Cartilage-derived morphogenetic proteins induce osteogenic gene expression in the C2C12 mesenchymal cell line
Abstract
Cartilage-derived morphogenetic protein-1, -2, and -3 (CDMP-1, -2, and -3) are members of the bone morphogenetic protein (BMP) family and have been shown to exhibit a variety of biological activities. In the present study, effects of these CDMPs on the temporal and spatial expression of genes in the pluripotent mesenchymal cell line C2C12 were examined. Cells cultured in the presence of CDMPs lost the characteristic elongated shape of myoblasts. At the molecular level, CDMP treatment did not change the mRNA expression of MyoD, aggrecan, Six1, and tendin. Scleraxis mRNA level was reduced by CDMP treatment. CDMP-1 and -3, but not CDMP-2, stimulated expression of osteogenic markers, such as alkaline phosphatase (AP), osteocalcin (OC), BSP, and type I collagen, in a dose- and time-dependent manner. With few exceptions, the three CDMPs changed, with different potencies, the expression profile of different members of the BMP family in a similar temporal pattern. Except at the late phase of treatment, CDMP treatment did not change the expression of ActR-IA, BMPR-IA, BMPR-IB, BMPR-II, and ALK-7 mRNAs. Based on the current data, the CDMPs appear to be able to stimulate the C2C12 cells to differentiate into the osteoblast pathway. © 2005 Wiley-Liss, Inc.
The cartilage-derived morphogenetic proteins (CDMPs) are members of a subgroup of the bone morphogenetic protein (BMP) family that belongs to the transforming growth factor-β (TGF-β) superfamily [Chang et al., 1994; Kingsley, 1994]. CDMP-1, -2, and -3 are also known as growth/differentiation factor-5, -6, and -7 (GDF-5, -6, and -7), respectively. The mature domains of CDMP-1 and -2 are equal in length, consisting of 120 amino acids, and CDMP-3 is shorter by 16 amino acids. They are about 80% identical in amino acid sequence with seven highly conserved cysteines. The mature domains of human and mouse proteins show 98%–99% amino acid identity. Though with different potencies, all three CDMPs have been shown to affect several skeletal processes, which include endochondral ossification [Tsumaki et al., 1999; Spiro et al., 2000], joint formation [Francis-West et al., 1999; Merino et al., 1999; Storm and Kingsley, 1999], and tendon/ligament repair [Polinkovsky et al., 1997; Wolfman et al., 1997; Aspenberg and Forslund, 1999, 2000; Clark et al., 2001; Lou et al., 2001; Mikic et al., 2001, 2002; Rickert et al., 2001]. CDMP-1 and -2 are capable of inducing cartilage and bone formation when implanted ectopically in intramuscular sites [Hotten et al., 1996; Erlacher et al., 1998; Spiro et al., 2000]. Cell culture studies also reveal that CDMPs stimulate osteogenesis of bone marrow stromal cells [Gruber et al., 2000, 2001]. Mutations in mouse GDF-5 and human CDMP-1 are responsible for the limb brachypodium phenotype and Hunter–Thompson syndrome, respectively [Thomas et al., 1996, 1997; Polinkovsky et al., 1997; Settle et al., 2003].
The pluripotent mesenchymal precursor cell line C2C12, a subclone of a mouse myoblastic cell line, has been widely used as a model to examine the early stages of osteoblast differentiation during bone formation in muscular tissues. Treatment of C2C12 cells with different members of the TGF-β superfamily produces distinct effects on the differentiation of these cells. For example, BMP-2 (300 ng/ml) inhibits myoblast differentiation of C2C12 cells and promotes osteoblastic cell differentiation [Katagiri et al., 1994]. C2C12 cells transfected with a human BMP-2 expressing adenoviral vector also display osteoblastic features [Okubo et al., 1999]. BMP-6 stimulates a dose-dependent increase in alkaline phosphatase (AP)-positive cells [Ebisawa et al., 1999]. OP-1 (BMP-7) inhibits myotube formation and induces osteoblastic cell formation [Yeh et al., 2002b]. In contrast, TGF-β, BMP-12, and BMP-13 inhibit myotube formation, but fail to induce the osteoblastic phenotype in C2C12 cells [Katagiri et al., 1994]. Both BMP-12 and -13 are much less potent than BMP-2 in inhibiting myotube formation.
These in vivo and in vitro observations led to our hypothesis that the CDMPs may affect the expression of a different set of genes or the same set of genes, but to different extents, than OP-1 or BMP-2 in C2C12 cells. Elucidation of these differences should provide a better understanding of the action of these BMPs in osteogenesis. In the present study, we focused on the effects of CDMP-1, -2, and -3 on cell morphology, the expression of characteristic osteoblastic, cartilage, and tendon/ligament biochemical markers and other members of the BMP family, as well as the BMP receptors in C2C12 cells.
MATERIALS AND METHODS
Materials
Hanks' balanced salt solution (HBSS), Dulbecco's modified eagle medium (DMEM), fetal bovine serum (FBS), penicillin/streptomycin, and trypsin-EDTA were purchased from Life Technologies (Grand Island, NY). Radioisotopes were purchased from ICN (Irvine, CA). TRI reagent was from Sigma (St. Louis, MO). Recombinant CDMP-1, -2, and -3 were provided by Stryker Biotech (Hopkinton, MA) and were dissolved in 47.5% ethanol/0.01% trifluoroacetic acid. All buffers were prepared with diethylpyrocarbonate-treated distilled water.
Cell Culture and Microscopic Examination
C2C12 cells (American Type Culture Collection, Rockville, MD) were cultured in DMEM containing 10% FBS and penicillin/streptomycin in a humidified 5% CO2 atmosphere at 37°C. Media were replenished every 3 days. Changes in cell morphology were monitored after 3, 7, 10, and 14 days of treatment using an Olympus CK2 inverted microscope equipped with a CCD camera.
Northern Blot Analysis
Northern analyses were conducted as previously described [Yeh et al., 1997]. Briefly, total RNAs (20 μg) were denatured, fractionated on 2.2M formaldehyde/1% GTG agarose gels, transferred onto a “Nytran Plus” membrane using a Turboblot apparatus (Schleicher & Schuell, Inc., Keene, NH), and covalently linked to the membrane using a UV Crosslinker (Stratagene, La Jolla, CA). The membranes were incubated at 42°C overnight with the radioactive cDNA probes. The radioactive signals were detected using the PhosphorImager and quantified using the ImageQuant software (Molecular Dynamics, Sunnyvale, CA). Before probing with another DNA probe, the blot was washed with Ambion's Strip-EZ Degradation and Reconstitution buffers following manufacturer's recommendation. The blots were finally probed with an 18S rRNA oligonucleotide probe to allow correction for loading variations.
The 440-bp cDNA probe for MyoD was isolated from pT7T3D-Pac, obtained from ATCC (IMAGE Clone ID 1064620) following PstI digestion. cDNA probes for osteocalcin (OC), rat bone sialoprotein (BSP), and type I collagen (TIC) were obtained as described previously [Yeh et al., 2000]. The Six1 and tendin probes were obtained by restriction endonuclease digestion of the DNA clones from ATCC (IMAGE Clone ID 1152512 and 2811628). The scleraxis and Runx2/Cbfa1 probes were generated by PCR as described previously [Tsai et al., 2003]. Probes were labeled with [32P]α-dATP using the Strip-EZ DNA labeling kit (Ambion, Austin, TX).
Alkaline Phosphatase (AP) Activity Assay
Cells were grown in 48-well plates in DMEM containing 5% FBS in the absence or presence of various concentrations of the growth factors. At the end of the indicated culture period, total cellular AP activity was measured using a commercial assay kit (Sigma Chemical Co., St. Louis, MO) as described previously [Yeh et al., 1997]. Protein was measured according to the method of Bradford [1976], using BSA as a standard. AP activity was expressed as nanomoles of p-nitrophenol liberated per microgram of total cellular protein.
Alcian Blue Staining
The quantity of sulfated glycosaminoglycan, representative of cartilage matrix, was measured by the amount of extractable dye from stained cultures. Cell cultures were stained with Alcian Blue using the modified procedure as described [Yeh et al., 2002a].
RNase Protection Assay (RPA)
The antisense RNA probe for aggrecan was prepared and labeled as described previously [Yeh et al., 2002a]. The aggrecan mRNA level was determined using the RPAII kit from Ambion. The RiboQuant RPA kits with the mBMP-1, the mGDF-1, and the mBMPR multi-probe template sets were purchased from BD PharMingen (San Diego, CA) and used according to the manufacturer's instruction. The mBMP-1 kit allows detection of mRNAs for BMP-1, -2, -3, -4, -5, -6, -7, -8A, and -8B with the protected fragment of 148, 160, 181, 226, 253, 283, 316, 353, and 133 nucleotides in length, respectively. The mGDF-1 kit allows detection of mRNAs for GDF-1, -3, -5, -6, -8, and -9 with the protected fragments of 148, 160, 181, 226, 283, and 316 nucleotides in length, respectively. The mBMPR kit allows detection of ALK-1, ALK-2 (ActR-I), ALK-3 (BMPR-IA), ALK-4, ALK-5, ALK-6 (BMPR-IB), ALK-7, AVR-2 (ActR-II), AVR2B (ActR-IIB), and MIS2R with the protected fragments of 430, 388, 349, 313, 280, 250, 223, 199, 178, and 161 nucleotides in length, respectively. All three kits also detect mRNA for ribosomal protein L32 and GAPDH as controls, allowing for correcting technique or sampling errors. The protected RNA fragments were separated on 8M urea/5% polyacrylamide gels. After electrophoresis, gels were fixed and dried. Radioactive signals were detected using the PhosphorImager and quantified with the ImageQuant software (Molecular Dynamics).
Statistical Analysis
Data are presented as the mean ± SEM. Statistical differences between means were determined by one-way ANOVA, followed by post-hoc Least Significant Difference Multiple Comparisons in the SIMSTAT3 software package (Normand Peladeau, Provalis Research, Montreal, Canada).
RESULTS
CDMPs Induced Changes in Cell Morphology
Effects of CDMP treatment on the morphology of C2C12 cells were examined as a function of protein concentration and duration of treatment. In control cultures, the C2C12 cells were elongated, resembling myoblastic cells. At low concentrations (<100 ng/ml), CDMPs did not induce noticeable morphological changes. At higher concentrations (>100 ng/ml), CDMP-1, -2, and -3, induced a change in cell morphology such that the cells no longer exhibited the characteristic elongated shape of myoblasts after 14 days. These cells became orthogonal like osteoblasts. Changes in cell morphology were time- and protein-concentration dependent (Fig. 1A–C).

Morphological changes in C2C12 cells cultured in the presence of (A) CDMP-1, (B) CDMP-2, and (C) CDMP-3. C2C12 cells were cultured in the absence or presence of 50, 100, or 200 ng/ml of CDMPs. Media were changed every 3 days. Cell morphology was monitored with a phase contrast microscope, and the images were captured on day 3, 7, and 14 with a CCD camera. Representative images (phase contrast with 100× magnification) are presented.
CDMPs Changed the Expression of Selected Transcription Regulatory Factors
To assess the molecular phenotype of the treated cell, effects of the various CDMPs on the mRNA expression of selected cell-type specific regulatory proteins were examined after 7 and 14 days of treatment. Representative Northern blots are shown in Figure 2A, and the quantitative data are shown in Figures 2B and 3. MyoD mRNA was detected in control cultures. After 7 and 14 days of treatment with any one of the CDMPs, MyoD mRNA expression did not change significantly, compared to the same day control. Six1 mRNA was expressed endogenously and its level was not significantly affected (compared to the same day control) by the three CDMPs examined (Fig. 2A,B). Scleraxis (scx) mRNA was detected in significant amounts in control C2C12 cells and its level was down-regulated to about 30% of the same day control value by CDMP-1, -2, or -3 by day 7 and remained low through day 14 (Fig. 2A,B). A significant level of endogenous tendin mRNA was observed and its level increased moderately during the culturing period, but its level was not changed by CDMPs (Fig. 2A,B). The mRNA expression of Runx2/Cbfa1 was low in control cells and was not altered by the CDMPs tested (Fig. 2A).
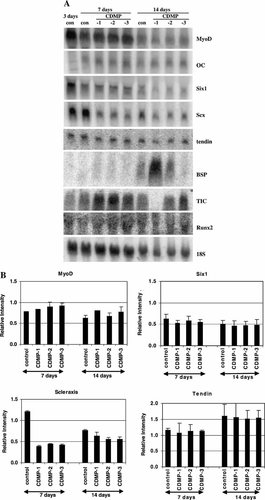
A: Northern blot analysis of mRNA expression encoding MyoD, osteocalcin, six1, scleraxis, tendin, BSP, type I collagen, and Runx2/cbfa1 in long-term cultures of C2C12 cells in the presence of CDMP-1, -2, and -3. Cells were treated with 200 ng/ml of CDMP-1, -2, -3, and vehicle for 7 and 14 days. mRNA expressions were measured by Northern blot analysis using 32P-labeled cDNA probes. The blots were also hybridized with the oligonucleotide probe for 18S rRNA to verify equal loading of mRNA. Representative PhosphorImages are presented. B: Quantitative analysis of MyoD, Six1, scx, and tendin mRNA levels. The intensity of the hybridized RNA species on Northern blots, as shown in (A), was analyzed by the ImageQuant software. The mRNA level was normalized to the 18S rRNA level. The normalized mRNA level was then compared to that in the day 3 control (as 1). Values represent mean ± SEM from three independent determinations.
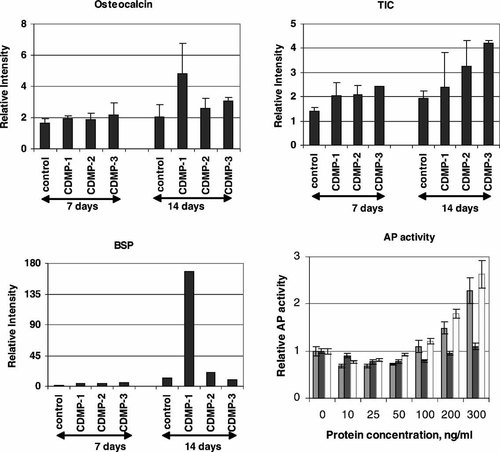
Quantitative analysis of osteocalcin (OC), bone sialoprotein (BSP), type I collagen (TIC), and AP activity in C2C12 cells treated with CDMP-1, -2, and -3. For the measurement of OC, BSP, and TIC mRNA levels, C2C12 cells were cultured in the absence or presence of 200 ng/ml of CDMP-1, -2, and -3. The intensity of the positive band on the Northern blots shown in Figure 2A was analyzed as described in Figure 2. Values were normalized to the solvent control (as 1) on day 3 and represent the mean ± SEM of three different measurements. For AP activity measurement, C2C12 cells were cultured in the absence or presence of 10, 25, 50, 100, 200, and 300 ng/ml of CDMP-1, -2, and -3 for 48 h. Total AP activity was measured as described in “Materials and Methods.” Light gray bars, CDMP-1; dark solid bars, CDMP-2; and open bars, CDMP-3. Values were normalized to the solvent control (as 1) and represent the mean ± SEM of three different measurements.
CDMPs Stimulated Expression of Osteogenic Markers
To determine whether CDMP-1, -2, and -3 promoted osteogenic differentiation of C2C12 cells, the expression of selected biochemical markers characteristic of osteoblastic cells was examined. CDMPs stimulated AP activity, an early-stage biochemical marker of osteoblastic differentiation. Both CDMP-1 and -3 stimulated AP activity dose-dependently, with a maximum 2.5-fold stimulation at 300 ng/ml (Fig. 3). In the concentration range tested, CDMP-2 did not stimulate AP activity in these cells.
The expression of OC, a late-stage osteoblastic differentiation marker, was studied. None of the CDMPs stimulated OC mRNA expression after 7 days of treatment. However, after 14 days, CDMP-1 and -3 stimulated OC mRNA by about 3- and 1.5-fold, respectively. CDMP-2 slightly elevated OC mRNA expression (Figs. 2A and 3). BSP mRNA level was elevated by 4- to 5-fold (compared to the same day control) after 7 days in cultures treated with any one of the CDMPs. By 14 days, the BSP mRNA level was elevated by 13-fold in CDMP-1-treated cultures but was not further affected significantly in CDMP-2- or -3-treated cultures (Figs. 2A and 3). Type I collagen mRNA levels were elevated significantly in a time-dependent manner in cells treated with CDMP-1, -2, or -3 (Fig. 3).
CDMPs did not Stimulate Aggrecan Expression
Since the CDMPs were first isolated as cartilage-derived morphogenetic proteins, their ability to promote synthesis of sulfated glycosaminoglycan, representative of cartilage matrix, in C2C12 cells was examined. Figure 4 shows the quantitative data of Alcian Blue staining of cultures treated with different concentrations of CDMP-1, -2, and -3 for different durations. The staining intensity increased in control cultures as a function of time. At the protein concentration range tested, none of the CDMPs tested stimulated to any significant extent the Alcian Blue staining during the entire 14 days of culture. Aggrecan mRNA was not detectable in C2C12 cells and its level was not stimulated to any significant extent even at the low CDMP concentrations at which the cells exhibited the elongated morphology that might be more characteristic of cartilage and tendon/ligament (data not shown).
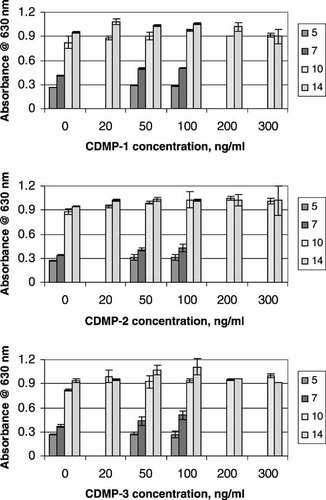
Quantitation of Alcian Blue stain of C2C12 cells after long-term culture. Cells were treated with different concentrations of CDMP-1, -2, or -3 for 5, 7, 10, and 14 days. Media were changed every 3 days. Cultures were stained with Alcian Blue and the amount of stain was measured at 630 nm after extraction of the stained cells with 1% SDS. Values represent mean ± SEM of two to three independent measurements.
CDMPs Altered BMP-1, -4, -5, -6, and -8A mRNA Expression
BMP mRNA expression in control cultures
Figure 5A is a representative PhosphorImage showing the protected fragments for five out of the nine possible BMPs detectable by the commercial kit in control and treated C2C12 cells. BMP-1, -4, -5, -6, and -8A mRNAs were observed. The mRNA expression level for each BMP was quantified and normalized to that of the ribosomal protein L32. The relative levels of mRNA for the different BMPs as a function of time are shown in Figure 5B. Based on the intensity of the protected band on the gel, the relative expression level of the detectable BMPs can be arranged in the following order: BMP-1 > BMP-4 > BMP-5 > BMP-6 > BMP-8A. During culturing, the BMP-1 mRNA expression decreased gradually by about 25% at day 14. The BMP-4 level did not change. The BMP-5 mRNA level also increased appreciably after 7 days and persisted at an elevated level after 14 days. The mRNA level of BMP-6 and -8 increased steadily as a function of time, reaching a 6- and 11-fold elevation, respectively, after 14 days.
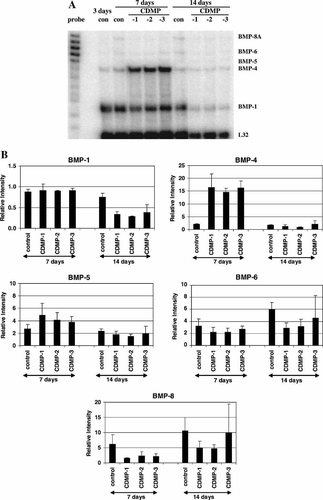
A: RNase protection analysis of the effect of CDMP-1, -2, or -3 on BMP mRNA expression in long-term cultures of C2C12 cells. Cultures were treated with vehicle, and 200 ng/ml of CDMP-1, -2, and -3 for 7 and 14 days. Twenty microgram of total RNA was used for the measurement of BMP mRNA expression by the RNase protection assay. The protected RNA fragments were fractionated on 5% polyacrylamide gels containing 8M urea and detected by PhosphorImaging. Positions of the labeled probes for the different BMPs and the ribosomal protein L32 gene as control are indicated on the left of the image. The protected fragments are indicated on the right. B: Quantitative analysis of the BMP mRNA levels in C2C12 cells. The intensity of the protected fragments as shown in (A) was analyzed, quantified using the ImageQuant Software, and normalized to the L32 expression level. The normalized mRNA levels were compared to that of the day 3 control (as 1). Values represent mean ± SEM from three different determinations.
BMP mRNA expression in treated cultures
Effects of continuous treatment with CDMPs on the mRNA expression of several BMPs as a function of culture time were also examined by RPA. These treatments were evaluated via comparison to their respective same-day controls. In cultures treated with CDMP-1, -2, or -3, BMP-1 mRNA expression was not affected after 7 days, but decreased significantly by about 50%–60% after 14 days (Fig. 5B). The BMP-4 mRNA expression was significantly stimulated by 8- to 10-fold in cultures treated with CDMP-1, -2, or -3 after 7 days, but returned to that of the same day control after 14 days. After 7 days, BMP-5 mRNA expression level was stimulated by the CDMPs by 2-fold, and returned to the control level after 14 days. All three CDMPs suppressed BMP-6 mRNA expression by 30%–50% after 7 and 14 days of treatment. Compared to the other BMPs, the BMP-8A mRNA expression was most extensively down-regulated (by 60%–70%) by CDMP-1 and -2, or -3 after 7 days. However, after 14 days, the BMP-8A mRNA level in CDMP-1 and -2 treated cultures remained suppressed; the level in the CDMP-3 treated cultures returned to that of the same day control.
CDMPs Affected GDF mRNA Expression
GDF mRNA expression in control cultures
Figure 6A is a representative PhosphorImage showing the protected fragments for five out of the six possible GDFs detectable by the kit in control and treated C2C12 cells. GDF-1, -5 (CDMP-1), -6 (CDMP-2), -8, and -9 mRNAs were detected, and the mRNA expression level for each GDF was quantified and normalized to the mRNA level of the ribosomal protein L32. The relative levels of mRNA for the different GDFs as a function of time are shown in Figure 6B. Based on the intensity of the protected band on the gel, the relative expression level of the detectable GDFs can be arranged in the following order: GDF-1 ≙ GDF-5 (CDMP-1) ≙ GDF-6 (CDMP-2)> GDF-8 ≙ GDF-9. The mRNA expression of GDF-1, -5 (CDMP-1), -6 (CDMP-2), and -9 increased gradually, reaching a maximum of about 3-, 6-, 4-, and 2-fold, respectively after 14 days. The GDF-8 mRNA expressions was very low initially, and remained relatively unchanged throughout the culture period examined.
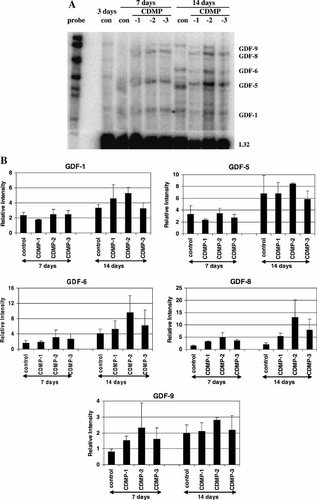
A: RNase protection analysis of the effect of CDMP-1, -2, or -3 on GDF mRNA expressions in long-term cultures of C2C12 cells. Experimental conditions are similar to those described in Figure 5. B: Quantitative analysis of the GDF mRNA levels in C2C12 cells. The intensity of the protected fragments as shown in (A) was analyzed and quantified as described in Figure 5B. Values represent mean ± SEM from three different determinations.
GDF mRNA expression in treated cultures
Effects of continuous CDMP treatment on the mRNA expression of several GDFs as a function of time in culture were also examined by RPA. Figure 6A,B shows the consequences of treatment with CDMPs on the expression of several GDF members. After 7 days, CDMP-1 did not significantly change GDF-1, -5 (CDMP-1), and -6 (CDMP-2) mRNA levels, but stimulated GDF-8 and -9 by about 2-fold above the same day control. After 14 days, CDMP-1 stimulated GDF-8 by about 3-fold without exerting a significant effect on the other GDFs. After 7 days, CDMP-2 did not change GDF-1 or -5 (CDMP-1) expression, but stimulated GDF-6 (CDMP-2), -8, and -9 by 2-, 3-, and 2-fold, respectively. After 14 days, CDMP-2 increased GDF-1, -5 (CDMP-1), -6, -8, and -9 mRNA expressions by 2-, 2-, 6-, 11-, and 1.5-fold, respectively. After 7 days, CDMP-3 did not affect mRNA expression of GDF-1, -5 (CDMP-1), and -6 (CDMP-2), but stimulated GDF-8 and -9 expression by about 2-fold. After 14 days, CDMP-3 stimulated significantly GDF-6 (CDMP-2) and -8 expression by about 2- and 6-fold, respectively, without any significant effect on GDF-1, -5 (CDMP-1), and -9.
CDMPs Altered BMP Receptor mRNA Expression
The expression of BMP receptors was also examined by RPA. Figure 7A shows a representative PhosphorImage and the quantitative data are shown in Figure 7B. In control cultures, the mRNAs for 5 out of the 10 possible receptors detectable by the kit were observed. ActR-I, BMPR-IA, BMPR-IB, BMPR-II, and ALK-7 mRNAs were detected, and the relative expression level (based on the intensity of the protected band on the gel) of the detectable BMPRs can be arranged in the following order: BMPR-IA > ActR-I > ALK-7 > BMPR-IB > BMPR-II. Their levels remained unchanged throughout the 14 days of culture. In CDMP-1 treated cultures, none of the three receptor mRNA expression was changed after 7 days, except BMPR-IA, which was down-regulated by about 25%. However, after 14 days, all detected receptor mRNA levels were reduced to about 40%–60% of the same day control. CDMP-2 did not alter the receptor mRNA expression after 7 days, but significantly down-regulated their expression after 14 days. After 7 days, CDMP-3 did not significantly change BMPR expression, but again down-regulated the mRNA levels of BMPR-IA, BMPR-IB, BMPR-II, and ALK-7 receptors after 14 days. However, CDMP-3 did not change the ActR-IA mRNA levels during the culturing period.
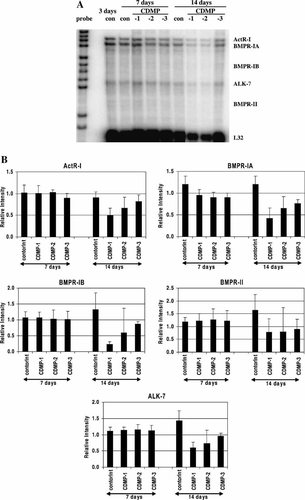
A: RNase protection assay analysis of BMP receptor mRNA expression in long-term cultures of C2C12 cells grown in the presence of CDMP-1, -2, and -3. C2C12 cells were treated for 7 and 14 days with 200 ng/ml of CDMP-1, -2, and -3. Total RNA was isolated and analyzed as described in Figure 5A. B: Quantitative analysis of the BMP receptor mRNA level. The intensity of the protected fragments as shown in (A) was analyzed and quantified as in Figure 5B. Values represent mean ± SEM from three different determinations.
DISCUSSION
Previously, BMP-2 and OP-1 (BMP-7) have been shown to inhibit myotube formation in a dose-dependent manner in the myoblastic C2C12 cell line [Katagiri et al., 1994; Yeh et al., 2002b]. CDMP-2 (GDF-6) and -3 (GDF-7) partially inhibits myotube formation [Inada et al., 1996]. The present results extend these observations by showing that CDMP-1 (GDF-5) also alters the morphology of the C2C12 cells.
At the molecular level, the present study reveals that none of the three CDMPs significantly suppresses mRNA expression of MyoD, a transcription regulator of myogenic differentiation. In contrast, BMP-2 [Katagiri et al., 1994] and OP-1 [Yeh et al., 2002b] significantly inhibited MyoD mRNA expression. Also, CDMP-2 (GDF-6) and -3 (GDF-7) treatments significantly suppressed the expression of myogenin, another transcription regulatory of myogenic differentiation in L6 myoblastic cells [Inada et al., 1996]. Although the reason for the discrepancy is not clear, the disparity may be partly due to the fact that different cell lines were used for these studies.
Previous in vivo data showed that CDMP-1 (GDF-5) induces formation of tendon/ligament-like tissues when implanted intramuscularly in rats [Aspenberg and Forslund, 1999]. However, our study shows that a higher concentration (200 ng/ml) of CDMP-1 (GDF-5) decreases the expression of scleraxis (scx) in C2C12 cells. At this protein concentration, cells changed to the cuboidal shape. Scx is a basic helix-loop-helix transcription factor that is reportedly a highly specific marker for tendons and ligaments [Salingcarnboriboon et al., 2003], and is found in progenitor cell populations for connective tissues (including tendons and ligaments) that mediate the attachment of muscle to bone in mouse and chick [Schweitzer et al., 2001]. Several additional studies also revealed that scx expression is suppressed by other BMPs. For example, the scx mRNA levels in BMP-2-treated C2C12 cultures [Liu et al., 1997] and in limb buds treated with recombinant BMP-2, -4, or -7 [Schweitzer et al., 2001] were down-regulated. In addition, aggrecan expression was not stimulated by low CDMP concentrations where the morphology of the cells is elongated and more resemble that of tendon/ligament cells. Even at higher CDMP concentrations where the cell morphology has changed, the CDMPs did not stimulate aggrecan expression.
Our findings indicate that the mouse pre-myogenic cell line C2C12 can be induced to become osteogenic when treated with CDMP-1, -2, and -3. The induction is dose-dependent. The osteogenic markers are stimulated in the expected temporal manner, such that CDMP-1, -2, and -3 induce AP activity, an early biochemical marker of osteoblastic cell differentiation, and OC, BSP, and type I collagen, late markers of osteoblastic cell differentiation. These findings indicate that the CDMPs examined are osteogenic and are consistent with the in vitro data that CDMP-1 and -2 stimulate osteogenic differentiation of bone marrow cells [Gruber et al., 2000]. However, the CDMPs appear to be less potent as osteogenic proteins compared to BMP-2, -6, and -7. Incubation of C2C12 cells with 300 ng/ml of BMP-2 for 6 days inhibited myotube formation and induced osteoblastic cell formation, with >90% of the cells stained positive for AP [Katagiri et al., 1994]. OP-1 is also more potent than the CDMPs in the up-regulation of AP activity as well as OC mRNA expression in C2C12 cells [Yeh et al., 2002b]. Our finding extends previous results that C2C12 cells can be induced to become osteogenic by BMP-2 [Katagiri et al., 1994], BMP-6 [Ebisawa et al., 1999], and OP-1 [Yeh et al., 2002b]. Relevant to this issue is the recent finding of Shea et al. [2003] that BMP-2 and -7 induce chondrogenesis and osteogenesis in the murine mesenchymal cell line C3H10T1/2. Our data suggest that the CDMPs tested do not induce chondrogenesis. Whether the difference lies in the different BMPs and/or the cell lines used in these studies remains to be answered experimentally.
Our study shows that even though CDMPs induced expression of several osteoblastic cell markers, they did not stimulate Runx2/Cbfa1 expression. In contrast, previous studies revealed that several BMPs could induce Runx2/Cbfa1 expression. For example, OP-1 elevates the mRNA expression of Runx2/Cbfa1, AP, and OC in C2C12 cells [Yeh et al., 2002b]. BMP-2 and TGF-β [Lee et al., 1999, 2000; Banerjee et al., 2001; Yeh et al., 2002b], as well as the BMP4/7 heterodimer (100 ng/ml) also induced Runx2/Cbfa1 mRNA expression [Tsuji et al., 1998].
Our finding that the CDMPs exert a differential effect on the mRNA expression of the different BMP family members is interesting. The current study shows that CDMP-1, -2, and -3 decreased BMP-1 significantly after 14 days, but previous studies reported that OP-1 elevated significantly the BMP-1 mRNA expression during the entire culture period [Yeh et al., 2002b].
Within the BMP-2/4 subgroup of BMPs, the BMP-2 mRNA level in C2C12 cells was below detection and none of the CDMPs tested changed its expression. Nevertheless, CDMPs considerably stimulated BMP-4 expression at the early stage of culture. Our observation is consistent with previous studies in transgenic mice that BMP-4 and GDF-5 (CDMP-1) may interact [Tsumaki et al., 1999], although the mechanism of interaction was not known. The recent finding that CDMPs enhanced the OP-1 action in C2C12 cells provides a mechanism for cross-talks between different BMPs, at least in this system [Yeh et al., 2004]. Specifically, the study reveals that the Smad5 level was elevated higher in cultures treated with the combination of OP-1 and CDMPs than in cultures treated with the individual proteins. The BMP-4 mRNA level was also elevated in C2C12 cells by OP-1 but was suppressed in FRC cells [Yeh et al., 2000]. BMP-4 was also inhibited initially but was stimulated during the mineralization phase in osteoblastic cells by BMP-2 [Chen et al., 1997].
Of the subgroup consisting of BMP-5, -6, and -7, only BMP-5 and -6 mRNAs were detected in control cultures. BMP-5 levels were stimulated by CDMPs but BMP-6 levels were suppressed. By comparison, BMP-5 mRNA expression was stimulated by OP-1, whereas BMP-6 mRNA expression was not affected in C2C12 cells [Yeh et al., 2002b]. mRNA expressions for both BMP-5 and -6 in FRC cells were down-regulated by OP-1, and BMP-6 mRNA expression in U2OS and SaOS-2 human osteosarcoma cell lines was increased by OP-1 [Honda et al., 1997].
Except BMP-4, the expression level of BMP-8A was changed most dramatically by all three CDMPs in a dose- and time-dependent manner. Conversely, BMP-8A mRNA levels were not significantly changed by OP-1 [Yeh et al., 2002b]. Taken together, the present results reveal a complex temporal interplay of the different members of the BMP family in osteogenesis. This supposition is further supported by the recent observation that the three CDMPs enhance OP-1 action in stimulating osteogenic cell differentiation in C2C12 [Yeh et al., 2004].
Various GDFs have been shown to participate in different skeletal tissue cell differentiation. For example, in vivo ectopic implantation studies with GDF-5 (CDMP-1) and -6 (CDMP-2) show that both proteins induced de novo cartilage and bone formation [Erlacher et al., 1998]. At OP-1 concentrations sufficient to induce osteoblastic phenotype in C2C12 cells, the GDF-6 (CDMP-2) and -8 mRNA levels were elevated significantly [Yeh et al., 2002b]. The present study shows that exogenous CDMP-1 did not exert any effects on its endogenous expression level. Exogenous CDMP-2, on the other hand, stimulated its own expression significantly at the late phase of treatment. Our study also reveals that the most dramatic change among the GDFs involves stimulation of GDF-8 expression, an observation that corroborates well with the published finding that GDF-8 (myostatin) is a negative regulator of skeletal muscle growth [Hamrick et al., 2000]. The result is consistent with the other observations reported here that the CDMPs suppress myogenic differentiation and promote osteogenic differentiation of C2C12 cells. Whether GDF-8 plays a direct role in osteogenesis must await further study.
Published studies suggest that BMPs may regulate their actions by affecting expression of the individual BMP receptors. Information about the receptors used by the GDFs is limited; Nishitoh et al. [1996] reported that GDF-5 (CDMP-1) binds to BMPR-IB, ActR-II and BMPR-II, but not BMPR-IA. Interestingly, the current study detected a high level of BMPR-IA mRNA and a very low level of BMPR-IB mRNA. All three CDMPs tested down-regulated the mRNA expression of these type I receptors after 14 days of treatment. Whether these observed changes in the steady-state mRNA levels are translated to the level of surface receptor proteins remains to be elucidated. The mRNA coding for ActR-II was below detection. The BMPR-II mRNA level was low but detectable and was not affected by CDMP treatment. By comparison, previous studies showed that OP-1 elevated ActR-IA mRNA level without affecting BMPR-IA, BMPR-IB, and BMPR-II mRNA expression in these cells [Yeh et al., 2002b]. BMP-2, which also is capable of converting C2C12 cells into the osteoblastic cells, mediates its effect through BMPR-IA [Namiki et al., 1997]. Taken together, it appears that different members of the BMP family exert their effects by employing different combinations of BMP receptors.
In conclusion, we demonstrate that CDMP-1, -2, and -3 are capable of inducing osteoblastic differentiation of the pluripotent mesenchymal cell C2C12 with different potencies. These proteins appear to have little effect on the expression of several characteristic markers for cartilage and ligament/tendons in these cells. Whether similar effects are observed on the expression of these markers in C2C12 cells under different culture conditions or in other cell lines will have to await for further experimental evidence. We also show a complex temporal pattern and interplay of different BMPs and GDFs in their actions in the induction of osteoblastic cell differentiation. In particular, the dramatic stimulation of BMP-4 expression in the early phase of osteogenesis induced by all CDMPs tested is intriguing. The finding provides a rationale for future study on the functional relationship between the CDMPs and other BMPs in osteoblastic cell differentiation.
Acknowledgements
The authors acknowledge the support by Stryker Biotech and thank Abra S. Yeh for her help in the preparation of this manuscript.




