid that contains the indicated HSV-1 gene promoter linked to luciferase and the indicated amounts of a plasmid (pXK1383) encoding SP100B. Cells were harvested 24 h post transfection. Luciferase activity in the absence of pXK1383 ranged from 2,200 U (VP5 promoter) to 15,800 U (UL26.5 promoter). Panel B: Effect of TSA on promoter activity. U2OS cells were transfected with a reporter plasmid that contains the indicated gene promoter linked to luciferase, maintained either in normal medium or in medium supplemented with 100 nM TSA for 24 h, and then harvested. For each reporter plasmid, the luciferase activity from the TSA-treated cells was divided by the luciferase activity from the untreated cells to calculate the fold change. Panel C: The procedure was the same as for panel A, except that transfected cells were maintained in 100 nM TSA for 24 h and then harvested. Luciferase activities in the absence of pXK1383 were approximately 21,000, 1,300,000, 900,000, 2,600,000, 1,500,000, and 530,000 U for the VP5, TK, UL26.5, ICP27, LAT, and gD gene promoters, respectively.
Expression of chromatin-associated genes is governed in part by posttranslational modifications of residues in the N-terminal tails of histones [Jenuwein and Allis, 2001]. In particular, acetylation of lysine residues in histones H3 and H4 is associated with actively transcribed DNA; removal of these acetyl groups by histone deacetylases (HDACs) results in gene silencing [Fischle et al., 2003]. It has been reported that transfected plasmid DNA is associated with hypoacetylated histones and, thus, is repressed [Mavromara-Nazos et al., 1986]. Inhibition of HDAC activity by trichostatin A (TSA) results in “derepression” of promoters associated with transfected plasmids [Laherty et al., 1997; Lea and Randolph, 1998]. Addition of TSA to U2OS cells transfected with reporter plasmids resulted in increased luciferase activity. We observed no differences in either cell morphology or monolayer density during the 18 h TSA incubation, compared to non-TSA-treated cells. The fold increase was promoter dependent. The HSV ICP27 gene promoter was derepressed 150-fold by TSA, whereas the HSV LAT gene promoter was derepressed only fourfold (Fig. 2B). Derepression was not specific for viral promoters; plasmids containing the cellular SRF or c-fos gene promoters were derepressed 30- and 140-fold, respectively, in the presence of TSA. There was no correlation between promoter strength (as defined by the activity of the promoter in the absence of TSA) and fold-derepression by TSA. For example, in the absence of TSA, the ICP4 and VP5 gene promoters are the strongest and weakest, respectively, in this series (data not shown), and both were derepressed to the same extent by TSA.
To determine whether TSA-derepressed promoters are more susceptible to repression by SP100B, we co-transfected cells with different reporter plasmids and a plasmid encoding SP100B and maintained the transfected cells in TSA. TSA treatment increased the susceptibility of all promoters to repression by SP100B (compare Fig. 2A,C), although not to the same degree and not in a pattern that correlated with promoter activity in the absence of TSA. The VP5 and ICP27 gene promoters were repressed less than twofold by SP100B in the absence of TSA (Fig. 2A), were derepressed 12- and 150-fold by TSA treatment (Fig. 2B), and were repressed 9- and 19-fold by SP100B in the presence of TSA (Fig. 2C). The TK and gD gene promoters were repressed three and twofold by SP100B in the absence of TSA, were derepressed approximately 75-fold by TSA treatment, and were repressed 15- and 40-fold by SP100B in the presence of TSA. We suspect that the lack of a consistent pattern in fold-derepression by TSA versus fold-repression by SP100B is a consequence of different transcription factors recruited by specific sequences in each promoter. These results reveal that repression of gene expression by SP100B occurs in the absence of ICP4 and is not promoter specific. Results using reporter plasmids encoding β-galactosidase revealed that repression by SP100B is not reporter specific (data not shown). Because all the data shown in Figure 2C were obtained on the same day with the same batch of cells, it is reasonable to conclude that the different response of each promoter to repression by SP100B is a consequence of events specific to that promoter rather than a consequence of an activity of SP100B that has a global effect on RNA processing, protein synthesis, or cell metabolism. These results suggest that SP100B represses transcription from plasmid-associated promoters in transfected cells and support the conclusion that the data shown in Figure 1 are a consequence of SP100B repression of the UL26.5 and ICP4 promoters rather than SP100B inhibition of ICP4 activity.
Identification of Functional Motifs in SP100B
Because the first 476 residues of SP100A and SP100B are identical (Fig. 3A), we concluded that the C-terminus of SP100B (aa 477–688), which includes the HMG interaction domain (aa 477–528) and the SAND domain (aa 590–688), is required for repression. In addition, other investigators have suggested that the HP1 interaction domain (aa 287–334) is required for repression [Lehming et al., 1998; Seeler et al., 1998]. The repressive activity of SP100B variants lacking each of these motifs was initially tested in U2OS cells co-transfected with the UL26.5-luciferase reporter plasmid and a plasmid encoding ICP4 and maintained in TSA (a similar pattern was seen in the absence of TSA). The ΔHMG1 variant (SP100BΔ477–514) was as active as wild-type SP100B and the ΔHP1 variant (SP100BΔ295–332) was actually more active than the full-length protein (Fig. 3B). It should be noted that the ΔHP1 variant also lacks the sumoylation site (K297) identified in SP100A [Sternsdorf et al., 1999]. In contrast, the variant (SP100BΔ631-659) with a deletion in the SAND domain was inactive (Fig. 3B). We conclude that the SAND domain is essential for SP100B to function as a repressor and that neither the HP1 nor HMG interaction domain is required for repression under the conditions of this assay. Immunoblot assays performed with extracts prepared from cells transfected with plasmids encoding each of the variants, all of which were 3XFLAG-tagged, revealed that the full-length and deleted variants of SP100B accumulated to similar levels (data not shown).
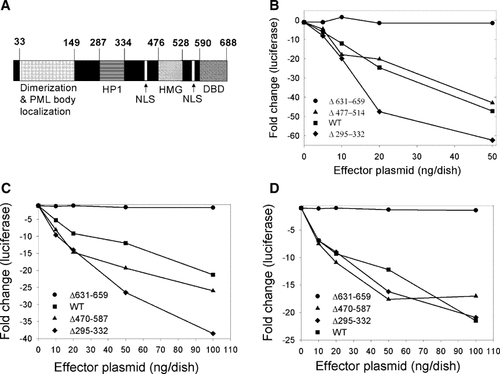
Activity of SP100B variants. Panel A: Schematic diagram of SP100B protein. The numbers correspond to amino acid positions. NLS, nuclear localization sequence; DBD, DNA binding domain. Panel B: U2OS cells were co-transfected with a reporter plasmid containing the UL26.5 gene promoter linked to luciferase (pXK859), an activator plasmid (10 ng/dish) encoding ICP4 (pXK1316), and an effector plasmid encoding the indicated SP100B variant. Transfected cells were maintained in TSA, harvested 24 h post transfection, and assayed for luciferase. Panel C: U2OS cells were co-transfected with a reporter plasmid (100 ng/dish) containing the LAT gene promoter linked to luciferase (pXK866) and an effector plasmid encoding the indicated SP100B variant. Transfected cells were maintained in TSA and harvested 24 h post transfection. Panel D: Same experimental procedure as for panel C, except that the reporter plasmid contained the c-fos promoter.
We also tested these SP100B variants with the HSV-1 LAT gene promoter (Fig. 3C) and the cellular c-fos promoter in TSA-treated cells but in the absence of ICP4 (Fig. 3D) to determine whether specific domains in SP100B are required for repression of other viral or cellular promoters. With these reporters, we tested an additional SP100B variant with a more extensive deletion in the C-terminus (aa 470–587); this variant is nearly equivalent to SP100A plus the SP100B SAND domain. Repression of the LAT promoter (Fig. 3C) was slightly higher with the Δ470–587 variant and significantly higher with the Δ295–332 variant compared to full-length SP100B. With the c-fos promoter (Fig. 3D), the repression activities of the Δ470–587 and Δ295–332 variants were similar to the full-length protein. The Δ631–659 variant failed to repress either promoter. These results confirm that the SP100B SAND domain is essential for repression and indicate that neither the HP1 nor HMG interaction domains are essential for repression.
Repression of Promoters Associated With Enhancers
In the experiments used to obtain the data shown in Figures 2 and 3, TSA was added to the transfected cells to derepress the reporter plasmids. An alternative approach to elevate promoter activity is to insert a transcriptional enhancer in a reporter plasmid with a “standard” (i.e., non-enhanced) promoter. We were curious to determine whether SP100B was effective as a repressor of an “enhanced” promoter. We inserted the enhancer derived from the SV40 (simian virus 40) genome into a reporter plasmid with the LAT promoter. The LAT promoter is repressed when ICP4 binds to a specific sequence that overlaps the transcription initiation site in the LAT promoter [Batchelor et al., 1994]. Reporter plasmids containing a standard or enhanced LATmut promoter (mutant ICP4 binding site) were constructed as controls. These reporter plasmids allowed us to compare repression of a TSA-derepressed or enhanced promoter by SP100B to repression of the same promoter by the sequence-specific repressor ICP4.
The activity of the LAT promoter was increased approximately 2.5-fold by TSA, 2.6-fold by the enhancer, and 18-fold by the combination of TSA plus enhancer (Fig. 4A). These results suggest that the mechanisms by which TSA or an enhancer elevate promoter activity are different. In TSA-treated cells, SP100B repressed the enhanced LAT or LATmut promoters approximately twofold less than the standard LAT or LATmut promoters (Fig. 4B). In contrast, under the same conditions ICP4 repressed the enhanced LAT promoter approximately twofold more than the standard promoter (Fig. 4C). As expected, the LATmut promoter was not repressed by ICP4 (Fig. 4C). The clear distinction between the effects of an enhancer on repression by SP100B versus ICP4 suggests that the mechanisms by which these two proteins repress transcription are fundamentally different. ICP4 binds directly to the transcription initiation site and thus blocks formation of the preinitiation complex. Apparently, SP100B functions by a different mechanism.
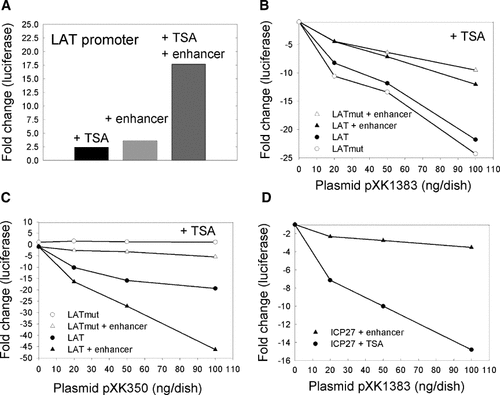
Effect of SP100B on enhanced promoter activity. Panel A: U2OS cells were transfected with a reporter plasmid (100 ng) containing the LAT promoter linked to luciferase (pXK866) and where indicated (+enhancer) the SV40 enhancer (pXK1244). Where indicated (+TSA), the transfected cells were maintained in 100 nM TSA. Cells were harvested 24 h post transfection. Luciferase activities were normalized to the activity (285,000 U) in cells transfected with pXK866 in the absence of TSA. Panel B: U2OS cells were transfected with the indicated reporter plasmid and the indicated amounts of a plasmid (pXK1383) encoding SP100B and maintained in 100 nM TSA for 24 h. The LATmut promoter in reporter plasmids pXK867 and pXK1232 (+enhancer) has a mutation that abolishes the ICP4 binding site. Panel C: Same experimental procedure as for panel B using an effector plasmid (pXK350) that encodes ICP4. Panel D: U2OS cells transfected with pXK554 (ICP27 promoter) and pXK1383 were maintained in 100 nM TSA for 24 h and then harvested. U2OS cells transfected with pXK1239 (ICP27 promoter + enhancer) and pXK1383 were maintained in the absence of TSA for 24 h and then harvested.
To directly compare the ability of SP100B to repress a promoter that was activated to a similar level either by TSA-treatment or by addition of an enhancer, we used a reporter plasmid with the ICP27 promoter. Under conditions used, the activity of the standard ICP27 promoter in TSA-treated cells (690,000 RLU) was similar to the activity of an “enhanced” ICP27 promoter in the absence of TSA (1,275,000 RLU). The results (Fig. 4D) revealed that the TSA-derepressed promoter was repressed 15-fold by SP100B, whereas the enhanced promoter was repressed less than fourfold. These results provide direct evidence that a TSA-derepressed standard promoter is much more susceptible to repression by SP100B than an enhanced promoter.
Effect of SP100B on Plasmid Structure
Some cellular gene promoters contain regions with an enriched CpG content. Methylation of the cytosines in these so-called CpG islands by DNA cytosine methytransferases (DNMT) results in recruitment of proteins that function as gene silencers [Ng et al., 1999]. To test the hypothesis that SP100B silences gene expression by inducing methylation of CpGs in transfected DNA, we constructed a reporter plasmid (pXK1231) in which the LAT promoter is immediately downstream of a 40 bp insert that contains CpGs within cleavage sites for the restriction enzymes AclI, BssHII, BsiWI, and BstBI. These enzymes fail to cleave sites that contain methylated cytosines. The 40 bp insert had no effect on the activity of the LAT promoter or on repression of the LAT promoter by SP100B (data not shown). U2OS cells were transfected in parallel with pXK1231 alone or in combination with a plasmid that expresses SP100B and the transfected cells were maintained in TSA until harvested. Nuclear DNA was extracted from the transfected cells, digested with selected restriction endonucleases, and analyzed by Southern blotting with a probe specific for the luciferase gene. The results revealed that SP100B had no effect on either the amount or topology of plasmid DNA recovered from transfected cells (Fig. 5). The undigested plasmid (5.1 kbp) was recovered as a mixture of supercoiled (approximately 45%), nicked circular (approximately 40%), and linear (approximately 15%) forms from cells that either did or did not overexpress SP100B. This result demonstrates that SP100B does not induce DNA degradation. To determine whether SP100B induces DNA methylation, the extracted DNA was digested with BstBI alone, which cuts within the 40 bp insert and near the 5′ end of the luciferase gene to generate a 4.5 kbp fragment that contains the luciferase gene, or with a methylation-sensitive enzyme (AclI, BsiWI, or BssHII) that cuts within the 40 bp insert and a methylation-insensitive enzyme (AlwNI or BamHI) that cuts at a unique site in the plasmid to generate a 2.9 or 2.2 kbp fragment that spans the luciferase gene. The transfected plasmid was digested to completion by all three methylation-sensitive enzymes (Fig. 5) following extraction from cells that either did or did not express SP100B. These results demonstrate that the ten CpGs in the 40-bp insert are not extensively methylated in cells that overexpress SP100B and suggest that SP100B does not repress transfected DNA by recruitment of a DNMT.
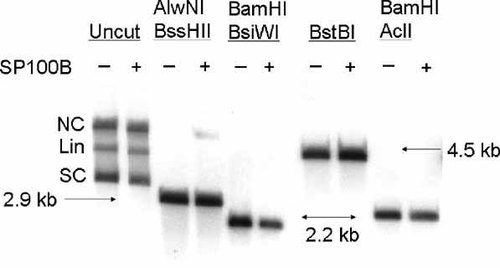
Effect of SP100B on plasmid structure. U2OS cells were transfected with a plasmid (pXK1231) containing a 40-nucleotide sequence with CpG dinucleotides within cleavage sites for BssHII, BsiWI, BstBI, and AclI followed by the LAT promoter, the luciferase gene, and unique cleavage sites for BamHI site and AlwNI. Where indicated, the U2OS cells were co-transfected with a plasmid (pXK1383) encoding SP100B. The transfected cells were maintained in 100 nM TSA. At 24 h post transfection, nuclear DNA was digested with the indicated restriction enzyme and subjected to Southern analysis. DNA fragments that hybridized to a [32P]-labeled probe for the luciferase gene were detected by autoradiography. Bands corresponding to nicked circular (NC), linear (LIN), and supercoiled forms (SC) of the plasmid are indicated.
DISCUSSION
The results presented here demonstrate that SP100B represses expression of luciferase from DNA transfected into either Vero or U2OS cells. Under similar conditions, no repression was observed with PML-1, PML-VI, SP100A, or an SP100B variant with a deletion in the C-terminal SAND domain. Because these results were obtained with native SP100B (rather than GAL4 fusions used for the analysis of SP100A [Seeler et al., 1998; Bloch et al., 1999]), it seems reasonable to conclude that SP100B has an intrinsic ability to mediate repression of gene expression. The immediate question raised by this conclusion is how does SP100B function as a repressor. For the three reasons discussed below, we favor the hypothesis that SP100B represses gene expression at the transcription level.
First, as shown in Figure 3, repression requires the SP100B SAND domain, which functions as a DNA binding domain in the AIRE, NUDR, DEAF-1, and GMEB-1 proteins, all of which are transcriptional regulators [Gross and McGinnis, 1996; Huggenvik et al., 1998; Kumar et al., 2001; Chen et al., 2002]. A highly conserved KDWK or KNWK sequence occurs at the protein:DNA interface in the SAND domain of each of these proteins. We have constructed an SP100B variant in which the KNWK motif at aa 653–656 in the SAND domain [Bottomley et al., 2001] was changed to ANWA. In contrast to wild-type SP100B, this variant does not bind DNA and does not repress expression from transfected DNA (data not shown). These results imply that repression requires association of SP100B with DNA.
Second, susceptibility to repression by SP100B differs among promoters. This suggests that repression is governed at the promoter level rather than at a downstream step such as RNA processing or translation. Although we have conducted tests with a number of promoters, no clear pattern has emerged, other than the observation that promoters with associated enhancers (such as the CMV major immediate early promoter or the LAT promoter plus SV40 enhancer) are relatively insensitive to repression by SP100B. Our data do not support a model in which SP100B represses only viral promoters. This is not surprising because, at least for most DNA viruses, viral promoters are not intrinsically different from cellular promoters.
The third observation in support of SP100B as a transcriptional repressor is that a transcriptional enhancer partially abrogates repression by SP100B. Transcriptional enhancers function by recruiting proteins that “open up” heterochromatin and facilitate formation of the preinitiation complex [Kadonaga, 1998]. Our observation (Fig. 4) that an enhancer reduces the susceptibility of the LAT and ICP27 promoters to repression by SP100B is consistent with a process that occurs at the level of transcription. Particularly noteworthy is the observation (Fig. 4D) that fold-repression by SP100B is quite different when the activity of the ICP27 promoter is elevated to nearly equivalent levels by either an enhancer or TSA-treatment. This result demonstrates that repression by SP100B can be differentially affected by factors that function at the level of transcription. We suspect that enhancers recruit one or more cellular factors to DNA that mitigate the effects of SP100B. If this is the case, then perhaps the presence of strong enhancers near the promoters of some immediate early viral genes provides a defense against repression by SP100B.
Although we favor transcriptional repression, there are other mechanisms by which SP100B might repress gene expression. One trivial explanation is that ectopic expression of SP100B in transfected cells is cytotoxic. We discount this explanation because we have not observed any gross differences in cell morphology or monolayer density up to 48 h after transfection with SP100B, compared to cells transfected with SP100A. Furthermore, if SP100B were cytotoxic, one would expect it to exert a similar reduction in luciferase expression from all promoters, which is not the case (Figs. 2 and 4). A more interesting hypothesis is that SP100B induces RNA degradation. By RT-PCR analysis, we observed that overexpression of SP100B resulted in decreased accumulation of luciferase transcripts from transfected reporter plasmids (data not shown), but these results do not distinguish between decreased RNA synthesis versus increased RNA degradation. However, the results shown in Figure 4D are not consistent with a model in which SP100B induces mRNA degradation. Under the experimental conditions used for this experiment, nearly equal amounts of luciferase (and presumably of luciferase mRNA) were produced in cells transfected with either pXK554 or pXK1239 in the absence of SP100B. However, in the presence of SP100B, the reduction in the amount of luciferase (and presumably of luciferase mRNA) was much greater with pXK554 compared to pXK1239.
Given the assumption that SP100B acts to inhibit transcription, what is the mechanism? The hypotheses that SP100B functions as, or recruits, a DNase or a DNA methyltransferase, though very appealing, are not supported by the data (Fig. 5). A simple hypothesis is that SP100B binds directly to core elements in promoters and sterically interferes with formation of the preinitiation complex, as is the case for ICP4. We are currently exploring this hypothesis by investigating the DNA binding properties of SP100B.
Based on the presence of the HP1 interaction site in SP100A and the observed repression by GAL4-SP100 fusion proteins of promoters with GAL4 binding sites, other investigators proposed that SP100 silences gene expression by recruiting HP1 and HMG to DNA, resulting in condensation of the DNA into a heterochromatin-like structure [Lehming et al., 1998; Seeler et al., 1998]. We considered this model to be attractive and thus were surprised to discover that neither the HP1- nor HMG-binding motif is required for repression by SP100B in our assays. One explanation for this apparent discrepancy is that SP100B may initially repress transcription in the absence of HP1 or HMG and subsequently recruit HP1 along with other factors that maintain the DNA in a long-term silenced state in the absence of SP100B. This long-term HP1-dependent silencing mechanism might not be in force in transiently-transfected cells. Stably-transfected cells that express SP100B could provide an experimental approach to investigate whether SP100B causes long-term silencing of transfected or infected DNA in an HP1-dependent manner.
As mentioned above, removal of acetyl groups from histones by HDACs is an early event in gene silencing. There are more than a dozen HDACs, of which some (but not all) are inhibited by TSA. It is especially noteworthy that SP100B functions as a potent repressor in the presence of TSA. This is in contrast to transcriptional repressors such as PML-IV, Blimp-1, SMRT, NCOR, mSin3A, and Mad that recruit TSA-sensitive histone deacetylases and thus do not function as repressors in the presence of TSA [Laherty et al., 1997; Nagy et al., 1997; Yu et al., 2000; Wu et al., 2001]. The fact that SP100B functions as a repressor in TSA-treated cells reveals that SP100B-mediated repression does not require a TSA-sensitive HDAC, but does not rule out histone deacetylation by a TSA-insensitive HDAC such as hSir2 [Vaziri et al., 2001] as a mechanism for SP100B-mediated repression.
The composition of nucleosomes associated with transfected DNA is dependent upon many factors, including transfection method, cell type, and whether the plasmid can replicate in transfected cells [Reeves et al., 1985; Jeong and Stein, 1994; Luo et al., 1998; Zhang et al., 2002]. The reporter plasmids used for this investigation cannot replicate in U2OS cells. Our preliminary results indicate that the majority of the plasmids in the nuclei of transfected U2OS cells are associated with histones. We have argued that the increased susceptibility of reporter plasmids to repression by SP100B in TSA-treated cells is because TSA inhibition of HDACs leads to histone hyperacetylation and consequently derepression of transfected DNA. Lea and Randolph [1998] reported that TSA-treatment leads to hyperacetylation of histones associated with transfected DNA in rat osteosarcoma cells. We are investigating the extent to which plasmid-associated histones in U2OS cells (human osteosarcoma) are acetylated in the presence or absence of TSA or SP100.
We would like to discuss briefly an issue that we believe is central to the significance of these findings, although it is not directly addressed by our experimental results. We and others have noted that the correlation between accumulation of SP100 isoforms in interferon-treated cells and degradation of SP100 isoforms in cells infected by members of the herpesvirus family is consistent with a role for these proteins in inhibition of productive infection. The results presented here demonstrate that SP100B can repress gene expression from transfected plasmids. We are currently testing the effects of SP100B expression on viral infection and on expression of endogenous genes. These experimental results will address the hypothesis that SP100B-mediated repression is an innate defense that specifically targets ectopic DNAs.
Acknowledgements
We thank Kay Nicholson and Heather Janscha for excellent assistance in all phases of this investigation. This research would not have been possible without the generous contributions of plasmids from G. Hayward, A. DeJean, E. Solomon, and R. Misra.




