Epigenetic regulation of Igf2/H19 imprinting at CTCF insulator binding sites†
Youwen Yang and Ji-Fan Hu contributed equally to this work.
Abstract
The mouse insulin-like growth factor II (Igf2) and H19 genes are located adjacent to each other on chromosome 7q11-13 and are reciprocally imprinted. It is believed that the allelic expression of these two genes is regulated by the binding of CTCF insulators to four parent-specific DNA methylation sites in an imprinting control center (ICR) located between these two genes. Although monoallelically expressed in peripheral tissues, Igf2 is biallelically transcribed in the CNS. In this study, we examined the allelic DNA methylation and CTCF binding in the Igf2/H19 imprinting center in CNS, hypothesizing that the aberrant CTCF binding as one of the mechanisms leads to biallelic expression of Igf2 in CNS. Using hybrid F1 mice (M. spretus males × C57BL/6 females), we showed that in CNS, CTCF binding sites in the ICR were methylated exclusively on the paternal allele, and CTCF bound only to the unmethylated maternal allele, showing no differences from the imprinted peripheral tissues. Among three other epigenetic modifications examined, histone H3 lysine 9 methylation correlated well with Igf2 allelic expression in CNS. These results suggest that CTCF binding to the ICR alone is not sufficient to insulate the Igf2 maternal promoter and to regulate the allelic expression of the gene in the CNS, thus challenging the aberrant CTCF binding as a common mechanism for lack of Igf2 imprinting in CNS. Further studies should be focused on the identification of factors that are involved in histone methylation and CTCF-associated factors that may be needed to coordinate Igf2 imprinting. © 2003 Wiley-Liss, Inc.
Abbreviations used:
Igf2, insulin like growth factor II; LOI, loss of imprinting; MOI, maintenance of imprinting; ChiP, chromatin immunoprecipitation; CTCF, CTCF insulating factor; H3-K9, histone 3 lysine 9; H3-K4, histone 3 lysine 9; H3-Ac and H4-Ac, histones 3 and 4 acetylation; DMR, differentially methylated region.
The mouse insulin-like growth factor II (Igf2) and H19 genes are clustered in an imprinting domain of chromosome 7, and are reciprocally imprinted such that Igf2 is expressed exclusively from the paternal allele [DeChiara et al., 1990; Hu et al., 1995] and H19 only from the maternal allele [Bartolomei et al., 1991]. These two genes are also coordinately expressed in many tissues, leading to the hypothesis that they are regulated by a common “enhancer competition” mechanism [Bartolomei and Tilghman, 1992; Zemel et al., 1992]. Several regulatory components have been identified in this imprinting domain, including two enhancers located downstream of H19 [Bartolomei and Tilghman, 1992; Ferguson-Smith et al., 1993; Leighton et al., 1995b], an upstream region of Igf2 [Constancia et al., 2000; Eden et al., 2001] that shows differential DNA methylation between the two parental alleles (DMR1) [Brandeis et al., 1993], a region in the last exon of Igf2 (DMR2) [Murrell et al., 2001], the H19 promoter [Ripoche et al., 1997; Vu et al., 2001], and a 2–4 kb upstream region of the H19 that contains parent-specific DNA methylation [Bartolomei et al., 1993; Tremblay et al., 1995; Khosla et al., 1999; Vu et al., 2000].
A recent breakthrough in our understanding Igf2/H19 imprinting is derived from the finding of CTCF protein insulation in an imprinting control region (ICR) (or barrier region) upstream of the H19 promoter [Bell and Felsenfeld, 2000; Hark et al., 2000]. The insulating effect of the zinc-finger protein CTCF is dependent upon the status of DNA methylation of four conserved CTCF binding sites in this barrier region [Bell and Felsenfeld, 2000; Hark et al., 2000]. The CTCF protein binds specifically to the unmethylated maternal allele, and thus insulates the maternal promoters of Igf2 to utilize two enhancers downstream of H19. This insulation leads to the silencing of Igf2 from its maternal allele, but leaves the enhancers able to access the maternal H19 promoter. The insulator protein, however, cannot bind to the methylated paternal allele and thus fails to insulate the paternal Igf2 promoters from accessing the enhancers. As a result, Igf2 is monoallelically expressed from the paternal allele and H19 is expressed from the maternal allele (Fig. 1).
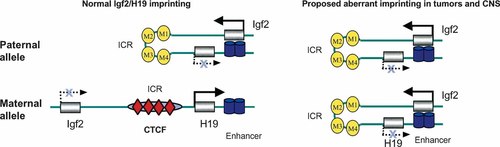
CTCF insulation model in the Igf2/H19 imprinting. ICR: imprinting control region, M1–M4; four methylated CTCF binding sites in the ICR. The active promoters for Igf2 and H19 are presented as solid arrows, and inactive promoters as broken arrows. In normal Igf2 imprinting, four paternal CTCF sites are methylated and thus prevent the binding of CTCF insulators, leaving the enhancers to be contacted by the Igf2 promoters for the paternal expression. The maternal CTCF sites, however, are unmethylated and are thus occupied by CTCF insulators. The CTCF binding prevents the Igf2 loci from accessing the enhancers, leading to the repression of the maternal allele (left panel). In CNS and tumors where Igf2 is biallelically expressed, the CTCF model predicts both parental alleles are methylated and there is no CTCF insulation at both alleles, leading to biallelic expression of Igf2 (right panel).
Previously, we [Hu et al., 1995] and others [DeChiara et al., 1991; Ohlsson et al., 1994 #306; Pedone et al., 1994] have demonstrated that murine Igf2 is monoallelically expressed in many tissues as previously reported [DeChiara et al., 1990], but is biallelically transcribed in CNS, sharing the same phenotype as commonly observed in human tumors [Feinberg, 1993; Ogawa et al., 1993; Rainier et al., 1993]. Later, we demonstrated that the human IGF2 was also biallelically expressed in fetal brains [Pham et al., 1998]. In addition, by using a most sensitive PCR method we showed that Igf2 imprinting was absent not only in leptomeninges and choroid plexus as reported by DeChiara et al. [1991], but also in other parts of the brain, including cerebrum, cerebellum, pons, and medulla [Hu et al., 1995]. Using PCR, Albrecht et al. [1996] also demonstrated the loss of IGF2 imprinting in human fetal cerebellum. Hemberger et al. [1998] also showed that the mouse Igf2 was biallelically expressed in cells derived from ventral midline region of both the hindbrain and spinal cord, where complete absence of Igf2 transcription was previously described [DeChiara et al., 1991]. Taken together, the tissue-specific imprinting in CNS has provided a natural model to study the mechanisms underlying the loss of Igf2 imprinting.
It is still not clear why Igf2 is biallelically expressed in CNS. In peripheral tissues, the three promoters of the mouse Igf2 gene are monoallelically expressed from the paternal allele. In the CNS, however, all three promoters drive Igf2 expression from both parental alleles [Hu et al., 1995]. Similarly, in Wilms' tumors with loss of IGF2 imprinting, all three upstream promoters (hP2-hP4) are also biallelically expressed [Vu and Hoffman, 1996]. In addition, we have found that the mouse H19 gene is still imprinted in mouse CNS [Hu et al., 1995]. Uncoupling of Igf2/H19 imprinting has also been observed in human tumors [Rainier et al., 1993, 1995; Ulaner et al., 2003]. Finally, aberrant DNA methylation in the DMR regions occurs in both CNS and in tumors with the LOI [Feil et al., 1994; Frevel et al., 1999; Cui et al., 2001, 2002; Nakagawa et al., 2001].
In this communication, we examined whether the lack of Igf2 imprinting in CNS is a result of aberrant CTCF insulation in the Igf2/H19 ICR. According to the CTCF insulator model [Bell and Felsenfeld, 2000; Hark et al., 2000], the two parental alleles at the Igf2/H19 imprinting center should be hypermethylated in CNS, where the two parental alleles of Igf2 are expressed. As a result, CTCF insulators should not bind to the methylated CTCF sites in CNS, allowing access of the Igf2 promoters to the enhancers downstream of H19. Addressing this mechanism in CNS will help us understand the regulatory control of Igf2 imprinting in peripheral tissues as well and may guide our further research examining aberrant control of IGF2 in human tumors.
MATERIALS AND METHODS
Animals
F1 generation mice, derived from breeding C57BL/6 female mice with M. spretus male mice (purchased from Jackson Laboratories, Maine), were used for this study. We focused on two peripheral tissues (liver and kidney) that show the monoallelic expression of Igf2, and brain that shows the biallelic expression of Igf2.
The animal experiments were approved by the Animal Care and Use Committee of the VA Medical Center and were conducted in accord with the procedures in “Guidelines for Care and Use of Experimental Animals.”
DNA and cDNA Preparation
Total RNA was extracted from tissues by TRI-REAGENT (Sigma, St. Louis, MO), according to the manufacturer's guide. To eliminate DNA contamination in cDNA synthesis, RNA samples were first treated with DNase I, and cDNA was synthesized with RNA reverse transcriptase [Hu et al., 1995, 1996; Vu and Hoffman, 1996]. Briefly, in a typical reaction mixture, aliquots of 2.0 μl RNA (200 μg/ml) under the evaporation barrier of 12 μl of liquid wax (MJ Research, Inc., Boston, MA) were treated with 1.0 μl of 0.4 U DNase I (Stratagene, La Jolla, CA) in 25 mM Tris (pH = 8.0), 25 mM NaCl, 5 mM MgCl2, and 0.15 U RNase inhibitor (5′Prime-3′Prime, Boulder, CO) at 37°C for 15 min, followed by enzyme denaturing at 75°C for 10 min. After DNA digestion, RNAs were reverse-transcribed into cDNAs with murine leukemia reverse transcriptase (Invitrogen, Carlsbad, CA) in the presence of random hexamers at 37°C for 25 min, followed by five cycles (50°C, 20 s and 37°C, 5 min) [Vu and Hoffman, 1994; Hu et al., 1995].
Allelic Expression of Igf2 and H19
Allelic expression of Igf2 and H19 was examined by PCR in cDNA samples as previously described. cDNA samples were amplified in a 3.0 μl reaction mixture in the presence of 50 μM dNTP, 1 nM primer, 0.125 U KT1 DNA polymerase [Hu et al., 1997b; Yao et al., 2003]. The cDNAs and primers were heated to 95°C for 2 min, then amplified by 35 cycles at 95°C for 20 s, 65°C for 40 s, and 72°C for 30 s.
As previously described [Hu et al., 1997a, 1998a], PCR primers used for Igf2 included #MII84 (5′-primer): CTT GTG CTG GAT CGC TGC TTA CGG and #MII219 (3′-primer): CTG CGA CGG TTG GCA CGG CTT GA. The 3′-primer (MII219) was end-labeled with γ-[32P]-ATP (Amersham Life Science, Arlington Heights, IL). PCR products were diluted and digested with 1 U DpnII (Biolab) in a 6 μl reaction and were electrophoresed on 5% polyacrylamide–urea gel. To examine allelic expression of H19 in F1 mice, cDNA samples were amplified with PCR primer set: #4025 (5′-primer): TAA GTC GAT TGC ACT GGT TTG GAG T and #4026 (3′-primer): TGA TGG AAC TGC TTC CAG ACT AG. PCR products were digested with 1 U FokI, which specifically cuts the M. musculus allele. After electrophoresis, the gel was scanned by PhosphoImage Scanner (Molecular Dynamics, Sunnyvale, CA).
Allelic DNA Methylation of CTCF Sites in Igf2/H19 Imprinting Center
To find a suitable polymorphism to study allelic DNA methylation, we first cloned and sequenced the CTCF binding region in M. spretus mice. Genomic DNA was extracted from the M. spretus liver and genomic DNA covering CTCF binding sites was amplified by PCR. Primers used to amplify the sequence containing CTCF site 1 and 2 were: #1428 (5′-primer): CCGAGAAAATAGCCATTGCCTACAGT and #1431 (3-primer): CGTTTTATCAAGGACTAGCATGAA; CTCF sites 3 and 4 were #1351 (5-primer): AGGTTGGAACACTTGTGTTTCTGGAG and #1354 (3-primer): GTCACAGCGGACCCCAACCTATG. PCR DNA products were cloned into TA vector (Invitrogen) and sequenced. Three polymorphisms were found within or near CTCF binding sites 1, 2, and 3 compared with the M. musculus DNA sequence available in Genebank (Fig. 2A). No polymorphisms are present within or near CTCF binding site 4.
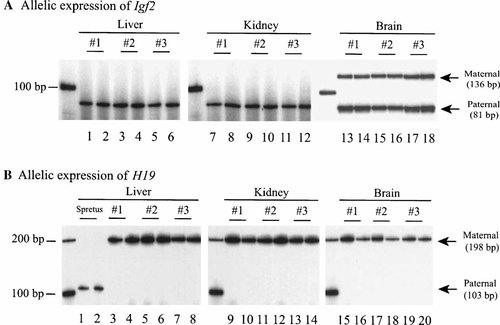
Differential allelic expression of Igf2 (A) and H19 (B) in F1 mice (M. spretus males × C57BL/6 females). Brain and peripheral tissues (liver and kidney) were collected from three F1 mice (1 month, 1 year, and 2 years old, respectively). Total RNA was extracted and converted into cDNA for allelic expression of Igf2 and H19 as described in Materials and Methods. A DpnII polymorphic restriction site was used to separate two parental alleles for Igf2 and FokI for H19. Note Igf2 is biallelically expressed in all three brains but monoallelically expressed in liver and kidney.
The status of DNA methylation at CTCF sites was then measured by the sodium bisulfite method as described by Frommer et al. [1992] with some modifications [Hu et al., 1998b; Li et al., 2002]. In brief, purified genomic DNAs (2 μg/17 μl) were denatured by adding 3 μl of 2 M NaOH at 37°C for 20 min. DNA samples were then overlaid with 60 μl of liquid wax (MJ Research) and treated with 220 μl of freshly prepared 3.5 M NaHSO3 containing 1 mM hydroquinone (pH 5.0) on ice overnight in the dark. To ensure a complete treatment with sodium bisulfite, the reaction was further incubated at 50°C for 8 h. Modified DNA was diluted with 250 μl of water and then purified with QIAEX DNA purification kit (QIAEX II kit, QIAGEN, Inc., Chatsworth, CA). DNA samples were treated with 5 μl of 2 M NaOH at room temperature for 15 min and further neutralized with 5 μl of 2 M HCl. After desalting by passing through a DNA purification column (Princeton Separations, Inc., Adelphia, NJ), DNA samples were diluted to 60 μl with sterile distilled water. Aliquots of 1 μl of modified DNA were used for PCR amplification using the conditions as described above.
Allele-specific primers were used to amplify bisulfite-modified DNA at CTCF sites 1, 2, and 3 based on three polymorphisms identified between M. spretus and M. musculus. The paternal allele at CTCF site 3A was amplified by the allele-specific 5′-primer #1444: TTG TGT TTT TGG AGG GGG TTT TTT GGT TT and a common 3′-primer #1443: AAA CCA CRA TAT ATA AAA ATA TAC TAC CAC. The maternal M. musculus allele was amplified by the allelic 5′-primer #1445: TTG TGT TTT TGG AGG GGG TTT TTT GGT TA and the 3′-primer #1443. For a second CpG site (3B) located at 156 bp upstream of CTCF site 3, the paternal allele was amplified by a common 5′-primer #1446: GGT TTG TTT ATG ATA ATG TTT AAG GGT TA and a M. spretus-specific 3′-primer #1447: AAA CCA CAC TAA CTA ATT TTT AAA ATT CAA A. The maternal allele at CTCF site 3B was amplified by #1446 (5-primer) and a M. musculus-specific 3′-primer #1448: AAA CCA CAC TAA CTA ATT TTT AAA ATT CAA T. Allele-specific primers were end-labeled with γ-[32P]-ATP in PCR amplification.
After sodium bisulfite treatment, unmethylated cytosine residues are converted to uracils, which will be amplified as T in PCR reaction. The methylated cytosine residues in CpG islands, however, will not be modified and will be amplified as C in the PCR reaction [Hu et al., 1997a; Yao et al., 2003]. The CTCF site 3A PCR products were digested by restriction enzyme BstUI which recognize a cutting sequence located on the third CTCF binding site to separate methylated and unmethylated DNA. For CTCF site 3B that is located at the 156 bp upstream of CTCF site 3, PCR products were digested with TaqI to separate the methylated and the unmethylated alleles.
Allelic CTCF Binding in Igf2/H19 Imprinting Center
Chromatin immunoprecipitation (ChIP) was used to assess the binding of CTCF to the Igf2/H19 imprinting control center. The ChIP assay kit was purchased from Upstate Biotechnology (Waltham, MA). The ChIP assay was carried out according to the protocol provided by the manufacturer with some modifications.
Frozen liver tissue (120 mg each), kidney tissue (180 mg each) and whole brain (360 mg each) were soaped in 30 ml PBS solution containing 1% formaldehyde and were chopped into small pieces with a razor blade in a ventilation hood. Tissues were homogenized and incubated at room temperature for 10–15 min and the crosslinking reaction was stopped by adding glycine to a final concentration of 0.125 M. After washing twice with PBS at 4°C, the crosslinked tissue was lysed in 5 ml SDS lysis buffer and sonicated to shear DNA to lengths between 200 and 1,000 bp by a Branson Sonifier 250 (Sonic Power Company, Danbury, CT) with the setting at 30–40% output, 90% duty cycle, 10 pulse for 18 cycles. The cell lysate was chilled on dry ice, spun-down briefly, and then kept in the ice-bath between each sonication. CTCF-bound chromatin fragments were immunoprecipitated with antiserum against CTCF (cat. #06–917, Upstate Biotechnology). After precipitation, the DNA was purified with MinElute PCR Purification Kit (Qiagen, Inc., #28004). DNA was eluted in 100 μl 2.5 mM Tris-CI (pH 8.0) and analyzed with PCR primers specific for CTCF binding sites. PCR conditions were the same as described above. PCR band density reflects the allelic binding of CTCF.
Primers used to quantitate CTCF binding at CTCF site 1 were #1428 (5′-primer): CCG AGA AAA TAG CCA TTG CCT ACA GT and #1429 (3′-primer): CAT GTT CCT TTG AGT CCT GGG TGT AT. This pair of primers amplified a DNA fragment that was 95 bp upstream of CTCF site 1. The sequences covering CTCF site 2 were amplified by primers #1430 (5′primer): CGGC AGTGAAGTCT CGTACATCGC and #1431 (3′primer): CGT TTT ATC AAG GAC TAG CAT GAA. CTCF site 3 was amplified with primers #1351 (5-primer): AGGTTGGAACACTTGTGTTTCTGGAG and #1415 (3′-primer): TGG GCC ACG ATA TAT AGG AGT ATG CT.
After PCR amplification, the two parental alleles at CTCF sites 1, 2, and 3 were separated, respectively, by the digestion of three polymorphic restriction enzymes: HpaII, NlaIII, and BsmA1. After restriction enzyme digestion, PCR products were run on 5% polyacrylamide–urea gel and were scanned by Phosphoimage Scanner (Molecular Dynamics) to quantitate allelic CTCF binding.
Allelic Histone Methylation at CTCF Binding Sites
Chromatin immunoprecipitation (ChIP) was used to assess the methylation status of histones at CTCF binding sites in Igf2/H19 imprinting center. The specific protocol was the same as that described above for CTCF binding, except the dimethylated histone-associated chromatin fragments were immunoprecipitated with anti-dimethylated H3-K4 (cat. #07–030) and anti-dimethylated H3-K9 (cat. #07–212) antisera (Upstate Biotech). As described in the CTCF binding section above, the immunoprecipitated chromatin DNA was amplified with PCR primers covering the CTCF site 3 (#1351 and #1415, Fig. 4A). PCR products were subject to the digestion of the polymorphic restriction enzyme BsmA1 to separate two parental alleles.
Allelic Histone Acetylation at CTCF Binding Sites
The status of histone acetylation at CTCF sites was examined by the ChIP method as described above. Antibodies specific for acetylated histone H3 (acetylated lysines 9, 14 cat. #06–599) and acetylated histone H4 (acetylated lysines 5, 8, 12, 16, cat. #06–866) were purchased from Upstate Biotech. Acetylated histone-associated DNA was amplified with primers covering the CTCF site 3 (#1351 and #1415) and digested with the polymorphic restriction enzymes BsmA1 to quantitate the allelic histone acetylation.
RESULTS
Biallelic Expression of Igf2 but Monoallelic Expression of H19 in CNS
Using two polymorphisms between M. spretus and M. musculus, we examined the allelic expression of Igf2 and H19 in F1 mice aged 1 month, 1 year, and 2 years. Igf2 cDNA was amplified by PCR primers #MII84 and #MII219 that cross the mouse Igf2 intron 4; this eliminates possible genomic DNA contamination. PCR products were then cut by DpnII to separate parental alleles.
As previously described [Hu et al., 1995], Igf2 is always monoallelically transcribed in liver and kidney (Fig. 2A, lanes 1–12), with mRNA transcripts detected only from the paternal allele detected. In CNS, however, both parental alleles were active, transcribing mRNA products (Fig. 2A, lanes 13–18). The presence of both Igf2 transcripts was observed in all F1 mice.
By using the in situ hybridization method, DeChiara et al. [1991] detected the expression of the maternally transmitted Igf2 primarily in choroid plexus and leptomeninges. However, when a most sensitive PCR method was used, it was found that Igf2 was also biallelically expressed in every part of CNS in addition to these two tissues [Hu et al., 1995; Albrecht et al., 1996; Hemberger et al., 1998]. Thus, the tissue-specific lack of Igf2 imprinting detected in whole brain represents a unique regulation of imprinting in CNS, rather than from choroid plexus and leptomeninges alone.
The mouse H19 gene, which is usually reciprocally imprinted and coordinately expressed with Igf2, was monoallelically expressed in CNS (Fig. 2B, lanes 15–20), showing no differences from the uniparental expression pattern in liver and kidney (Fig. 2B, lanes 3–14). This differential imprinting of Igf2 between CNS and peripheral tissues in the same animal thus provides an ideal model to study the mechanism underlying genomic imprinting.
Allelic DNA Methylation in CTCF Binding Sites in Igf2/H19 Imprinting Center
As a first step to study this mechanism, we examined the status of DNA methylation at CTCF binding sites in the Igf2/H19 ICR, hypothesizing that an aberrant epigenetic modification in the imprinting center would contribute to the lack of the allelic expression of Igf2 in CNS.
To distinguish the two parental alleles, we used PCR to amplify M. spretus genomic DNA covering all four CTCF sites in the imprinting center. After comparing this sequence with the sequence of M. musculus in Genbank, we found three polymorphic sites located at or near CTCF binding sites 1–3. These polymorphisms were then used to separate allelic PCR products (Fig. 3A). No polymorphisms were found near CTCF binding site 4. Thus, we cannot distinguish potential allelic epigenetic differences at CTCF site 4.
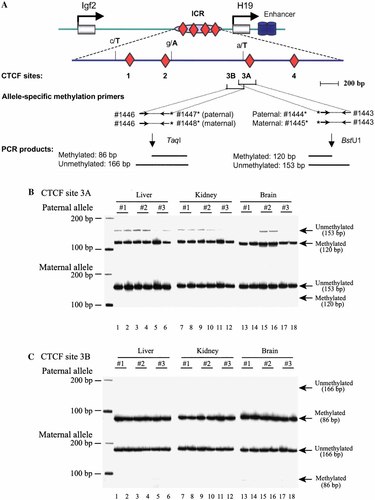
Allelic DNA methylation of Igf2/H19 imprinting center in CNS and peripheral tissues. A: CTCF sites and allele-specific methylation primers. Four CTCF sites were depicted as gray diamond. Three polymorphisms used to separate two parental alleles were depicted as the vertical line. Single nucleotide variations in M. spretus were presented in large bold letters to distinguish sequences in M. musculus (lower letters). B: Allelic DNA methylation at CTCF site 3A. C: Allelic DNA methylation at CTCF site 3B. Genomic DNA was extracted from brain, liver, and kidney and was treated with sodium bisulfite. Treated DNA was amplified with PCR primers covering two regions (3A and 3B) around the third CTCF binding site of the Igf2/H19 imprinting center. As described in Materials and Methods, one primer in each pair was polymorphic for two parental alleles and thus amplified either paternal or maternal allele. PCR products were then digested with BstUI (A) and Taq1 (B) to separate methylated and unmethylated DNA. Note in both regions the paternal allele was methylated and the maternal allele was unmethylated. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Genomic DNA extracted from CNS and peripheral tissues was treated by sodium bisulfite and the genomic DNA covering the CTCF binding sites was amplified by two sets of allelic PCR primers designed specifically for the A/T polymorphism near CTCF site 3 (Fig. 3A). These PCR products covered two CpG sites that were located, respectively, upstream (site 3B) and downstream (site 3A) of the A/T polymorphism. Parent-specific PCR products were then digested by restriction enzymes (BstU1 for site 3A and Tag1 for site 3B) to distinguish the unmethylated and methylated CpG as described in a COBRA method [Xiong and Laird, 1997]).
In liver and kidney, where Igf2 is monoallelically expressed, the expressed M. spretus paternal allele was completely methylated (Fig. 3B, lanes 1–12) and the imprinted M. musculus maternal allele was unmethylated at CTCF site 3 (Fig. 3B,C, lanes 1–12), in agreement with the CTCF insulator theory [Bell and Felsenfeld, 2000; Hark et al., 2000]. Surprisingly, we found that in CNS, where Igf2 was biallelically expressed, we could not find a correlation with Igf2 allelic expression as predicted from the CTCF insulation model. In CNS, the two parental alleles were also differentially methylated (Fig. 3B,C, lanes 13–18), showing the same pattern as seen in peripheral tissues (Fig. 3B,C, lanes 1–12).
Similarly, by using a G/T polymorphism near CTCF site 1 and a G/A polymorphism near CFCT site 2, we also demonstrated that genomic DNA at these two CTCF sites was also methylated in the expressed paternal allele and unmethylated in the imprinted maternal allele. There were no differences in CTCF site methylation between the non-imprinted CNS and the imprinted peripheral tissues (data not shown). Thus, the allelic DNA methylation at these CTCF binding sites are not sufficient to account for the lack of allele-specific silencing at the Igf2/H19 locus.
Allelic CTCF Binding in Igf2/H19 Imprinting Center
It has been proposed that CTCF insulators bind to CTCF sites at the unmethylated maternal allele and thus insulate the Igf2 promoters from accessing two enhancers that are shared with H19, leading to the monoallelic expression of Igf2. Because these CTCF sites were also differentially methylated in CNS as in liver and kidney, we were interested in whether CTCF binding in CNS would differ from that seen in the periphery.
CTCF-bound chromatin fragments were immunoprecipitated with anti-CTCF antisera and the associated genomic DNA was amplified by PCR. With the aid of polymorphic restriction enzymes, we were able to accurately quantitate the CTCF binding at the two parental alleles (Fig. 4A). Using a HapII polymorphism at CTCF site 1, we found that CTCF bound only to the maternal allele in liver and kidney (Fig. 4B, top panel, lanes 5–10). Interestingly, in CNS where Igf2 is not imprinted, CTCF factors also bound only to the maternal allele (Fig. 4B, top panel, lanes 11–16). No CTCF insulator binding was found in the paternal allele. Thus, in agreement with the allele-specific DNA methylation shown above, there was no differential CTCF binding between the non-imprinted CNS and the imprinted peripheral tissues.
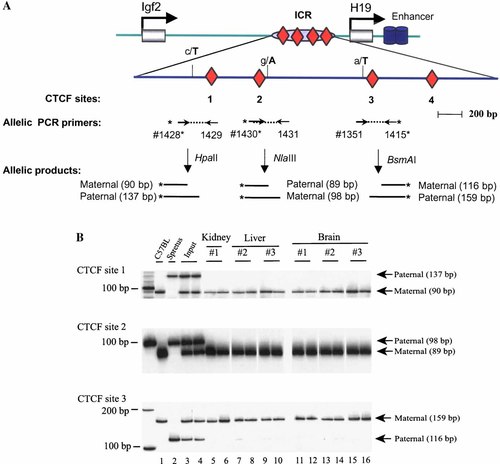
CTCF insulator binding in the Igf2/H19 ICR. A: Schematic presentation of four CTCF sites in the ICR and PCR primers used to amplify CTCF DNA sequences. Three polymorphic restriction enzymes are shown near three CTCF sites as in A. B: Allele-specific binding of CTCF at sites 1–3. As described in Materials and Methods, chromatin DNA was crosslinked with 1% formaldehyde and was sheared with sonicator. CTCF-bound chromatin fragments were immunoprecipitated with anti-CTCF antisera. The precipitated DNA was amplified with PCR primers covering CTCF sites 1–3 and was digested with HpaII or NlaIII or BsmA1 II polymorphic restriction enzymes to separate two parental alleles. Note CTCF insulator bound only to the maternal allele in both CNS and peripheral tissues. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
To confirm the above findings, we also used another two polymorphisms (NlaIII and BsmA1) to examine the CTCF binding for CTCF sites 2 and 3 (Fig. 4B, middle and bottom panel). Similarly, in both CNS (lanes 11–16) and peripheral tissues (lanes 5–10), CTCF insulators bound only to the unmethylated maternal allele, but not to the methylated paternal allele. Thus, binding of CTCF insulators at these CTCF sites in ICR cannot completely account for the lack of Igf2 imprinting in CNS.
It has been reported that a repressive domain created by the recruitment of the Sin3-Rpd3 histone deacetylase complex can be highly localized over a limited range of one to two nucleosomes [Kadosh and Struhl, 1998]. CTCF sties 1 and 2 are clustered together within a short distance in ICR (Fig. 4A). It is thus very hard to distinguish CTCF binding at these two sites by our ChIP method, which usually shears DNA into the size of 200–1,000 bp. However, CTCF sites 1 and 3, which are 1,287 bp apart from each other, still showed the same binding pattern (Fig. 4B). Thus, these four CTCF sites may act coordinately to mediate the allelic insulation of Igf2 as previously reported [Bell and Felsenfeld, 2000; Hark et al., 2000].
Histone Acetylation at CTCF Binding Sites
Core histones are epigenetically modified by acetylation, generating dynamic transitions between transcriptionally active or silent chromatin states. Differential allelic histone acetylation occurs frequently in several imprinted genes, in which histone hyperacetylation is always associated with the expressed allele.
We thus examined the status of histone acetylation of CTCF binding sites in F1 mice, hypothesizing that histone acetylation at these sites might contribute to differential Igf2 imprinting in CNS. With chromatin immunoprecipitation, we found that at the third CTCF binding site, histones H3 and H4 were acetylated predominantly but not exclusively at the maternal allele (Fig. 5A,B). However, we did not find any differences in the histone acetylation pattern between the imprinted liver and kidney and the non-imprinted CNS. We also obtained similar results at CTCF site 1 when immunoprecipitated DNA was amplified with the primer set of #1428 and #1429 (data not shown). These data thus suggest that this particular epigenetic modification at CTCF binding sites, although related to gene activity, may not be the primary mechanism underlying the lack of Igf2 imprinting in CNS.
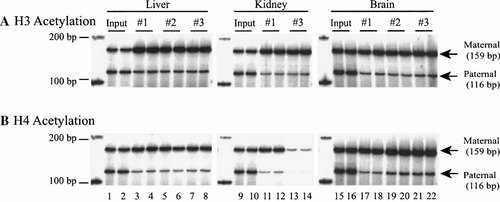
Status of histone acetylation of CTCF binding sites in Igf2/H19 imprinting center (duplicate for each sample). A: Histone H3 acetylation; (B) histone H4 acetylation. Chromatin fragments carrying acetylated histones were immunoprecipitated with anti-acetylated histones H3 and H4 antisera. Genomic DNA is the imprinting center was amplified with PCR. Two parental alleles were separated with a polymorphic BsmA1 restriction enzyme in M. spretus mice and scanned by PhosphoImager Scanner (Molecular Dynamics, Sunnyvale, CA).
Histone Methylation at CTCF Binding Sites
Core histones are also modified by methylation at lysines 4 and 9 of histone 3 (H3-K4 and H3-K9). These two epigenetic modifications also serve as a mechanism to regulate gene expression. We were thus interested to learn whether these histone modifications at CTCF sites might also serve as epigenetic marks to regulate the allelic expression of Igf2 and H19 between CNS and peripheral tissues.
Chromatin fragments containing methylated H3-K4 histones were immunoprecipitated with specific antisera. Genomic DNA at CTCF binding sites in the immunoprecipitates was amplified by PCR and digested with polymorphic restriction enzymes to distinguish the two parental alleles. As seen in Figure 6A, the methylated H3-K4 was predominantly associated with the maternal allele at CTCF site 3. However, there were no significant differences in allelic H3-K4 methylation between the imprinted peripheral tissues and non-imprinted CNS. Similar results were also obtained at CTCF site 1 when immunoprecipitated DNA was amplified with the primer set of #1428 and #1429 (data not shown).
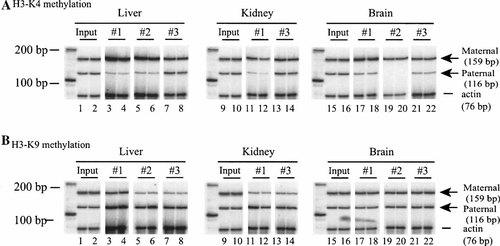
Allelic methylation of histone-3 lysines 4 (A) and 9 (B) in Igf2/H19 imprinting center (duplicate for each sample). The method was similar to that described in the figure, but chromatin fragments carrying methylated histones were immunoprecipitated anti-methylated histone-3 lysines 4 (H3-K4) and 9 (H3-K9) antisera and anti-methylated lysines 9 (K9) and 4 (K4) antisera, respectively. Note the differential histone methylation at H-K9 site between brain and peripheral tissues.
We then examined the status of H3-K9 methylation at CTCF site 3 (Fig. 6B). In liver and kidney, H3-K9 at CTCF site 3 was predominantly methylated at the paternal allele (paternal/maternal enrichment ratio of 3.9–6.7). In CNS, however, H3-K9 at CTCF site 3 was equally methylated at both of the parental alleles (paternal/maternal enrichment ratio of 0.9–1.1). The allelic methylation of H3-K9 is in concord with the pattern of Igf2 allelic expression. However, methylated H3-K9 is usually linked with the suppressed chromatin structure. It is thus not clear whether and how this biallelic H3-K9 methylation in CNS can be mechanically linked to the loss of Igf2 imprinting.
DISCUSSION
The current model suggests that the allelic expression of the two reciprocally imprinted genes Igf2/H19 is regulated by a common “enhancer competition” mechanism [Bartolomei et al., 1993]. DNA methylation at the 5′-end of H19, established in the male gamete in a paternal allele-specific manner [Bartolomei et al., 1993; Brandeis et al., 1993; Ferguson-Smith et al., 1993], serves as an imprinting signal or imprinting center [Brandeis et al., 1993]. The methylated paternal imprint functions to block the interaction of the paternal H19 promoter with the enhancers, which are then accessible to the paternal Igf2 gene. As a result, Igf2 is expressed and H19 is suppressed on the paternal chromosome. On the maternal chromosome, in contrast, unmethylated H19 competes with Igf2 for expression due to its access to the downstream enhancers. Deletion of the H19 enhancers alone [Leighton et al., 1995b] or the H19 gene plus enhancers [Leighton et al., 1995a] changes allelic expression of both genes, indicating that H19 and Igf2 utilize the same endoderm enhancers, but on different parental chromosomes.
The recent finding of the “boundary insulator” [Bell and Felsenfeld, 2000; Hark et al., 2000] between H19 and Igf2 has greatly extended the above enhancer competition theory. The insulator marks the boundary (ICR) between the H19 enhancer and the Igf2 promoter, and is differentially methylated on two parental alleles. A ubiquitous DNA binding factor, CTCF, can insulate the Igf2 promoter from the influence of a remote enhancer downstream of H19. The maternally derived copy of Igf2 is silenced by the binding in cis of CTCF to the ICR. Allelic methylation of the paternal ICR, however, abrogates the binding of CTCF, and thus allows the expression of H19 from the maternal allele and Igf2 from paternal allele (see reviews [Mann et al., 2000; Wolffe, 2000; Bell et al., 2001; West et al., 2002]). When the ICR is deleted in mice, the normally silent maternal allele of Igf2 is expressed [Thorvaldsen et al., 1998; Kaffer et al., 2000]. Biallelic hypermethylation of these CTCF sites in the human gene is correlated with the loss of IGF2 imprinting in colorectal cancers [Nakagawa et al., 2001]. A recent study by Tilghman's group [Schoenherr et al., 2003] has suggested that binding of CTCF is necessary to maintain, but not establish the imprint in the ICR.
CTCF insulators may also set the boundary effect on X-inactivation in mammals. X-inactivation silences one of two female X chromosomes and depends upon two overlapped yet oppositely transcribed genes (the sense Xist and the antisense Tsix) identified within the X-inactivation center (Xic). Xist, the gene that initiates the inactivation process, is only transcribed from the inactive X chromosome. The antisense Tsix, however, is transcribed only from the active X chromosome. CTCF binds to Tsix and coordinates the epigenetic switch for X-inactivation [Chao et al., 2002].
We [Hu et al., 1995; Pham et al., 1998] and others [Ohlsson et al., 1994; Pedone et al., 1994; Albrecht et al., 1996; Hemberger et al., 1998] have previously demonstrated that in both mouse and human, the Igf2 gene is biallelically expressed in CNS. In this tissue-specific loss of imprinting, all three mouse Igf2 promoters drive Igf2 expression from both parental alleles [Hu et al., 1995], showing a similar pattern as that observed in Wilms' tumors [Vu and Hoffman, 1996]. This finding would suggest that the aberrant regulation in the CTCF barrier region may be one of the mechanisms accounting for the loss of Igf2 imprinting in CNS. We hypothesized that in the CNS, the cis CTCF binding elements in the ICR would be epigenetically modified by DNA methylation differentially from those in peripheral tissues. According to the CTCF insulating model [Bell and Felsenfeld, 2000; Hark et al., 2000], we predicted that both parental alleles of this cis element would be hypermethylated (Fig. 1). As a result, the highly conserved zinc finger DNA binding protein CTCF would not bind to the four CTCF binding sites in the ICR and thus it would fail to insulate the utilization of common enhancers by Igf2 promoters, leading to the biallelic expression of Igf2 in CNS.
We were surprised to find that CTCF sites were not biallelically methylated in CNS as expected. After sodium bisulfite treatment, we found CTCF sites in the ICR were only methylated in the paternal allele, showing no differences from those in liver and kidney (Fig. 3). To confirm this finding, we also examined the in vivo binding of CTCF to the insulating element. In agreement with above finding, we found that CTCF proteins bound only to the unmethylated maternal allele in CNS (Fig. 4B). Again, we found no differences in CTCF protein binding in the ICR between the CNS and peripheral tissues. These data strongly suggest that, at least in CNS, the epigenetic regulation of Igf2/H19 imprinting may not be explained by CTCF insulation as simply as previously proposed. CTCF protein binding alone may not be sufficient to insulate the Igf2 promoters and to control the allelic expression of Igf2. We have also confirmed this finding in the human Igf2/H19 ICR (Ulaner et al, unpublished data). In addition, we found that in CNS where Igf2 was biallelically expressed, H19 was still monoallelically transcribed as in peripheral tissues (Fig. 2). Loss of the coordinated imprinting of Igf2 and H19 in CNS is also difficult to explain using the CTCF insulating model. Thus, lack of Igf2 imprinting can occur in CNS in a manner that is independent of H19 allelic expression and CTCF epigenetic modification.
Our finding has been supported by the report from Ishizaki et al. [2002], who recently discovered that the loss of Igf2 imprinting in tumor cells was not necessarily linked to abnormal DNA methylation. They isolated four cell clones from mouse hepatic tumors showing loss of Igf2 imprinting and found H19 imprinting was well maintained. Interestingly, these cell lines still maintained normal differential DNA methylation at the Igf2 DMRs and at H19 and Kvlqt1 DMRs as well. Similarly, Feinberg et al. [Cui et al., 2001, 2002] also found that methylation of the CTCF core consensus site was not sufficient to account for the LOI of IGF2 in Wilms' tumors and colorectal cancers. They found that tumors with normal IGF2 imprinting were also biallelically hypermethylated or hypomethylated at CTCF sites. Interestingly, some tumors with loss of IGF2 imprinting showed a normal semi-methylated pattern as those in normal tissues.
The explanation for these exceptions to the CTCF insulation model is unknown. One factor might be that binding of CTCF insulators to the ICR is not sufficient to insulate the Igf2 promoters. Other factors, such as putative CTCF-associated proteins may be needed to coordinate the Igf2 insulation. A recent report by Lutz et al. [2000] has shown that one of the zinc-finger clusters of the CTCF protein binds directly to the co-repressor Sin3A, suggesting that CTCF-driven gene repression may be mediated partly by the recruited histone deacetylase activity. Another possible explanation may be that CTCF may interact with factors that bind to a region close to Igf2 promoters and thus it may prevent the enhancers from activating Igf2 promoters. Differential loss of these CTCF-associated factors may be the real cause accounting for the biallelic expression of Igf2 in CNS and in tumors.
To test this possibility, we thus used a gel-retardation assay to examine other putative factors that may be involved in Igf2 imprinting. We prepared the probe containing CTCF binding site 3 and incubated with nucleic proteins extracted from tissues that showed differential Igf2 imprinting, hypothesizing we might see different CTCF binding patterns between these tissues due to the presence of CTCF-associated proteins. We amplified a 159 bp fragment covering the third CTCF site with a γ-[32P]-ATP labeled primer. After purification, the γ-[32P]-ATP labeled probe was incubated with nuclear proteins extracted form both CNS and peripheral tissues. The interaction of the CTCF-associated protein with CTCF would lead to the retardation of the probe. If these putative CTCF-associated factors are differentially expressed, we should be able to see a differential retardation pattern between the imprinted peripheral tissues and the non-imprinted CNS.
However, data from the gel retardation assay did not support this hypothesis (data not shown). We found that a single shifted band that represents CTCF binding was observed in peripheral tissues (data not shown) in complete agreement with previous reports [Bell and Felsenfeld, 2000; Hark et al., 2000]. In CNS, we also found that there was a single band that shifted to the same level as that in peripheral tissues (data not shown). These results thus provide no evidence for the presence of CTCF-associated proteins that account for the lack of genomic imprinting in CNS, although we cannot completely eliminate the possibility of the interaction of small proteins that cannot be detected by the assay.
Another possibility is that CTCF binding in ICR is just one of many key components of the Igf2 imprinting regulatory machinery. After binding to ICR, CTCF insulators may directly interact with other DNA regions (e.g., DMR). Differential binding at those DMR regions may be the underlying mechanism for the lack of Igf2 imprinting in CNS. It could be also possible that Igf2/H19 ICR as the primary imprint, although being critical for the establishment of Igf2 imprinting as previously demonstrated [Thorvaldsen et al., 1998; Kaffer et al., 2000; Schoenherr et al., 2003], may actually not be essential for the maintenance of Igf2 imprinting. Mechanisms other than aberrant CTCF binding, which maintains Igf2 imprinting, may contribute to the lack of Igf2 imprinting in CNS and in some human tumors. This hypothesis has been supported by the data provided by Feinberg and his colleagues [Cui et al., 2002], who failed to link enhancer competition directly to the LOI of IGF2 in colorectal cancers. Instead, they found a strong correlation of IGF2 LOI with the methylation status in IGF2 DMR0 a region that is methylated specifically on the maternal allele in both human [Sullivan et al., 1999]and mouse [Moore et al., 1997]. Loss of allele-specific DNA methylation at DMR0 is related to the LOI of IGF2 in Wilm's tumors [Sullivan et al., 1999]. Genomic deletion covering the DMR0 region leads to the activation of the suppressed maternal allele [Hu et al., 1997b; Constancia et al., 2000]. A brain-specific enhancer at the 5′ of the ICR is also important for the allelic expression of Igf2 [Jones et al., 2001].
Histones are also modified by methylation at lysines 4 and 9 of histone 3 (H3-K4 and H3-K9). These two epigenetic modifications also serve as a mechanism to regulate gene expression. The methylated H3-K9 interacts with heterochromatin-associated regions [Bannister et al., 2001; Nakayama et al., 2001; Noma et al., 2001; Nielsen et al., 2002] and correlates with gene silencing [Bannister et al., 2001; Nielsen et al., 2001] and X chromosome inactivation [Heard et al., 2001; Boggs et al., 2002]. In contrast, H3 methylation at lysine 4 (H3-K4) is specific to the surrounding euchromatic regions [Noma et al., 2001] and is associated with gene expression. We were thus interested in whether this epigenetic mark is related to the differential imprinting of Igf2 between CNS and peripheral tissues.
Using a chromatin-immunoprecipitation assay, we found histone methylation at H3-K9, but not H3-K4 at CTCF binding sites was closely related to the allelic expression of Igf2. In liver and kidney where Igf2 is mono-allelically expressed, H3-K9 was methylated primarily at the paternal allele (Fig. 6B), showing a similar methylation pattern as its interacting DNA (Fig. 3). In CNS where Igf2 is biallelically expressed, H3-K9 was equally methylated at both parental alleles in all samples tested (Fig. 6B). The methylated H3-K9 usually correlates with gene silencing. However, whether and how this histone epigenetic modification at these CTCF sites plays an important role in regulating the allelic expression of the distant Igf2 is not clear. It would be interesting to test whether such a single epigenetic modification at H3-K9 alters the local chromatin structure at CTCF binding sites and thus overrides the insulating effect of CTCF proteins on Igf2 expression.
The methylated H3-K9 is usually associated with the suppressed gene. However, we found that the expressed β-actin was associated with H3-K9 methylation (Fig. 6B). We also observed the same pattern for β-actin in a separate study with the imprinted Igf2r (Yang et al, unpublished data). Similarly, Igf2 is biallelically expressed in CNS but is also linked with H3-K9 biallelic methylation (Figs. 2 and 6B). It would be expected that H3-K9 should be biallelically unmethylated while H3-K4 be biallelically methylated in CNS, where Igf2 is biallelically expressed. Thus, H3-K9 methylation may also be associated with the expressed gene depending upon the local chromatin structure.
Histones can be modified by lysine mono-, di-, or tri-methylation. There is a concern that antibodies raised against the dimethylated H3 tail may share little cross-reactivity with antibodies raised against the equivalent trimethylated H3, and vice versa [Orlando and Jones, 2002]. In this communication, we used an antibody directed against dimethylated H3 in the ChIP assay. It is not clear whether antibodies against tri-methylated H3 will give the same results. In a separate study of two DMR regions in Igf2r, however, we found that there were no differences in histone methylation when antibodies against mono-, di-, or tri-methylated H3-K4 and H3-K9 were used with the ChIP method (Yang et al, unpublished data).
Nucleosome core histones are dynamically modified by acetylation, which in turn dictate dynamic transitions between transcriptionally active or silent chromatin states. Proteins that specifically bind to methylated DNA, like MeCP2, MBD2, and MBD3, are associated with complexes that include histone deacetylases (HDACs). Recent studies have focused on the role of histone hypoacetylation in the maintenance of centromeric structure, X-inactivation, and genomic imprinting. Differential allelic histone acetylation occurs frequently in several imprinted genes, including H19 and Igf2 [Pedone et al., 1999; Grandjean et al., 2001], Igf2r [Hu et al., 2000; Fournier et al., 2002], SNRPN [Saitoh and Wada, 2000; Fournier et al., 2002], and Xist [Keohane et al., 1999; O'Neill et al., 1999]. However, we did not demonstrate a significant alteration of histone acetylation at these CTCF sites between the imprinted and non-imprinted tissues (Fig. 6). Thus, histone H3 and H4 acetylation at these CTCF binding sites, although correlated with the transcriptional activity of parental promoters of Igf2, is not a major determinant accounting for the loss of Igf2 imprinting in CNS.
It is also interesting to examine H3-K9 methylation and acetylation in the brain. As seen in Figure 5, the maternal imprinting center in brain is predominantly associated with acetylated H3. The antibodies against methylated H3-K9 used in the ChIP assay also co-immunoprecipitated with the maternal ICR (Fig. 6B). Recently, we (Yang et al, unpublished data) and others [Fournier et al., 2002] have observed similar results for the Igf2r DMR regions. Thus, these two types of epigenetic modification may not necessarily function antagonistically but rather occur on nearby H3 molecules in nucleosomes associated with the maternal ICR, as observed by Maison et al. [2002] in pericentric heterochromatin. However, whether the co-existence of histone methylation and acetylation at H3-K9 is mechanically linked to the regulation of genomic imprinting awaits further investigation.
In summary, our data suggest that allele-specific DNA methylation and binding of CTCF insulators in the ICR may not be the sole factor to determine the imprinting status of Igf2. Instead, factors that are involved in histone epigenetic modifications as well as CTCF-associated proteins may be equally important in the regulation of genomic imprinting. Determining what histone modifications (“histone code”) are allele-specific in all examples will bring us closer to an understanding of the regulation of the imprinting process.




