Identification of a novel protein from glial cells based on its ability to interact with NF-κB subunitsr
Abstract
Nuclear factor κB (NF-κB) represents a family of inducible DNA-binding transcription factors whose activity is critical for expression of the HIV-1 genome in a broad range of cells. In addition to its interaction with the κB DNA sequence, the association of NF-κB subunits with other cellular proteins plays an important role in stimulation of HIV-1 gene transcription in astrocytic cells. Here, we utilized a yeast two-hybrid system to screen a cDNA library from a human astrocytic cell line and were able to isolate a partial cDNA belonging to a gene with an open reading frame of 1,871 amino acid residues which binds to both the p50 and p65 subunits of NF-κB. This gene, named NF-κB-binding protein (NFBP) is located on chromosome 10q24.2-25.1 and hybridized to a single transcript of nearly 6 kb in size. It is localized to the nucleus, specifically the nucleolus of cells. Extensive computer analysis was performed with the sequence of the full length NFBP and significant homology was found between NFBP, and yeast and mouse proteins. A discussion of the potential roles of NFBP in normal and viral infected cells is included. © 2003 Wiley-Liss, Inc.
The nuclear factor κB (NF-κB) family of DNA-binding proteins are a group of well-studied transcription factors that are involved in the regulation of several cellular and viral genes. The five well-characterized members of this family include p105-p50 (NF-κB1), p100-p52 (NF-κB2), c-rel p65, (Rel A) and Rel B [Ghosh and Karin, 2002]. All of these members share a 300 amino acid region that facilitates their DNA-binding as well as their interaction with each other forming homo- and hetero-dimers. In addition to interfamily communication, NF-κB members have been shown to interact with other cellular proteins including the IκB family [Phelps et al., 2000], the TATA-binding protein (TBP) [Schmitz et al., 1995], C/EBP [Stein et al., 1993], glucocorticoid receptor [Ray and Prefontaine, 1994], and YB-1 [Raj et al., 1996]. The ability of these NF-κB subunits to interact with various cellular proteins expands the scope of NF-κB-mediated gene regulation. The HIV-1 regulatory sequence spanning the long terminal repeat (LTR) contains two copies of κB motifs spanning nucleotides between −117 to −80, that is the ideal target for NF-κB interaction. Previous studies by several laboratories have established that the interaction of NF-κB subunits, such as p50 and p65, with the HIV-1 κB motif is critical for basal and Tat-induced transcriptional activation of the viral promoter [Nabel and Baltimore, 1987; Taylor and Khalili, 1994]. Tat is a potent HIV-1 transcriptional activator that upon interaction with the TAR RNA sequence positioned within the leader of the viral transcripts tremendously enhances viral gene transcription in lymphoid and several non-lymphoid cells. In earlier studies we and others demonstrated that in human astrocytic cells as well as in lymphoid cells under conditions where NF-κB is stimulated, Tat has the ability to augment transcription of a TAR-negative viral promoter. The TAR-independent regulatory event seems to be mediated, at least in part, through the κB motif of the LTR [Harrich et al., 1990; Taylor et al., 1992]. Results from several biochemical studies suggested that the association of Tat with the NF-κB subunits either directly or indirectly through other cellular proteins may mediate the activation of a TAR-negative LTR [Taylor et al., 1992; Yang et al., 1997]. To gain more insight into the modulation of the LTR via cellular proteins from human astrocytes that communicate with NF-κB as well as Tat, we utilized a yeast two-hybrid system using the p50 subunit of NF-κB as bait to identify a novel cellular protein that physically interacts with NF-κB
METHODS AND EXPERIMENTAL PROCEDURES
Yeast Two-Hybrid System
cDNA for the p50 subunit of NF-κB was cloned into the pAS2-1 plasmid, containing the GAL-4 DNA-binding domain. A cDNA library made from U-87MG cells was cloned into pGAD-10, containing the GAL-4 DNA-activation domain. The procedure used followed the manufacturer's protocol (BD Biosciences Clontech, Palo Alto, CA). Briefly, colonies that grew after co-transforming yeast cells with both plasmids were further screened with a ß-galactosidase filter assay. To eliminate false positives, positive colonies were assayed using cyclohexamide counterselection and those positive colonies were further tested by a yeast-mating assay.
Glutathione S-Tranferage (GST)-Pulldown Assays
p50 and p65 proteins were radiolabeled with an in vitro coupled transcription/translation kit (Promega, Madison, WI). Five microgram GST-NFBP (NF-κB-binding proteins)/f or GST on glutathione sepharose beads were incubated with radiolabeled p50 or p65 protein for 2 h. The sepharose bead pellets were washed several times and the radiolabeled proteins were detected using SDS–PAGE.
RNA Isolation and Northern Blotting
RNA was isolated using guanidine isothiochanate according to Chomczynski and Sacchi [1987]. Twenty micrograms of total RNA were incubated 15 min at 60°C in MOPS buffer containing 50% deionized formamide and 16% formaldehyde, and resolved on an agarose gel containing MOPS buffer and 2.5% formaldehyde. The RNA was transferred to a nylon membrane (Amersham, Piscataway, NJ) using a Turboblotter system (Schleicher & Schuell, Keene, NH) according to the manufacturer's instructions. The DNA probes were labeled using a random primed labeling reaction with Klenow enzyme and α32P dCTP. Unincorporated labeled nucleotides were removed from the DNA labeling reaction using a MicroSpin G-50 column (Amersham) according to the manufacturer's instructions. The RNA blots were prehybridized and hybridized in ULTRAhyb solution (Ambion, Austin, TX) according to the manufacturer's instructions. The 1.5 × 106 cpm/ml of probe was hybridized to each blot overnight at 42°C. The blots were washed once in 2× SSC/0.1% SDS and then two- to three-times more in 0.1× SSC/0.1% SDS until background counts were less than 1,000 cpm.
FISH Mapping
Plasmid DNA was prepared using a Maxiprep kit (Qiagen, Valenica, CA), following the manufacturer's protocol. Twenty micrograms of NFBP in pcDNA3 vector was sent to SeeDNA Biotech, Inc. (Windsor, Ont., Canada) for FISH analysis. Lymphocytes isolated from human blood were cultured in α-minimal essential medium (α-MEM) supplemented with 10% fetal calf serum and phytohemagglutinin (PHA) at 37°C for 68–72 h. The lymphocyte cultures were treated with BrdU (0.18 mg/ml; Sigma, St. Louis, MO) to synchronize the cell population. The synchronized cells were washed three-times with serum free medium to release the block and recultured at 37°C for 6 h in α-MEM with thymidine (2.5 μg/ml; Sigma). Cells were harvested and slides were made using standard procedures including hypotonic treatment, fixation, and air-dry. The DNA probe was biotinylated with dATP using Gibco BRL BioNick kit (15°C, 1 hr) [Heng et al., 1992]. The procedure for FISH detection was performed according to Heng et al. [1992] and Heng and Tsui [1993]. Briefly, slides were baked at 55°C for 1 h. After RNase A treatment, the slides were denatured in 70% formamide in 2 × SSC for 2 min at 70°C followed by dehydration with ethanol. Probes were denatured at 75°C for 5 min in a hybridization mix consisting of 50% formamide and 10% dextran sulfate. Probes were loaded in the denatured chromosomal slides. After overnight hybridization, slides were washed and detected as amplified. FISH signals and the DAPI banding pattern was recorded separately by taking photographs, and the assignment of the FISH mapping data with chromosomal bands was achieved by superimposing FISH signals with DAPI banded chromosomes [Heng and Tsui, 1993].
Cellular Localization
U-87MG cells were transfected with the fluorescent reporter vector pEYFP-C1 (BD Biosciences Clontech) or with the pEYFP-C1 fused with NFBP, and were visualized with fluorescent microscopy. NFBP in pBluescript vector was obtained from the Kazusa Research Institute. The full length NFBP was also cut from pBluescript with SalI and DraI, and cloned into the pEYFP-C1 vector at the SalI and SmaI sites.
RESULTS AND DISCUSSION
A yeast two-hybrid system was utilized to examine new potential NF-κB binding proteins. Briefly, the transcriptional activator GAL-4 DNA-binding and -activation domains were separated and cloned into two different plasmids. The p50 subunit of NF-κB was fused to the GAL-4 DNA-binding domain and used as the “bait.” A cDNA library was generated from the human astrocytic cell line, U-87MG and fused to the GAL-4-activation domain. The two plasmids were cotransformed into yeast and plated onto medium lacking histidine. Growth without histidine indicated that the unknown protein and p50 bound to each other, bringing the GAL-4 DNA-binding and -activation domains within close proximity activating GAL-4 responsive genes, including production of histidine necessary for growth. Any positive colonies were further screened by a filter assay for β-galactosidase activity, another GAL-4 responsive gene. Using this system a novel human gene was isolated. The cDNA clone (Clone 45) was approximately 1,200 bp long with an open reading frame of 383 amino acids. To assess the binding of amino acid peptides encoded by Clone 45, herewith called NF-κB-binding protein fragment (NFBP/f), with the p50 and p65 subunits of NF-κB, a GST-NFBP/f fusion protein was immobilized on glutathione beads and incubated with radiolabeled p50 or p65 proteins. The proteins that bound to GST-NFBP/f or the control GST were analyzed by SDS–PAGE. As shown in Figure 1A, a band corresponding to p50 was detected in the fraction obtained from GST-NFBP/f but not GST. Similar results were seen with p65 (Fig. 1B).
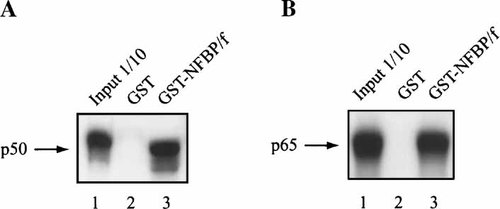
GST-pulldown. Five microgram GST-NFBP/f or GST on glutathione sepharose beads were incubated with radiolabeled p50 (A) or p65 (B) protein for 2 h. The sepharose bead pellets were washed several times and the radiolabeled proteins were detected using SDS–PAGE. Lane 1 contains 0.5 μg radiolabeled p50 and p65, lane 2 contains GST alone on glutathione beads, and lane 3 contains GST-NFBP/f on glutathione beads.
Results from a National Center for Biotechnology (NCBI) BLAST database analysis revealed 100% identity of the NFBP/f DNA sequence with the 3′ region of a 5.8 kb cDNA labeled KIAA0185 [Nagase et al., 1996]. KIAA0185 was obtained from the Kazusa DNA Research Institute Foundation and used for some of the studies presented here. It has been renamed NFBP. This cDNA encompasses a 1,871 amino acid open reading frame in which 131 amino acids corresponding to Clone 45 resides on its C-terminus region, suggesting that the KIAA0185 may represent a full-length cDNA for Clone 45. This notion was verified by results from Northern blot analysis when both cDNA probes corresponding to full-length and partial NFBP were hybridized to a single mRNA species of approximately 5.5 kb in several cell lines, including the human astrocytic cell line, U-87MG, rat glial-origin cells (C6), and monkey kidney cells (CV-1, Fig. 2). The transcript was also seen in Daoy cells, a human meduloblastoma cell line (data not shown), and human neurons cultured from fetal tissue (data not shown). Interestingly, the transcript is seen in much lower levels in T98G cells (data not shown), a human glioblastoma cell line, which originates from a more aggressive tumor than either U-87MG or Daoy.
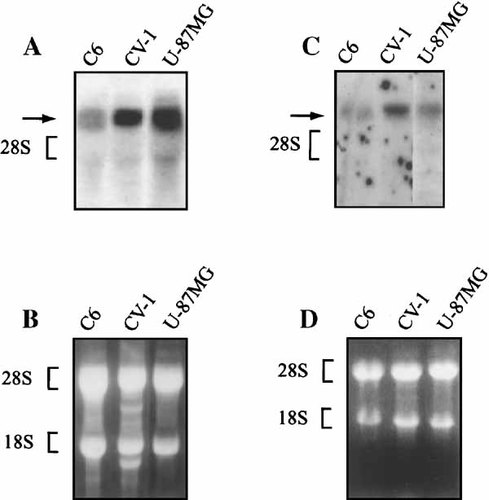
Northern blot. Total RNA from various cells were isolated from cells using guanidine isothiocyanate according to the procedure described previously. Hybridization of RNAs with NFBP and NFBP/f cDNA is shown in panels A and C, respectively. Panels B and D show ethidium bromide stained gels. The position of the NFBP transcript in each blot is shown by an arrow.
Using the NCBI BLAST web site, the cDNA of NFBP was mapped to its chromosomal location (Fig. 3A). The genomic DNA is complex, containing more than 35 exons. NFBP/f maps to the 3′ end of the full length NFBP cDNA. The protein encoded by full length NFBP contains 1,871 amino acid residues, while NFBP/f is 131 amino acids long.
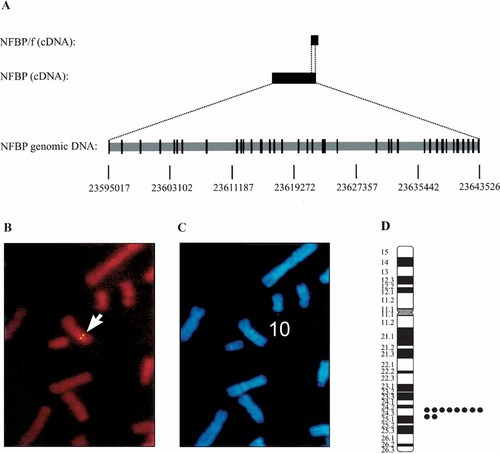
DNA mapping and FISH analysis. A: The organization of the NFBP genome. Exons of the genomic DNA are indicated with dark boxes. NFBP cDNA and NFBP/f cDNA are drawn on the same scale as the genomic DNA. B–D: Chromosomal mapping using FISH. Example of FISH mapping with NFBP is shown in panel B and mitotic figures as determined by DAPI are shown in panel C. This picture shows the FISH signals on a human chromosome. D: Diagram of FISH mapping result. Each dot represents double FISH signals in human chromosome 10.
NFBP was used as a probe for FISH mapping. The FISH detection efficiency was approximately 87% and combined with DAPI staining summary from 10 photos showed that NFBP mapped to human chromosome 10, region q24.2-q25.1 (Fig. 3B–D). Information from the Mitelman Database of Chromosome Aberrations in Cancer [2001] showed that translocations and deletions in this region are associated with lymphoblastic leukemia, non-Hodgkin's lymphoma (NHL), adenocarcinoma, and other forms of leukemia and lymphoma. Abnormalities in chromosome bands 10q23-25 are commonly seen in cases of NHL. Speaks et al. [1992] found these genetic abnormalities in up to 67% of patients with NHL, depending on the disease subtype, with an overall occurrence for all NHL cases examined of 10%. In a more recent report, describing the karyotype of patients with follicular adenoma, a form of NHL, deletions in the region, 10q22-24, can be seen in greater than 10% of cases [Horsman et al., 2001]. In children with T-cell lineage acute lymphoblastic leukemia (T-ALL), t(10;14)(q24:q11) can be seen in 3–6% of cases [Schneider et al., 2000], while in adults with T-ALL, the occurrence of t(10;14)(q24:q11) is higher, at 14% [Group Francais de Cytogenetique Hematologique, 1996]. While none of these genetic abnormalities have been mapped to their precise locations on the chromosome, it is apparent that this region is a hotspot for genetic abnormalities seen in cases of human cancer.
Using both the PFAM [Bateman et al., 2000] and NCBI databases, the NFBP protein sequence was analyzed for common motifs and domains. Seven S1 RNA-binding domains with significant E values were found as was one Histone Acetyl Transferase (HAT) repeat (Fig. 4C). Proteins with HAT activity transfer the acetyl group from acetyl-CoA to lysine residues in the N-terminal tails of core histone proteins. The typical result of hyperacetylation of histones is gene activation [for review see Kuo and Allis, 1998]. While the exact mechanism of this activation is unknown, it is known that the acetyl group neutralizes the positive charge of the lysine residue and acetylation of histones may change the native configuration of the proteins, loosening their binding to chromosomal DNA. Both effects of acetylation may be necessary for cellular transcription factors to gain access to gene promoters.
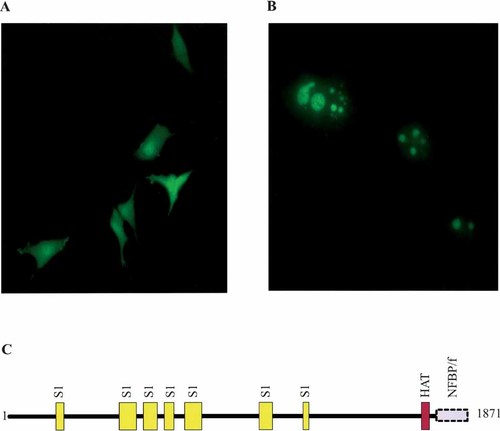
Protein mapping and cellular localization of NFBP. A and B: U-87MG cells were transfected with the (A) fluorescent reporter vector pEYFP or with the (B) fluorescent reporter vector pEYFP fused with NFBP and were visualized with fluorescent microscopy. C: The deduced amino acid sequence of NFBP was analyzed using the PFAM and the NCBI databases. The NFBP amino acid sequence was analyzed using the PFAM (pfam.wustl.edu) and National Center for Biotechnology (NCBI) databases (www.ncbi.nlm.nih.gov). For the PFAM analysis, domain regions with e values less than or equal to 0.05 are shown. The positions of RNA-binding region (S1), HAT, and NFBP/f are depicted.
The S1 RNA-binding domain originates from the S1 bacterial protein and is found in many other RNA-binding proteins. The S1 protein is important during bacterial initiation and translation. The S1 domain is found in many other RNA-binding proteins, including eukaryotic translation initiation factor eIF2α [Gribskov, 1992], bacterial RNase E, which aids in the degradation of mRNA [Carpousis et al., 1994], RNase II, another bacterial protein which degrades mRNA [Nierlich and Murakawa, 1996] and NusA protein, which is important in transcription elongation in prokaryotes [Bycroft et al., 1997].
In order to identify its cellular localization, full-length NFBP was cloned into a fluorescent reporter vector, pEYFP-C1. The plasmid was transfected into U-87MG cells and visualized with a fluorescent microscope. NFBP is always nuclear, appearing as one to several granular aggregates in the nucleus (Fig. 4B). Little or no fluorescence was seen in the cytoplasm. As a control, the YFP vector alone was also transfected into U-87MG cells and there was uniform fluorescence through out the entire cell (Fig. 4A). The bacterial S1 ribosomal RNA-binding protein is also located in the nucleus as small aggregates [Hügle et al., 1985]. Given that NFBP contains several S1 RNA-binding domains and that the cellular distribution of S1 and NFBP are so similar, it is likely NFBP also binds to nucleic acid. Further evidence that NFBP is localized to the nucleus comes from a recently published report by Anderson et al. [2002]. Proteins were isolated from the nucleoli from HeLa cells and analyzed using mass spectrometry. NFBP was one of the proteins found to be in the nucleolus.
Using the BLAST database, a gene from Saccharomyces cerevisiae was identified which shares significant homology with NFBP (26% identity), named RRP5. RRP5 is involved in processing of the 35S yeast pre-rRNA to the mature 18S and 5.8S rRNA species [Venema and Tollervey, 1996; Torchet et al., 1998; Eppens et al., 1999]. Like NFBP, RRP5 contains many S1 RNA-binding motifs [Torchet et al., 1998; Eppens et al., 1999] and these RNA-binding motifs are important for assembly of the RNaseMRP processing complex necessary for cleavage of a precursor rRNA species to the eventual mature 5.8Ss rRNA [Eppens et al., 1999]. The strong similarity between NFBP and RRP5 suggests that they perform a common function in the cell, namely, processing of pre-rRNA species to the mature and active rRNA forms.
Another BLAST database search revealed a mouse gene, originally named ALG-4 and later renamed programmed cell death protein 11, Pdcd11, which shares 82% identity with NFBP. Pdcd11 protein induces activation of NF-κB and the FasL promoter, leading to apoptosis in Jurkat cells [Lacana and D'Adamio, 1999]. NFBP is found primarily in the nucleus, where like, Pdcd11, it could activate transcription of the NF-κB promoter to induce apoptosis. Additionally, it has been shown that NF-κB activates the p105 gene, the precursor to p50, in human monocytic cells [Paya et al., 1992], which leads to a perpetual high level of p50/p65 dimers which can continually activate the HIV-1 LTR in chronically infected monocytes. If, like Pcdc11, NFBP upregulates NF-κB, this could help sustain HIV-1 infection in cells of monocytic origin, including brain macrophages.
NFBP is a normal human protein found in many cell types. We have shown that it localizes primarily to the nucleus and contains several S1 RNA-binding sites. While the normal function of NFBP is still unclear, the potential RNA-binding ability of the protein and its cellular localization are logical in relation to one another, especially in light of the high degree of homology it shares with the yeast RRP5 rRNA biogenesis protein. One probable role of NFBP could be processing of a rRNA precursor to its more mature and active forms, which occurs in the nucleolus. Additionally, NFBP was isolated due to its ability to bind to the p50 subunit of NF-κB, which when activated, is localized to the nucleus where it acts as a transcription factor, activating and suppressing numerous cellular and viral genes. Homologous to the mouse protein, Pcdc11, NFBP may activate one or more NF-κB promoters and then in turn bind to NF-κB to increase the ability of NFBP and NF-κB to activate viral promoters, including the HIV-1 LTR. Finally, computer analysis has shown that NFBP contains a HAT repeat, which may also enable it to activate both cellular and viral genes. It is known that the HIV-1 LTR, once integrated, is associated with histone proteins and is therefore inaccessible to both cellular and viral factors necessary for transcription [Verdin et al., 1993]. Like P300 and CREB-binding protein, both of which contain HAT repeats and interact with the nucleosomes surrounding the LTR to increase viral activation [Marzio et al., 1998], NFBP may aid in HIV-1 gene activation by acetylating the histone proteins surrounding the integrated promoter.




