An inactive X specific replication origin associated with a matrix attachment region in the human X linked HPRT gene
Abstract
Early in female mammalian embryogenesis, one of the two X chromosomes is inactivated to compensate the gene dosage between males and females. One of the features of X chromosome inactivation (XCI) is the late replication of the inactivated X chromosome. This study reports the identification, by competitive PCR of nascent DNA, of a replication origin in intron 2 of the human X-linked HPRT gene, that is functional only on the inactive X. Features frequently associated with replication origins, including a peak of enhanced DNA flexibility, a perfect match to the yeast ACS sequence, a 14/15 match to the Drosophila topoisomerase II consensus, and a 20/21 match to an initiation region consensus sequence, were identified close to the replication origin. The origin is located approximately 2 kb upstream of a matrix attachment region (MAR) and also contains two A:T-rich elements, thought to facilitate DNA unwinding. © 2005 Wiley-Liss, Inc.
Sex determination in mammals is determined by an XY system where females have two X chromosomes and males a single X and a single Y chromosome. Dosage compensation of X linked genes is achieved by the transcriptional silencing, early in embryogenesis, of genes on one of the two X chromosomes in female cells [Lyon, 1961]. In eutherian mammals the X chromosome to be inactivated is initially chosen randomly, but once chosen, is epigenetically inherited throughout succeeding generations [Monk, 1986]. X chromosome inactivation (XCI) requires a cis-acting locus for the initiation and propagation of the silencing signal. The XIST gene (X inactive specific transcript) encodes a large noncoding transcript that coats the X chromosome to be inactivated [Brown et al., 1992] and accumulation of XIST RNA initiates the events associated with XCI. Numerous studies based on the in vitro differentiation of embryonic stem cells (ES cells) have shown that XCI events occur in a stepwise manner [reviewed Brockdorff, 2002]. Accumulation of XIST RNA is the earliest event, starting about day 1 and occurring in most cells by day 5 [Sheardown et al., 1997]. Late replication and transcriptional silencing of X linked genes is first seen at day 2, reaching its maximal levels by day 6 [Keohane et al., 1996]. Global hypoacetylation of histone H4 is first seen at day 4 [Keohane et al., 1996] and is followed later (day 7) by incorporation of the variant histone macroH2A1.2 [Mermoud et al., 1999]. Methylation of CpG dinucleotides is not detectable until day 21 [Keohane et al., 1996]. DNA replication is initiated at discrete regions (replication origins) which replicate at defined times and at defined locations of the nucleus during S phase [Spector, 1992]. Active genes usually replicate within the first half of S phase [Goldman et al., 1984; Holmquist, 1987] whilst permanently silenced chromatin, such as inactive X linked genes, are replicated in late S phase [Boggs and Chinault, 1994]. The molecular model for the initiation of DNA replication is based on the bacterial replicon model where replication is initiated by a cis-acting replicator element bound to a trans-acting initiator protein. In yeast, replication origins were initially identified as regions (autonomously replicating sequences, ARS) that allowed plasmids to be maintained extrachromosomally. Yeast ARSs contain an 11 bp sequence, the autonomously replicating core consensus sequence (ACS) that is essential for replication origin function [Van Houten and Newlon, 1990] and that binds a six subunit protein origin replication complex (ORC) both in vivo and in vitro [Bell and Stillman, 1992; Diffley and Cocker, 1992]. Human homologues to five of the six yeast ORC subunits have now been identified [Tugal et al., 1998] indicating that the mechanism for initiation of DNA replication has been conserved from yeast to human. Replication origins identified thus far appear to have several common sequence features that may contribute to origin function [reviewed in Dobbs et al., 1994; Zannis-Hadjopoulos and Price, 1998]. For example matrix attachment regions (MARs) are frequently located in proximity to replication origins and may be required to connect the replication origin to an appropriate nuclear region or at an appropriate time in S phase [Boulikas, 1996]. DNA unwinding elements (DUEs), which are stretches of DNA that are easily unwound to allow initiation of strand synthesis, appear to be absolutely required for origin function [Natale et al., 1993]. Other features associated with replication origins include sequences with homology to the yeast ACS, Drosophila topoisomerase II cleavage consensus, Pur protein binding sites, and pyrimidine tracts [Dobbs et al., 1994]. A 21 bp consensus sequence (initiation region consensus, IRC) has also been identified within several initiation regions, although its function is unknown [Dobbs et al., 1994]. Peaks of significantly enhanced flexibility have also been demonstrated at or close to some replication origins, suggesting that enhanced DNA flexibility is functionally important for replication initiation [Toledo et al., 2000].
Despite the identification of sequence features associated with replication origins, it is not known how eukaryotic replication timing is controlled or how this may relate to the activity state of the associated gene. The frequent association of replication origins with MARs led us to search, using a PCR based nascent strand abundance assay, for the presence of a replication origin in the vicinity of a nuclear MAR-3 that we had previously identified in intron 3 of the human X linked HPRT gene [Chong et al., 1995]. To enable discrimination of replication origin activity on the active versus the inactive X, the experiments were performed on DNA from hamster: human hybrid cells containing either an active (Y162/11C) or an inactive (X86T2) human X chromosome [Hansen et al., 1988]. The results indicate the presence, approximately 2 kb upstream of MAR-3, of a replication origin, which is utilized on the inactive X chromosome only. Immediately 5′ to the inactive X specific replication origin is a peak of enhanced DNA flexibility, which is co-incident with a perfect match to the yeast ACS and a 14/15 match to a topoisomerase II consensus. A 20/21 match to a consensus sequence previously noted in association with replication origins [Dobbs et al., 1994] is also located just upstream of the region of enhanced DNA flexibility.
MATERIALS AND METHODS
Cell Culture
The hamster: human hybrid cell lines X86T2 (inactive X) and Y162/11C (active X) [Hansen et al., 1988] were grown in glutamine-containing RPMI medium supplemented with 10% fetal bovine serum and, for the Y162/11C cell line, 5 × 10−5M hypoxanthine; 2 × 10−7M aminopterin; and 8 × 10−4M thymidine (HAT) to maintain the human active X chromosome. The maintenance of the inactive X in the X86T2 cell line was tested at regular intervals by addition of HAT to the media, which induced death in the cells due to lack of an active HPRT gene. All cell lines were grown at 37°C under 95% air/5% CO2.
Extraction and Purification of Nascent DNA
Cells grown to 40%–50% confluence in three 420 cm2 multi-floor tissue culture flasks (Techno Plastics Products, South Australia, Australia) were harvested with trypsin, collected by centrifugation (1,000g × 10 min, 4°C) and resuspended in 9 ml of ice cold 0.32M sucrose containing 5 mM MgCl2, 10 mM Tris pH 7.6 (RT), and 1% triton X100. Nuclei were pelleted by centrifugation (10,000g × 10 min, 4°C), then resuspended in 4.5 ml ice-cold 0.1M Tris pH 8.0 (RT) containing 20 mM EDTA, and 0.15M NaCl and, following addition of SDS, proteinase K, and RNAse A to final concentrations of 2% and 100 μg/ml respectively, were left at room temperature overnight. The DNA was then extracted with an equal volume of phenol (Tris buffered, pH 7) at least twice, then re-extracted once with phenol:chloroform:isoamyl alcohol (25:24:1, v:v:v), once with chloroform:isoamyl alcohol (24:1, v:v), then precipitated with two volumes of absolute ethanol, washed in 70% ethanol, and dried before resuspending in TE (10 mM Tris, 0.1 mM EDTA, pH 8.0 at RT).
Isolation of nascent DNA was achieved by nascent strand extrusion and sucrose gradient fractionation as described in Tao et al. [1997] with the modifications described below. DNA (approx. 350 μg in 2 ml) was incubated at 50°C for 18 h to allow extrusion of the nascent strands [Zannis-Hadjopoulos et al., 1981] then layered onto 8 ml of a 5%–30% (w/v) sucrose gradient containing 0.2M NaCl, 10 mM TE pH 8.0, and 0.02% sodium azide, and fractionated by centrifugation at 164,000g in a Sorvall TH-641 rotor at 4°C for 20 h. After centrifugation, 20 fractions of 500 μl, collected from the top, were ethanol precipitated then resuspended in TE and electrophoresed on a 1% agarose gel with molecular weight markers. The gel regions containing nascent DNA of approximate size 0.6–1.2 kb were excised and combined, and the DNA purified using a Bresa Clean Purification kit (Bresatec, South Australia, Australia) to give a final volume of purified nascent DNA of 450 μl.
Construction of Competitors for Quantitative PCR of Selected Regions
For each of the regions selected for replication origin localization, competitor DNA molecules that shared the same sequence as the genomic targets except for an additional 20 bp insert were constructed. The genomic targets were amplified in reactions containing 200 ng genomic DNA, 1× PCR buffer, 0.5–1.0 μM of forward and reverse primers (shown in Table I), 200 μM dNTPs, 1.5 mM MgCl2, and 0.625 U Taq polymerase (Biotech International, Queensland, Australia). Primers were chosen from the known sequence of the human HPRT gene [Edwards et al., 1990] (GeneBank accession no. M26434) and named according to their approximate distance from the 5′ end of MAR-3. Cycling conditions consisted of denaturation at 94°C for 1 min × 1 cycle; 92°C for 40 s, 60°C (58°C for the −1.1 primer set) for 40 s, 75°C for 1.5 min for a total of 30 cycles; and a final extension of 75°C for 5 min in an MJ research PTC-100 cycler. The PCR reactions were run on 4% agarose gels and the corresponding products for each region purified from the gel using the Bresa Clean procedure (Bresatec) then subcloned into pGEM-T vector for confirmatory sequencing.
| Primera | 5′ nucleotide | Sequence 5′–3′b |
|---|---|---|
| −5.1F | 12,954 | GAGAGGCTGTTTGGATTTGG |
| −5.1R | 13,175 | AAAATTAGCCAGGCATGTGG |
| −5.1CU | 13,057 | UT-AGTGCAGTGGTGTGATCTCG |
| −5.1CL | 13,056 | LT-CCAGCCTGGGTGACACAGCA |
| −3.7F | 14,313 | GCCATCTCAGCTCAGAGACA |
| −3.7R | 14,593 | CACACGTGTAATCCCAGCAC |
| −3.7U | 14,433 | UT-CAAGTAGCTGGGACTACAGG |
| −3.7L | 14,432 | LT-GGAGGCTGAGGCAGGAGAAT |
| −1.8F | 16,266 | AGGGCAAAGGATGTGATACG |
| −1.8R | 16,376 | AACTGTGATTGTGCCACTGC |
| −1.8U | 16,326 | UT-CAAGGTCTTGCTCTATTGTC |
| −1.8L | 16,325 | LT-TCTCAAAAAAATGCATTAAA |
| −1.1F | 17,021 | GGAATTGCTGTTGGGACTTG |
| −1.1R | 17,303 | CAGGCAGACTGTGGATCAAAA |
| −1.1U | 17,190 | UT-TTGGCGAATTGAAGAAATCC |
| −1.1L | 17,189 | LT-TACTCTAGCTCTCCATGTGC |
| −0.1F | 17,965 | TGGCCAGCCTTTAATACATTG |
| −0.1R | 18,120 | ATTCTGCACAATCCCCAAAG |
| −0.1U | 18,043 | UT-TATAGCCTCCTTCCCCATCC |
| −0.1L | 18,042 | LT-GAAGACCAAAAAGATAAATC |
| +1.5F | 19,388 | ATGCCCAGCCTGAAGTAGC |
| +1.5R | 19,616 | GGATTAGTACGGATCAGCCAG |
| +1.5U | 19,540 | UT-TGTCAGTATGATTTTTACAT |
| +1.5L | 19,539 | LT-TCAATGTTATTGATGCTGCA |
- a F, forward primer; R, reverse primer.
- b UT, internal forward primer tail (5′GTCGACGGATCCCTGCAGGT); LT, internal reverse primer tail (5′ACCTGCAGGGATCCGTCGAC).
The PCR products specific to each region of interest were then used as templates to construct the corresponding competitors. For each of these regions internal primers were designed to construct competitors that shared the same sequence as the genomic targets except for an additional 20 nucleotides in the middle. The internal primers used for each region (Table I) had complementary 20 nucleotide tails, unrelated to the genomic DNA, added to their 5′ ends. The tail added to internal reverse primers was 5′ACCTGCAGGGATCCGTCGAC, and to internal forward primers 5′GTCGACGGATCCCTGCAGGT [Diviacco et al., 1992]. Half competitor molecules were amplified using the relevant genomic outer primer and tailed inner primer and the same conditions used for the genomic amplification. After purification from polyacrylamide gel slices by elution, the two competitor half molecules were co-amplified with the corresponding genomic outer primers as described in Diviacco et al. [1992]. Briefly this involved, denaturation at 94°C for 1 min, then slow reduction of temperature to 50°C over 10 min. The annealed products were then extended at 72°C for 5 min and amplified with 5 cycles of 94°C for 1 min, 37°C for 30 s, 72°C for 30 s, 5 cycles of 94°C for 1 min, 42°C for 30 s, 72°C for 30 s and 20 cycles of 94°C for 1 min, 55°C for 30 s, and 72°C for 30 s. Competitor DNA molecules were purified by elution and re-amplified (20 ng/25 μl reactions) to increase yield, using the outer primers and standard conditions for amplification of each genomic region described earlier. Multiple PCR reactions were combined, and competitor DNA isolated from 4% agarose gels using Bresa Clean, followed by re-precipitation with ethanol to reduce UV absorbing contaminants. After confirmatory sequencing of the PCR products, competitor concentrations were determined by spectrophotometry at 260 and 280 nm and checked by running in an agarose gel against markers of known concentration.
Quantitative PCR of Selected Regions in Nascent DNA
Fixed amounts of nascent DNA were co-amplified with known increasing amounts of competitor molecules using the corresponding shared outer primers for each region. Amplification reactions (25 μl) consisted of 1× PCR buffer, 0.5–1.0 μM primers, 200 μM dNTPs, 1.5 mM MgCl2, and 0.625 U of Taq polymerase (Biotech International). The cycling profiles for co-amplification involved an initial denaturation at 94°C for 1 min × 1 cycle; 92°C for 40 s, 60°C (for primer sets −5.1, −3.7, −1.8, −0.1, +1.5)/62°C (for primer set −1.1) for 40 s, 75°C for 1.5 min for a total of 30 cycles; with a final extension cycle of 75°C for 5 min. A negative control with no added DNA was included with each set of PCR reactions. The amount of nascent DNA needed to produce a detectable amount of PCR products for each primer set was determined empirically in preliminary ranging experiments without added competitor. For each preparation of nascent DNA tested, 1 μl of a 1/10 dilution was typically sufficient for amplification of nascent DNA from the X86T2 cells with the −1.8 primer set, however, for all other primer sets with X86T2 DNA and all primer sets with Y162/11C DNA at least 5 μl of purified nascent DNA was needed. Range finding experiments were also carried out to determine the appropriate amounts of competitor DNA required to compete effectively with the nascent DNA for each primer set. Experiments varying amount of template and cycle numbers were also carried out to ensure that the conditions used maintained amplification in the exponential phase. Reactions were electrophoresed on polyacrylamide gels, stained with ethidium bromide (0.5 μg/ml) and images captured. The amount of nascent and competitor DNA produced was determined using the NIH image analysis program and the ratio of competitor to nascent PCR products plotted against the known amount of competitor molecules added.
Sequence Analysis
Sequence data was compiled and aligned using the Multisequence Alignment and ‘Pile-up’ programs available from the Australian National Genomic Information Service (ANGIS). General sequence manipulations were performed using the MacVector program. The Findpatterns program in the Genetics Computer Group (GCG) sequence analysis software package (accessed through ANGIS) was used to identify matches to the various consensus sequences of interest. DNA flexibility was measured as described previously [Toledo et al., 2000] using the FlexStab program available through the Hebrew University of Jerusalem web site (http://leonardo.ls.huji.ac.il/departments/genesite/faculty/bkerem.htm). The entire 56,736 bp of published HPRT sequence [Edwards et al., 1990; GeneBank accession no. M26434] was analyzed for flexibility in overlapping windows of 100 bp with a shift increment of 1 bp. For each window, the DNA flexibility value at each bp is added along the window and averaged by the window length. The mean value plus 2.8 standard deviations (SD) for the entire HPRT sequence was then subtracted from the values for each window within the sequence 13,021–20,061 bp and the relative values plotted against the first nucleotide position of each window in the sequence.
RESULTS
Replication Origin Mapping
We have previously identified a MAR-3 with an associated Alu at its 3′ end in intron 3 of the human HPRT gene [Chong et al., 1995]. Since MARs are frequently found near replication origins we tested for replication origin activity around MAR-3 using a method based on quantification of newly replicated (nascent) DNA [Giacca et al., 1994], isolated by extrusion from replication bubbles by branch migration [Zannis-Hadjopoulos et al., 1981]. If nascent DNA of small size is selected, PCR products originating from sites closest to the origin will be more highly represented than those produced from further distant sites. The number of molecules of various regions in the isolated nascent DNA is quantified by competitive PCR.
Five regions, named according to their location relative to the MAR in intron 3, were initially chosen for nascent strand analysis. These were located approximately 5.1 kb (−5.1R), 1.8 kb (−1.8R), and −0.1 kb (−0.1R) upstream of MAR-3 and approximately 1.5 kb (+1.5R) and 5 kb (+5R) downstream of the 5′ end of MAR-3. Nascent DNA was extruded from genomic DNA isolated from X86T2 (inactive X) and Y162/11C (active X) human: hamster hybrid cell lines and nascent DNA of size approximately 0.6–1.2 kb purified to provide the nascent sample for amplification and quantification by competitive PCR. The relative amounts of target and competitor products were determined for each competitor amount added and the number of target DNA molecules of each region present in the nascent DNA read from the graph as equal to the amount of competitor required to be added to produce an equal amount of target and competitor product. The results indicated the presence of a replication origin approximately 1.8 kb upstream of MAR-3, but only in nascent DNA isolated from the inactive X-containing X86T2 cells (results not shown).
To map the replication origin more closely two additional primer sets were chosen at approximately 3.7 kb (−3.7R) and 1.1 kb (−1.1R) upstream of MAR-3 and competitors constructed for inclusion in analysis of a second nascent DNA preparation from each cell line. The results for the experiments performed on X86T2 nascent DNA are shown in Figure 1 and those for Y162/11C nascent DNA in Figure 2. The amount of each target region present per microliter of nascent DNA from each cell line was determined from this data and is summarized in Figure 3A. The results show that in the X86T2 (inactive X) DNA, the abundance of the region (−1.8R) located approximately 1.8 kb upstream from MAR-3 was 127 times higher than that of region −1.1R and 152 times higher than region −3.7R. Hence the 0.6–1.2 kb nascent DNA from X86T2 cells is highly enriched in the region centered at −1.8R, confirming the findings obtained on the previous nascent preparation. In the case of the Y162/11C cell line (active X) the abundance of nascent DNA was low (<30 molecules/μl nascent DNA, Fig. 3B) for each of the six different regions. Between cell lines, region −1.8R was 55 times more enriched in nascent DNA in the X86T2 cell line than in the Y162/11C cell line. Assuming bi-directional replication of similar efficiency from the origin we infer that the inactive X specific replication initiation point is most likely located within approximately 600 bp upstream or downstream of the −1.8R region. However since the −1.1R region was not enriched in the nascent DNA preparation we also infer that the replication origin is greater than 600 bp from the most 3′ end of −1.1R. This would place the replication origin between 15,685 and 16,703 bp, or, 1.4–2.4 kb upstream of MAR-3 in cells containing the human inactive X (X86T2).
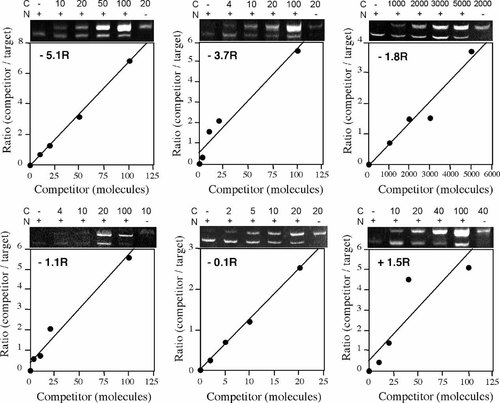
Competitive PCR of X86T2 cell line nascent DNA. A known increasing number of competitor DNA molecules (C) specific to each region examined (region indicated on graphs) was co-amplified with a fixed amount of nascent DNA (N) isolated from X86T2 cells. The ratios of amounts of competitor DNA to nascent DNA products after amplification (as visualized on the Ethidium bromide stained gels shown) were plotted against the corresponding number of competitor molecules used. The number of nascent DNA molecules containing the relevant region before amplification was determined by reading from the graphs the number of competitor molecules that would have produced a ratio of 1.
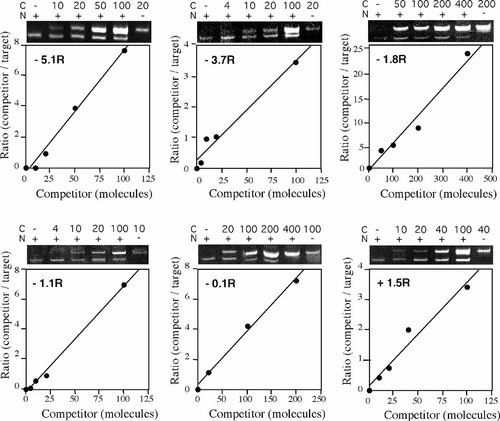
Competitive PCR of Y162/11C cell line nascent DNA. A known increasing number of competitor DNA molecules (C) specific to each region examined (region indicated on graphs) was co-amplified with a fixed amount of nascent DNA (N) isolated from Y162/11C cells. The ratios of amounts of competitor DNA to nascent DNA products after amplification (as visualized on the Ethidium bromide stained gels shown) were plotted against the corresponding number of competitor molecules used. The number of nascent DNA molecules containing the relevant region before amplification was determined by reading from the graphs the number of competitor molecules that would have produced a ratio of 1.
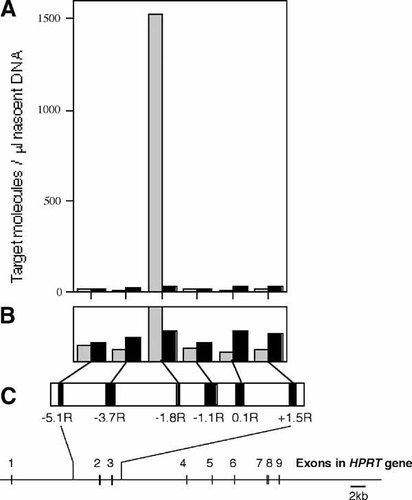
Mapping of the replication origin. A: The number of nascent target molecules containing the relevant region per microliter of nascent DNA from inactive X containing X86T2 cells (shaded boxes) and active X containing Y162/11C cells (solid black boxes) was determined from the data shown in Figures 1 and 2 and plotted for each region examined. B: Shows an expansion of the scale to visualize the lower amounts measured. C: Shows the position in the human HPRT gene of each region examined.
Sequence Analysis
Since an association between replication origins and enhanced DNA flexibility has been noted previously [Toledo et al., 2000] we searched the region from the −5.1R to the +1.5R PCR products for peaks of enhanced DNA flexibility using the computer program FlexStab. Regions of enhanced flexibility were defined as having a mean flexibility value greater than 2.8 SD above the mean of the entire HPRT sequence [Toledo et al., 2000]. The mean flexibility value and SD of the entire HPRT gene sequence (56,736 bp) was 10.72° and 0.74°, respectively. As shown in Figure 4, several closely spaced major peaks of flexibility were observed between bp 15,440–15,455, with bp 15,450 having the highest value, being 0.30 U greater than the mean plus 2.8 SD. The region of enhanced flexibility spreads from 15,440 to 15,555 since each analysis window extends 100 bp from the first base pair of each window. This zone of enhanced flexibility is located just upstream of the inactive X specific region most enriched in nascent DNA as measured in our experiments.
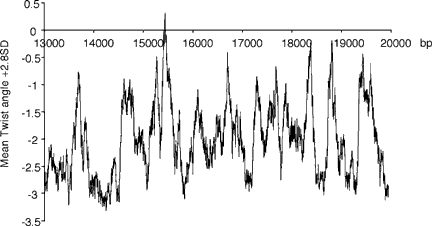
Flexibility analysis. Helix flexibility within the region was calculated with the FlexStab program. The horizontal axis indicates nucleotide position and the vertical axis shows mean twist angle per window plus 2.8 SD from the mean of the entire HPRT sequence.
Analysis of the region between −5.1R and +1.5R also revealed several matches (shown in Fig. 5) to sequence elements noted in association with previously characterized eukaryote replication origins [Dobbs et al., 1994]. These include one perfect match to the yeast autonomously replicating consensus sequence (ACS); sites with one mismatch to the Drosophila topoisomerase II consensus sequence (TopoII); multiple tracts of 12 or more pyrimidines and a 20/21 match to an initiation region consensus sequence (IRC). Two of the pyrimidine tracts located within the replication origin region are A:T rich elements comprising thymidines on one strand and adenines on the other. Figure 5 also shows the locations of the region of enhanced flexibility identified via the FlexStab program; MAR-3; the regions examined for the presence of newly replicated DNA; the region within which the replication origin is localized; and the location of Alu elements and CpG sites. While Pur motifs have been reported to be associated with replication origins [Bergermann and Johnson, 1992] the best Pur motif found here (15/16) was located relatively remote from the replication origin region (Fig. 5). Pur motifs with two mismatches were more common, but no clustering was evident (results not shown). Similarly additional ACS and topoisomerase II consensus sites with lower homology were present throughout the region examined, but since these consensus sequences are themselves fairly degenerate, only the highest matches obtained are shown in Figure 5. However an observation that may be significant, was the clustering of six overlapping 19/21 matches to the IRC localized within the 69 bp footprint sequence containing multiple TATTT motifs that we previously identified in MAR-3 [Chong et al., 1995].
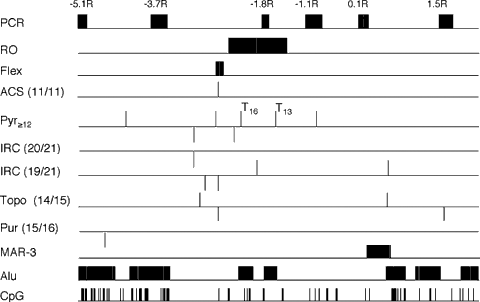
Sequence features of the inactive X specific replication origin region. The locations, within bp 13021 to 20061 of the HPRT gene, of sequences with homology to the yeast ACS; the IRC; the Drosophila topoisomerase II consensus and the Pur consensus are shown. The degree of match to the relevant consensus is indicated in parentheses. Also shown are the locations of stretches of at least 12 pyrimidines (Pyr≥12) including T-tracts (T16, T13). Sequence homologies on the plus strand are indicated by lines above the sequence line, and on the minus strand by lines below the sequence line. The positions of the regions examined for enrichment in nascent DNA (PCR); the inactive X specific replication origin region (RO); the region of enhanced flexibility (Flex); MAR-3; CpG sites and the locations of Alu elements are also indicated.
DISCUSSION
The results described here identify an inactive X specific replication origin located approximately 2 kb upstream of a MAR in intron 3 (MAR-3) in the human X linked HPRT gene. The replication origin region contains two A:T rich elements, which may facilitate DNA unwinding [DePamphilis, 1999] and a zone of enhanced flexibility with a perfect match to the yeast ACS sequence, 12/12 pyrimidine tract and a 14/15 match to the topoisomerase II consensus almost coincident with the major peak of enhanced flexibility (Fig. 5). Topoisomerase II is involved in controlling the topological structure of DNA [Earnshaw and Heck, 1985] and associates with the nuclear matrix [Christensen et al., 2002]. Pyrimidine tracts may serve as preferred sites for strand synthesis initiation [Benbow et al., 1992]. A 20/21 match to an initiation region consensus sequence (IRC), previously noted in association with replication initiation regions [Dobbs et al., 1994] is located just 5′ to the replication origin window and almost coincident with an additional 12/12 pyrimidine tract and 14/15 topoisomerase II match. Additional 19/21 IRC matches are located within the replication origin and enhanced flexibility regions. Six 19/21 IRC matches located outside the replication origin window, but in a previously identified footprint in MAR-3 [Chong et al., 1995] may also be significant.
In addition to the above possible sequence requirements, replication origins may also be dependent upon an accessible chromatin structure. Studies of rapidly cleaving Drosophila and Xenopus embryos show that embryonic replicons are much smaller than somatic replicons, indicating that DNA contains many potential replication initiation sites which may, in the presence of large amounts of initiation proteins and a permissive chromatin structure, be activated [Hyrien et al., 1995; Sasaki et al., 1999]. As development proceeds and chromatin becomes progressively more constrained the number of replication origins decrease and the replicons correspondingly increase in size, indicating an influence of chromatin structure on origin utilization. Thus whether a potential replication origin will be selected in different cell types or on different chromosomes may be determined by its accessibility, which for gene associated replication origins may vary with the expression status of the linked gene.
Consistent with this possibility, a shift in replication timing of the IgH region during mammalian B cell development is reportedly accompanied by activation of a novel replication origin [Zhou et al., 2002]. In contrast, Kitsberg et al. [1993] reported that the same replication origin, located approximately 2 kb upstream of the β-globin gene, was functional in both expressing (early replicating) and non-expressing (late replicating) cells treated with emetine, an inhibitor of lagging strand synthesis. However a subsequent study of non-expressing HeLa cells indicated that replication initiates at several locations within the β-globin domain, with the most frequent being approximately 20 kb upstream of the β-globin gene [Kamath and Leffak, 2001], suggesting that emetine may have biased initiation to one of the various possible origins. Thus it remains possible that the spectrum of replication origins used in late replicating non-expressing cells differs from that in early replicating β-globin expressing cells.
When considering replication origin usage a distinction may also need to be made between ‘non-expressing’ genes and ‘silenced’ genes, such as those on the inactive X. The naturally occurring, Hispanic thalassemia deletion, which removes 40 kb from 92 to 52 kb upstream of the β-globin locus, results in replication of the locus from a different origin, a shift in replication timing to late S phase, and silencing of globin gene expression with associated heterochromatinization [Forrester et al., 1990; Aladjem et al., 1995]. The Hispanic deletion removes four of five DNase I hypersensitive sites in the globin LCR, plus approximately 27 kb of additional upstream sequence. However a smaller, targeted deletion, which removes the same four DNase I hypersensitive sites caused a loss of β-globin gene expression without altering the replication origin or timing (early), or the (open) chromatin conformation [Cimbora et al., 2000]. Thus β-globin replication origin selection and timing correlated with chromatin conformation, but not with gene transcription per se.
Our identification of an inactive X specific replication origin is a clear demonstration of naturally occurring differential origin selection on different alleles. In a previous study, in which 15 possible replication origins were identified on the long arm of the human X chromosome, 1 was shown to be present in nascent DNA isolated up to 2 h after release from a synchronizing block of human active X containing hybrid cells, but not from human inactive X containing hybrids [Rivella et al., 1999]. Whilst this indicates that the replication origin was specific to an early replicating gene on the active X, it does not exclude the possibility that the same replication origin was used by the inactive X during late S phase, since samples were only taken for 2 h after release of the block. Interestingly, all of the 15 identified replication origins were localized in a 200 kb region close to constitutive genes actively expressed in the studied cell lines. No origins were found close to four other genes that were not expressed in the cells examined, suggesting this method is not suitable for isolation of replication origins on inactive loci.
Two recent studies have reported the identification of replication origins at the promoters of the human X linked HPRT [Cohen et al., 2002] and G6PD [Cohen et al., 2003] genes in male fibroblasts. These origins were reported to be used equivalently in hybrid hamster cells carrying either a human active X or a human inactive X, thus the authors concluded that replication was initiated from these promoter origins independent of activity status. However the enhancement of the HPRT promoter replication origin in nascent DNA from either the active or inactive X in the latter experiments was at most two-fold compared to regions approximately 6 kb distant [Cohen et al., 2003]. Similarly the enhancement of the G6PD promoter replication origin was only two- to four-fold compared to regions approximately 3 kb distant [Cohen et al., 2003]. In contrast our mapping results show greater than 100 fold enrichment of nascent strands in the region containing the intron 2 replication origin on the inactive X compared to regions at either side of the origin (1.9 and 0.7 kb distant), suggesting a much stronger origin of replication than those reported at the promoter of the human HPRT and G6PD genes.
Promoter CpG island associated replication origins have also been reported recently for the mouse X linked genes: Agg1, MeCP2, Mtml, Mtm1r, Xist, and Tsix [Gomez and Brockdorff, 2004]. However identification of the Agg1, MeCP2, Mtml, and Mtm1r replication origins was not performed by competitive PCR, but by visual inspection of products formed after 35 PCR cycles with primer sets from various regions. Since the degree of enrichment of the putative replication origins in nascent DNA was not measured, the status of these sites as replication origins remains undetermined. Competitive PCR was performed for the Xist site giving an enrichment of approximately five-fold in both XX and in XY cells, where it is not expressed. SNuPE analysis of interspecific XX mouse fibroblast lines showed that nascent strand preparations contained equivalent amounts of the Xist replication origin from both active and inactive X chromosomes. However competitive PCR of the Tsix proximal site indicated an enrichment in nascent DNA of approximately 10 fold in XX cells but no enrichment in XY cells. Due to the absence of polymorphisms it was not possible to determine the allelic contribution in nascent DNA of the Tsix replication origin, thus this origin may also be inactive X specific, as is the one we report here. None of the studies reporting activity independent CpG island promoter associated replication origins [Cohen et al., 2002, 2003; Gomez and Brockdorff, 2004] have demonstrated an enrichment as high as we found for the inactive X specific replication origin in intron 2 of the human HPRT gene. It is noteworthy that if the putative mouse Tsix replication origin proves to be inactive X specific its enrichment in nascent DNA would be approximately 20 fold, again higher than for the activity independent origins reported. The enrichment difference between these various types of replication origins could reflect replication origin diversity, with some origins stronger and specific to a particular chromatin state, while others having a more generic or different role.
Our demonstration of an inactive X specific replication origin leads to the question of why this origin is selected on the inactive allele and not the active allele. An association between hypomethylation and an open chromatin structure has been demonstrated numerous times in other contexts. This may be significant in this regulatory context with an emphasis on critical CpG sites, rather than CpG islands. As shown in Figure 5, the inactive X specific HPRT replication origin region is flanked by four CpG rich Alu repeats at the 5′ end and three (including the MAR-3 Alu) at the 3′ flank. Two other Alus are located closer to the replication origin. Most Alus are transcriptionally silent and highly methylated in normal somatic tissues [Rubin et al., 1994]. Alus have been implicated in establishing regional methylation states with methylation spreading from Alus into surrounding regions when these regions do not have bound regulatory factors to protect them from methylation [Graff et al., 1997]. Thus the cluster of Alus around the replication origin may maintain the region methylated and inaccessible on the active X where the replication origin is not used, but methylation spreading may be prevented on the inactive X by the binding of replication associated complexes or other factors to the region. Consistent with this possibility we have demonstrated by bisulphite genomic sequencing, significant inactive-X-specific hypomethylation of specific CpG sites in the Alu associated with MAR-3 [Gal'Lino and Piper, 2001]. Studies to extend these methylation studies to sites throughout the region are currently underway.
Replication, timing may also be determined by nuclear matrix interactions. Late replication timing of the hamster β-globin locus is established in G1 phase, coincident with its positioning in a peripheral sub-nuclear compartment compared to the early replicating DHFR locus, which is more internally localized [Li et al., 2001]. Late replicating origins in yeast are enriched at the nuclear periphery in G1 phase, whilst early origins are randomly located within the nucleus [Heun et al., 2001]. Sequences flanking late origins appeared to target these yeast origins to the periphery of the G1 phase nucleus where it is suggested a modified chromatin structure is established [Heun et al., 2001]. In this context differential methylation in the MAR-3 Alu associated sites could regulate matrix attachment or the binding of other proteins to the matrix attached DNA. Studies probing in vivo chromatin accessibility and matrix attachment at late and early replication origins throughout the cell cycle should prove instructive in distinguishing between these possibilities. Our demonstration here of a differentially utilized replication origin provides an excellent model with which to carry out such experiments.
Acknowledgements
We thank Scott Hansen for providing the hamster: human hybrid cell lines.




