COLI-Net: Deep learning-assisted fully automated COVID-19 lung and infection pneumonia lesion detection and segmentation from chest computed tomography images
Isaac Shiri and Hossein Arabi contributed equally to this study.
Funding information: Swiss National Science Foundation, Grant/Award Number: SNRF 320030_176052; WOA Institution: Universite de Geneve; Blended DEAL: CSAL
Abstract
We present a deep learning (DL)-based automated whole lung and COVID-19 pneumonia infectious lesions (COLI-Net) detection and segmentation from chest computed tomography (CT) images. This multicenter/multiscanner study involved 2368 (347′259 2D slices) and 190 (17 341 2D slices) volumetric CT exams along with their corresponding manual segmentation of lungs and lesions, respectively. All images were cropped, resized, and the intensity values clipped and normalized. A residual network with non-square Dice loss function built upon TensorFlow was employed. The accuracy of lung and COVID-19 lesions segmentation was evaluated on an external reverse transcription-polymerase chain reaction positive COVID-19 dataset (7′333 2D slices) collected at five different centers. To evaluate the segmentation performance, we calculated different quantitative metrics, including radiomic features. The mean Dice coefficients were 0.98 ± 0.011 (95% CI, 0.98–0.99) and 0.91 ± 0.038 (95% CI, 0.90–0.91) for lung and lesions segmentation, respectively. The mean relative Hounsfield unit differences were 0.03 ± 0.84% (95% CI, −0.12 to 0.18) and −0.18 ± 3.4% (95% CI, −0.8 to 0.44) for the lung and lesions, respectively. The relative volume difference for lung and lesions were 0.38 ± 1.2% (95% CI, 0.16–0.59) and 0.81 ± 6.6% (95% CI, −0.39 to 2), respectively. Most radiomic features had a mean relative error less than 5% with the highest mean relative error achieved for the lung for the range first-order feature (−6.95%) and least axis length shape feature (8.68%) for lesions. We developed an automated DL-guided three-dimensional whole lung and infected regions segmentation in COVID-19 patients to provide fast, consistent, robust, and human error immune framework for lung and pneumonia lesion detection and quantification.
1 INTRODUCTION
The recent pandemic of severe acute respiratory syndrome coronavirus 2 disease (COVID-19) is posing great health concerns globally.1, 2 The COVID-19 pandemic has resulted in loss of lives, health, and economic issues.3 Although a large number of trials have been conducted to produce vaccines and/or treat COVID-19, a specific vaccine or therapy is still lacking.4, 5 For the diagnosis of COVID-19, reverse transcription-polymerase chain reaction (RT-PCR) is a high sensitive molecular test, but bears inherently a number of limitations.6, 7 Furthermore, previous studies have indicated that thoracic computed tomography (CT) is a fast and highly sensitive approach for COVID-19 detection and management.8, 9 In this regard, dedicated ultralow-dose CT scanning protocols were recently devised.10
In connection with the use of CT in COVID-19 management, a wide range of qualitative and quantitative studies have been carried out for diagnostic, prognostic and longitudinal follow-up of patients.11-13 In these studies, whole lungs or infectious lesions were analyzed and several patterns and features were found to have high diagnostic and prognostic value.13-18 However, accurate segmentation of lungs and infectious pneumonia lesions remains challenging.19 Hence, segmentation is the main issue impacting the outcome of both qualitative and quantitative studies.12, 19, 20 Although several segmentation approaches including manual delineation, semiautomated21 and fully automated20 techniques have been applied to CT images for COVID-19 management, they are still facing serious challenges to produce robust and dependable outcomes.
In medical image segmentation, particularly whole three-dimensional (3D) volumes definition and big data analysis, manual delineation requires experienced trained radiologists, is time consuming, labor-intensive, and suffers from interobserver and intraobserver variability concerns.22, 23 Whole lung segmentation is a pivotal step for further analysis, including extraction of the percentage of infection, well aerated portion of the lung, and enabling radiomics and deep learning (DL) analysis of COVID-19 patients.14, 17 Conventional algorithms, including rule-based and atlas-based, performed relatively well on normal and mild disease chest CT, but might fail in COVID-19 patients lung segmentation because of different stages of disease with different levels of severity.19 Furthermore, developing a fully automatic tool for lung and pneumonia COVID-19 lesions is highly desired owing to rapid changes in appearance and manifestation at different stages of the disease.13, 19
Artificial intelligence (AI) algorithms, particularly its two major subcategories, machine learning (ML), and DL, have been widely used for medical image analysis24-31 and more recently in the segmentation of lung and pneumonia infectious lesions from chest CT images of COVID-19 patients.15 These studies reported that AI improved the accuracy of lesion detection/segmentation and reduced the bias associated with conventional approaches. In a study by Zheng and coworkers,32 a weakly supervised DL algorithm was applied to chest CT images for automatic COVID-19 detection. Fan et al.33 presented a COVID-19 lung infection segmentation deep network (Inf-Net) based on semisupervised learning. Furthermore, a number of DL algorithms, namely UNet, UNet++, V-Net, Attention-UNet, Gated-UNet, and Dense-UNet were used for COVID-19 lesion detection and segmentation from chest CT images.34, 35
CT images are commonly acquired on various scanner models using different imaging protocols, and as such, the resulting datasets are heterogeneous, which might lead to inaccuracy in the developed models. Training a robust and generalizable DL model requires a large clean annotated dataset.36 Owing to the relatively recent outbreak of COVID-19 pandemic, producing a large labeled COVID-19 image dataset is impractical. Transfer learning (TL) has received attention to address the lack of large datasets for the implementation of machine/DL-based algorithms.37, 38 Various TL-based strategies were used for transferring knowledge from different domains, including natural images to medical images to develop more robust and generalizable models.38
In the present study, we developed a DL-based automated detection and segmentation of lung and COVID-19 pneumonia infectious lesions (COLI-Net) from chest CT images. In this work, large lung and COVID-19 lesions datasets and TL used to train a residual network (ResNet) for lung and pneumonia infectious lesions segmentation.
2 MATERIALS AND METHODS
2.1 Clinical studies
For lung and COVID-19 lesions segmentation, we prepared 2368 (347 259, 2D slices) and 190 (17 341, 2D slices) multicentric and multivendor volumetric CT images with lung and COVID-19 lesion segmentations.
2.2 Lung datasets
For lung segmentation training, we used 2298 chest CT exams (328′205, 2D slices) with different pathologies from different centers, including 800 exams of normal subjects without any lung abnormalities from Iran Center#1 (81′347, 2D slices); 400 images of non-small cell lung carcinoma patients from Cancer Imaging Archive (TCIA)39-41 (48′568, 2D slices); 200 non-COVID-19 pneumonia (49′465, 2D slices); and 898 (148′825, 2D slices) RT-PCR positive COVID-19 patients from Iran Center#2. All lung segmentations were performed using a region-growing algorithm followed by manual verification and amendment by an experienced radiologist.
2.3 COVID-19 lesions datasets
For COVID-19 lesions segmentation training, we used 120 (9557, 2D slices) RT-PCR positive image datasets, including 90 (8338, 2D slices) datasets from three different centers in Iran (Centers#1, #2, #3) where the infectious lesions were manually segmented by experienced radiologists, in addition to 30 (1250, 2D slices) CT exams from Russia.42 Lesions were segmented manually for the local dataset whereas these segmentations were provided by data providers for the external dataset.42-44
2.4 Image preprocessing
Prior to network training, all images were cropped without losing important information (parts of lungs) and resized to 296 × 216 matrix size. In the first step of intensity normalization, voxel intensities in the entire dataset were clipped between −1024 and 300 HUs to reduce the dynamic range of the intensity of CT images. This range of HUs covers air, lung tissue, fat, soft-tissue, and calcifications in the lung. Only bony structures will be suppressed, which are irrelevant to lung and lesion segmentations. Hence, we found this range of HUs optimal for ML-based lung and lesion segmentation. Moreover, to further reduce the dynamic range of voxel intensities, CT images were normalized with an empiric factor equal to 1000 to keep the original dynamic intensity range and put the bulk of CT intensity within the range of 0–1 HU.
2.5 Residual neural network
The ResNet proposed by Li et al.45, 46 built upon TensorFlow was used for lung and COVID-19 lesions segmentation. The ResNet is composed of 20 convolutional layers where different dilation factors were used for different levels of feature extraction (zero dilatation factor for low-level, two dilatation factors for medium-level, and four dilatation factors for high level). Every two layers were linked together with residual connections (Figure 1). Non-square Dice was used as loss function, and Figure 1 provides descriptive detail of ResNet.
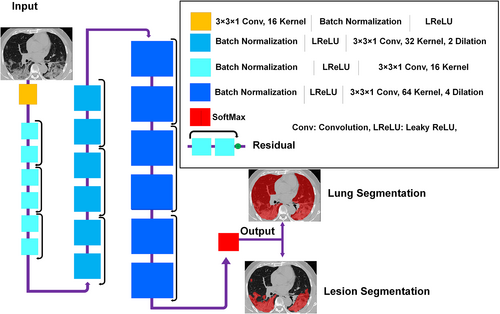
2.6 Training and evaluation
Lung and COVID-19 lesions training was performed on 2D slices owing to the wide variability in slice thicknesses across the datasets from the different centers. We used the following hyperparameters for model training: loss function = non-square Dice, learning rate = 0.001, optimizer = Adam, decay = 0.0001, batch size = 32, and weights regression type = L2norm, drop out = 0.5, and number of epochs = 300. For lung segmentation training, we used 2178 3D CT images (347 259, 2D slices). For COVID-19 lesions segmentation, we used pretrained lung segmentation network as initial weights followed by fine-tuning for lesion segmentation of 120 3D CT images (9557, 2D slices). Body fine-tuning approaches were used for TL where all pretrained weights of lung segmentation were used as initial weights for lesion segmentation. The quantitative assessment of segmentations was performed independently on RT-PCR positive COVID-19 datasets from different centers, including 20 CT exams (2214, 2D slices) from Center#1 (Iran1); 10 exams (2552, 2D slices) from Center#2 (Iran2); 20 exams (1250, 2D slices) from center#3 (Russia)42; 10 exams (939, 2D slices) from Center#4 (China)43, 44; and 10 exams (829, 2D slices) from center#5 (Italy*).44 Training datasets were split into training (80%) and validation (20%) sets. Overall, the evaluation was performed on 7333 2D slices from different centers. Data splitting into training and test sets was performed based on 3D image of patients without overlap between the training and test sets. All evaluations were performed in 3D mode.
2.7 Evaluation
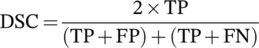 (1)
(1) (2)
(2) (3)
(3) (4)
(4)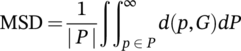 (5)
(5)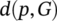 is the distance between a point p belonging to the surface of a 3D surface predicted image (P) and its closest distance between the two surfaces P and G (ground truth).
is the distance between a point p belonging to the surface of a 3D surface predicted image (P) and its closest distance between the two surfaces P and G (ground truth).
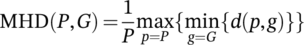 (6)
(6)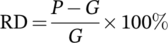 (7)
(7)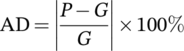 (8)
(8)3 RESULTS
Figures 2 and 3 compares visually in 2D and 3D views for different external validation sets of lungs and lesions delineated manually by experienced radiologists and automatically by the DL model. Additional results from the external validation sets are provided in Supplemental Figures 1–13 (2D views) and 14–17 (3D views). Overall, there is good agreement between manual and predicted lung and infectious lesions segmentation in the different datasets. Despite the variability of the subjects among the different centers, COLI-Net performed consistently well in multicentric and multiscanner setting. What stands out from these results is that COLI-Net can detect and segment infectious regions (within lesion segmentation) while excluding arteries and tracheae in lung segmentation.
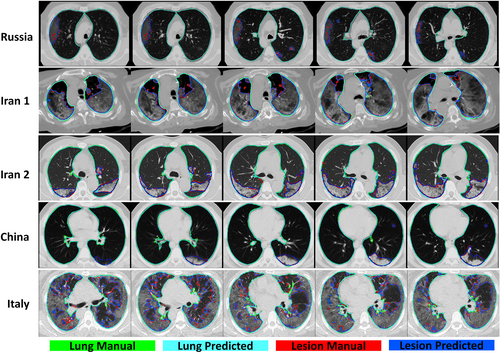
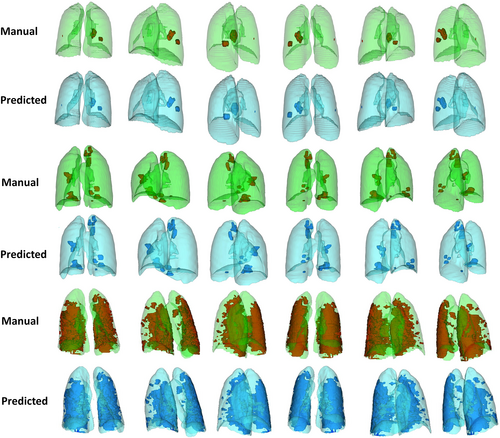
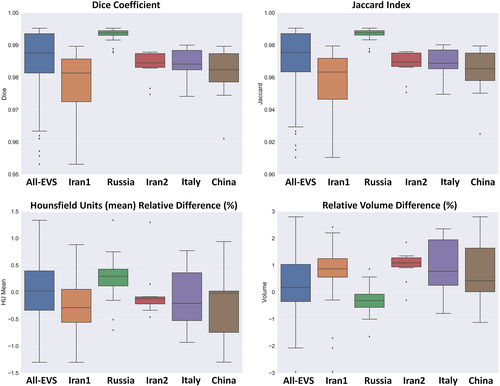
Table 1 summarizes segmentation quantification metrics for lungs and COVID-19 lesions. It can be seen that the mean Dice coefficients were 0.98 ± 0.011 (95% CI, 0.98–0.99) and 0.91 ± 0.038 (95% CI, 0.90–0.91) for lung and lesions segmentation, respectively. The mean Jaccard index was 0.97 ± 0.022 (95% CI, 0.97–0.97) and 0.83 ± 0.062 (95% CI, 0.82–0.84) for lung and COVID-19 lesions segmentation, respectively. Lung segmentation in Russia datasets exhibited better results compared to the other centers/datasets. This might be attributed to the homogeneity and mild severity of the lesions in this dataset. Supplemental Tables 2–7 summarize lung and lesion segmentation quantification metrics for different external validation sets.
| Metric | Min | Max | Mean ± SD | 95% CI | |
|---|---|---|---|---|---|
| Lung | Dice | 0.92 | 0.99 | 0.98 ± 0.011 | 0.98–0.99 |
| Jaccard | 0.86 | 0.99 | 0.97 ± 0.022 | 0.97–0.97 | |
| False negative | 0.003 | 0.086 | 0.013 ± 0.011 | 0.011–0.015 | |
| False positive | 0.002 | 0.073 | 0.017 ± 0.014 | 0.014–0.019 | |
| Average Hausdorff distance | 0.005 | 0.14 | 0.022 ± 0.026 | 0.018–0.027 | |
| Mean surface distance | 0.005 | 0.17 | 0.026 ± 0.028 | 0.021–0.031 | |
| Lesions | Dice | 0.8 | 0.98 | 0.91 ± 0.038 | 0.9–0.91 |
| Jaccard | 0.66 | 0.96 | 0.83 ± 0.062 | 0.82–0.84 | |
| False negative | 0.015 | 0.23 | 0.086 ± 0.044 | 0.078–0.094 | |
| False positive | 0.024 | 0.32 | 0.098 ± 0.055 | 0.088–0.11 | |
| Average Hausdorff distance | 0.043 | 5.6 | 0.42 ± 0.73 | 0.29–0.55 | |
| Mean surface distance | 0.046 | 6.1 | 0.45 ± 0.79 | 0.31–0.59 |
Table 2 summarizes the impact of lung and lesions segmentations on mean Hounsfield unit and volume calculation. Mean relative HU differences (%) of 0.03 ± 0.84 (95% CI, −0.12 to 0.18) and −0.18 ± 3.4 (95% CI, −0.8 to 0.44) were achieved for lungs and lesions, respectively. The relative volume difference for the lung was 0.38 ± 1.2 (95% CI, 0.16–0.59) whereas it was 0.81 ± 6.6 (95% CI, −0.39 to 2) for lesions. The results obtained from the mean Hounsfield unit and volume calculation for lung and infectious lesions for the different external validation sets are presented in Supplemental Tables 8–11.
| Metric | Min | Max | Mean ± SD | 95% CI | |
|---|---|---|---|---|---|
| Lung | Relative mean HU diff (%) | −4.2 | 3.9 | 0.03 ± 0.84 | −0.12 to 0.18 |
| Absolute relative mean HU diff (%) | 0.006 | 4.2 | 0.52 ± 0.66 | 0.4–0.64 | |
| Relative volume diff (%) | −3.1 | 6.4 | 0.38 ± 1.2 | 0.16–0.59 | |
| Absolute relative volume diff (%) | 0.004 | 6.4 | 0.89 ± 0.88 | 0.73–1 | |
| Lesions | Relative mean HU diff (%) | −9.8 | 10 | −0.18 ± 3.4 | −0.8 - 0.44 |
| Absolute relative mean HU diff (%) | 0.026 | 10 | 2.4 ± 2.5 | 1.9–2.8 | |
| Relative volume diff (%) | −14 | 21 | 0.81 ± 6.6 | −0.39 to 2 | |
| Absolute relative volume diff (%) | 0.018 | 21 | 4.8 ± 4.6 | 4–5.6 |
Figures 4 and 5 depict the Dice similarity index, Jaccard, mean Hounsfield unit, and volume difference box plots for lung and lesions segmentation, respectively. Supplemental Figures 18 and 19 show box plots of Hounsfield unit absolute relative difference (%), absolute relative volume difference (%), false negative, false positive, average Hausdorff distance, and mean surface distance for lung and lesions.
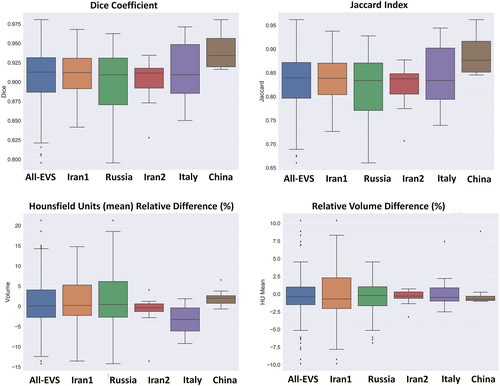
Descriptive statistics of relative volume (lesion/lung) indices are presented in Table 3. A relative error of 0.22 ± 6.3 (95% CI, −0.95 to 1.4) and absolute relative error of 4.7 ± 4.2 (95% CI, 3.9–5.5) were achieved for relative volume (lesion/lung). Supplemental Tables 12 and 13 summarize the results obtained for the relative volume (lesion/lung) index for different external validation sets. Figure 6 depicts boxplot of manual and predicted relative volume lesion/lung differences (%) and absolute/relative error of lesion/lung relative volume errors (%) for different external validation sets.
| Metric | Min | Max | Mean ± SD | 95% CI |
|---|---|---|---|---|
| Manual segmentation relative volume (lesion/lung) | 0.001 | 0.82 | 0.13 ± 0.19 | 0.095–0.16 |
| Predicted segmentation relative volume (lesion/lung) | 0.001 | 0.84 | 0.13 ± 0.19 | 0.094–0.16 |
| RE volume diff lesion/lesion (%) | −14 | 16 | 0.22 ± 6.3 | −0.95 to 1.4 |
| ARE volume diff lesion/lesion (%) | 0.004 | 16 | 4.7 ± 4.2 | 3.9–5.5 |
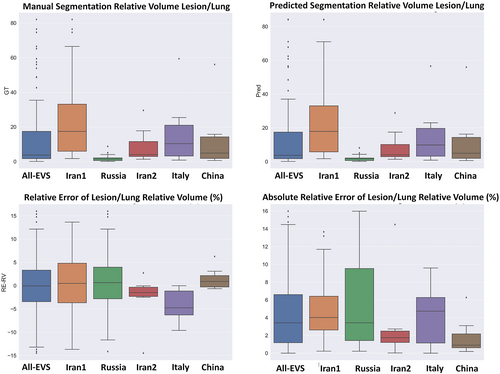
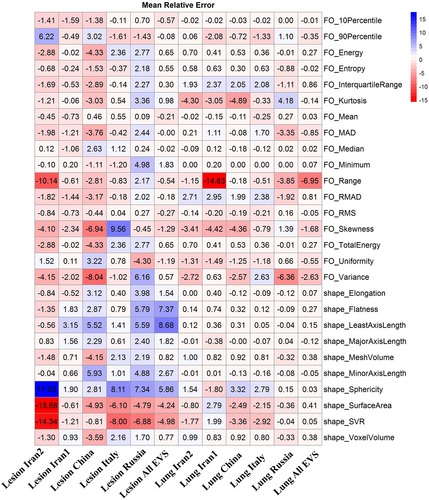
Figure 7 presents heatmap of the mean relative error of first-order and histogram shape radiomic features in the lung and lesions for different validation sets. Most radiomic features exhibited a mean relative error less than 5% with the highest mean relative error for the lung being −6.95% for range first-order feature and least axis length shape feature (8.68%) in lesions. The heatmap of the mean absolute relative error is depicted in Supplemental Figure 20.
4 DISCUSSION
Chest CT imaging has emerged as a complementary tool for COVID-19 early diagnosis and longitudinal follow-up.8 However, a number of challenges still need to be addressed for the accurate diagnosis of COVID-19 and its differentiation from other lung diseases, such as viral and bacterial pneumonia and other respiratory diseases.17 In this regard, several AI-based solutions exhibiting different levels of accuracy and robustness were proposed and evaluated.14, 17
Another challenging problem that arises in the domain of quantitative analysis of CT images in clinical practice is lung and pneumonia infectious lesions segmentation.19 At the outset, different complex manifestations (appearance, size, location, boundaries and contrast) of infectious lesions, including consolidation, reticulation, and ground-glass opacity at different stages of the disease (longitudinal changes in the same patients) have been observed.13 Furthermore, providing ground truth segmentation for infectious lesion segmentation is challenging owing to interobserver/intraobserver variability, noisy annotations, and the long processing time.19
Previously developed atlas,47 rule,48 and hybrid (atlas and rule)49 based algorithms for lung segmentation have shown acceptable performance on normal lungs and in the presence of mild pathogens (low density), such as emphysema.50 However, they presented limited performance in severe conditions (high density), including pleural effusion, atelectasis, consolidation, fibrosis, and pneumonia.51 Recent developments in the field of ML have led to a renewed interest in automatic lung segmentation. However, most seminal works in this area used a limited training dataset, predominantly containing normal cases or focusing on one class of pathogeneses, which could impact generalizability for unseen/non-diagnosed test datasets.52 In the present study, we applied DL algorithms and TL on CT images obtained from different imaging centers to detect and segment the whole lung and pneumonia infected regions in COVID-19 patients.
A number of previous works attempted to develop automated segmentation algorithms for lung and infectious lesions in COVID-19 CT images. Hofmanninger et al.51 developed models for lung segmentation and reported a Dice coefficient of 0.98 ± 0.01 for different pathological states (atelectasis, fibrosis, mass, pneumothorax, and trauma). They concluded that diversity in the training dataset is more important than the DL algorithms. Müller et al.53 implemented a 3D U-Net using data augmentation for generating image patches during training for lung and lesion segmentation on 20 annotated CT volumes. They achieved Dice coefficients of 0.950 and 0.761 for lung and lesions, respectively. A modified 3D U-Net (feature variation and progressive atrous spatial pyramid pooling blocks) proposed by Yan et al.34 was developed for lung and infectious lesion segmentation on 861 patients, reporting a Dice similarity index of 0.987 for lung and 0.726 for lesions segmentation. Moreover, comparisons were performed with a dense fully convolutional network (lung: 0.865, lesions: 0.659)54; U-Net (lung: 0.987, lesions: 0.688)55; V-Net (lung: 0.983, lesions: 0.625)56; and U-Net++ (lung: 0.986, lesions: 0.681).57 The mean Dice coefficient for lung and lesions segmentation for different external validation sets used in our work were 0.98 ± 0.011 and 0.91 ± 0.038, respectively.
Chen et al.58 used the residual attention U-Net for multi-class segmentation of CT images, achieving a Dice coefficient of 0.94 for infectious lesions segmentation. Zhou et al.35 used a modified U-net network through spatial and channel attention mechanisms along with focal Tversky loss in the training process for improving small lesions segmentation. The results were evaluated on 427 slices achieving a Dice coefficient of 0.83. Elharrouss et al.59 adopted an encoder-decoder for infectious lesions segmentation using 20 clinical studies from the Italian Society of Medical and Interventional Radiology to report a Dice coefficient of 0.786. They compared the results with U-Net (Dice: 0.439),60 Attention-UNet (Dice: 0.583),61 Gated-UNet (Dice: 0.623),62 Dense-UNet (Dice: 0.515),63 U-Net++ (Dice: 0.422),57 and Inf-Net (Dice: 0.739).33 Wang et al.64 proposed a robust algorithm for COVID-19 infectious lesions segmentation from CT images (COPLE-Net) designed to learn from noisy labeled data. The algorithm relies on noise-robust Dice loss and mean absolute error loss for generalized Dice loss for robust segmentation of noisy datasets and a modified version of U-Net to better handle infectious lesion segmentation with various manifestations and scales. The best results achieved by COPLE-Net were 0.807 ± 0.099 and 0.160 ± 0.171% as Dice coefficient and relative volume error (RVE [in %]) respectively. Wang et al.64 evaluated different DL algorithms, including modified 3D U-Net (3D New-Net U-Net, Dice: 0.704 ± 0.187, RVE: 25.41 ± 24.73%),65 modified 2D U-Net (2D New-Net U-Net, Dice: 0.791 ± 0.129, RVE: 18.37 ± 17.43%),65 spatial attention gate U-Net (Attention U-Net, Dice: 0.772 ± 0.123, RVE: 19.77 ± 18.41%),61 spatial and channel “squeeze and excitation” blocks with U-net (ScSE U-Net, Dice: 0.780 ± 0.125, RVE: 18.85 ± 16.69%),66 and light-weight power efficient and general purpose CNN (ESPNetv2, Dice: 0.698 ± 0.148, RVE: 23.69 ± 20.26%).67 Our proposed COLI-Net approach showed good performance compared to previous studies with a Dice coefficient of 0.91 ± 0.038 (95% CI: 0.90–0.91) and RVE of 0.38 ± 1.2% (95% CI: 0.16–0.59) for pneumonia infectious lesions.
A large labeled dataset is required to build a robust and generalizable model while avoiding overfitting. Previous studies attempted to transfer the knowledge from natural to medical imaging domain, leading to improved accuracy by addressing the issue of limited datasets.37, 38 TL was recently applied for the detection and classification of COVID-19 using chest x-ray and CT images.68, 69 More recently, Wang et al.70 applied four TL methods on COVID-19 CT images for the segmentation of infectious lesions using 3D U-Net. The information was transformed from cancer and pleural effusion data to COVID-19 lesion segmentation. The Dice coefficient increased from 0.673 ± 0.22 to 0.703 ± 0.20 after TL.70 They concluded that the transferability of non-COVID-19 data improved the quality of COVID-19 lesion segmentation to build a robust segmentation model. In our study, we exploited TL from a large multicentric lung labeled dataset with various pathologies to overcome the shortcomings of infectious lesion segmentation.
Li et al.71 used thick-section chest CT images of 531 COVID-19 patients for automatic segmentation of lesions using 2.5D U-net to achieve Dice coefficients of 0.74 ± 0.28 and 0.76 ± 0.29 with respect to manual delineation performed by the two radiologists. The interobserver variability measured by the Dice metric was 0.79 ± 0.25 between two radiologists. They calculated two imaging biomarkers, including the percentage of infection and average infectious HU for severity and progression assessment, resulting in AUC of 0.97. Thick-section CT imaging was recommended for high-pitch scans to decrease the acquisition time and motion artifacts (due to breath holding) and reduce radiation doses to patients.10, 72 In our dataset, various slice thicknesses (1–8 mm) have been included to train a robust network against this parameter, which highly impacts image manifestations. The relative error of volume difference for the percentage of infections (lesion/lung) and relative mean HU Diff (%) were 0.22 ± 6.3% (95% CI: −0.95 to 1.4%) and −0.18 ± 3.4% (95% CI: −0.8 to 0.44%), demonstrating the high accuracy of COLI-Net for biomarker generation.
Potential foreseen applications are not limited to the detection and segmentation but could be useful in providing diagnostic and prognostic parameters calculated using lung and infections segmentation to estimate the percentage of infections, and enabling advanced image processing in COVID-19 patients. The existing body of research on pneumonia suggests that the pneumonia severity index (PSI) can potentially be used as a severity marker.73 A recent study classified COVID-19 patients into severe and nonsevere patients based on PSI calculated using CT images.74 Different DL algorithms and radiomics analysis approaches using CT images have been examined recently for developing diagnostic (discriminating COVID-19 from bacterial/viral pneumonia) and prognostic (survival, hospital stay, intensive care unit [ICU] admission, risk of outcome) models, which require lung and lesion segmentation.17 Moreover, calculating the percentage of infection and well-aerated regions in the lung are frequently performed through visual assessment or by simply calculating HU values in the lungs, which is not only time-consuming but also lacks accuracy.
The established model exhibited noticeable performance variation across different COVID-19 patients collected from different countries, centers, with different patient backgrounds, and stages of the disease. Since the quality of CT images depends directly on the scanner model, imaging protocol (tube voltage, tube current, pitch factor, etc.), and reconstruction algorithm, we employed various datasets from different centers to cover a large variability.10, 72 Although the proposed algorithm was evaluated using a multicenter, multiscanner, multinational dataset and patients with a diverse background, stages of the disease, a full-scale adaptation of this model requires further clinical investigation and fine-tuning to the specific image acquisition parameters of a center. This framework provides multiple imaging biomarkers for COVID-19 patients to facilitate the assessment of their clinical relevance in diagnostic (discriminating COVID-19 from bacterial/viral pneumonia) and prognostic (survival, hospital stay, ICU admission, risk of outcome) applications. Further development should involve implementing lung lobes segmentation to calculate all potential imaging biomarkers at the lobes level. In this study, we used only the ResNet architecture for model evaluation. However, further evaluations should be conducted to compare different models, including UNet, VNet, and GAN architectures.
5 CONCLUSION
We set out to develop an automated algorithm capable of segmenting 3D whole lung and infected regions in COVID-19 patients from chest CT images using DL techniques to enable fast, consistent, robust, and human error immune framework for lung and pneumonia lesion detection and delineation. Owing to the complex nature of the problem and high variability in lesion manifestation, TL from whole lungs to pneumonia infection lesions was proposed and implemented to enrich specific COVID-19 pneumonia features identification from clinical studies. Moreover, a multicentric and multiscanner dataset was collected for the development of the DL model to establish an automated and generalizable platform for efficient COVID-19 patients management. The developed AI model was evaluated using a wide range of COVID-19 patients of diverse populations with different stages of the disease from multiple centers around the world to enable big data analysis of COVID-19 for automated progression/regression assessment of pneumonia lesions in follow-up studies, provide diagnostic and prognostic metrics, and enable further advanced image processing.
ACKNOWLEDGMENT
This work was supported by the Swiss National Science Foundation under grant SNRF 320030_176052. Open access funding provided by Universite de Geneve.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTIONS
Isaac Shiri and Hossein Arabi are the co-first authors of this paper. Mostafa Ghelich Oghli and Habib Zaidi contributed to the study conception and design. Isaac Shiri and Hossein Arabi designed, implemented, and evaluated the image segmentation and ML framework. Yazdan Salimi, Amirhossein Sanaat, Azadeh Akhavanallaf, Ghasem Hajianfar, Dariush Askari, Shakiba Moradi, Zahra Mansouri, Masoumeh Pakbin, Saleh Sandoughdaran, Hamid Abdollahi, Amir Reza Radmard, and Kiara Rezaei-Kalantari collated the datasets. Habib Zaidi contributed to the study conception and design, initial draft of the manuscript and supervision and funding of this work. All authors contributed to the data preparation and revision of the manuscript for important content.
Endnote
Open Research
DATA AVAILABILITY STATEMENT
The data supporting the findings of this study are not available.