Characterization of a possible founder synonymous variant in TECTA in multiple individuals with autosomal recessive hearing loss
Abstract
Synonymous variants have been shown to alter the correct splicing of pre-mRNAs and generate disease-causing transcripts. These variants are not an uncommon etiology of genetic disease; however, they are frequently overlooked during genetic testing in the absence of functional and clinical data. Here, we describe the occurrence of a synonymous variant [NM_005422.4 (TECTA):c.327C>T, p.(Gly109=)] in seven individuals with hearing loss from six unrelated families. The variant is not located near exonic/intronic boundaries but is predicted to impact splicing by activating a cryptic splicing donor site in exon 4 of TECTA. In vitro minigene assays show that the variant disrupts the reading frame of the canonical transcript, which is predicted to cause a premature termination codon 48 amino acids downstream of the variant, leading to nonsense-mediated decay. The variant is present in population databases, predominantly in Latinos of African ancestry, but is rare in other ethnic groups. Our findings suggest that this synonymous variant is likely pathogenic for TECTA-associated autosomal recessive hearing loss and seems to have arisen as a founder variant in this specific Latino subpopulation. This study demonstrates that synonymous variants need careful splicing assessment and support from additional testing methodologies to determine their clinical impact.
Hearing loss is a common disorder, with an estimated genetic etiology in more than 60% of pediatric patients (Morton & Nance, 2006). More than 100 genes have been implicated in nonsyndromic sensorineural hearing loss (NSHL) and its mimics. Despite comprehensive genetic testing using high-throughput DNA sequencing, a genetic diagnosis is currently only made in 40%−50% of patients with hearing loss (Guan et al., 2018; Sloan-Heggen et al., 2016). In a significant portion of cases, the testing remains inconclusive with the reporting of variants of uncertain clinical significance (VUS) and/or monoallelic variants in genes associated with autosomal recessive hearing loss. Additional evidence for further variant interpretation or supplementary genetic testing to identify variants not covered by comprehensive assays is often needed to establish a genetic diagnosis.
TECTA is a gene commonly involved in hearing loss and variants in this gene account for at least 4.13% of autosomal recessive NSHL (Mahdieh et al., 2010). TECTA encodes alpha-tectorin, one of the major non-collagenous components of the tectorial membrane in the inner ear. Disease-causing alterations in TECTA cause allelic hearing loss conditions with two different modes of inheritance. Heterozygous, primarily missense, pathogenic variants cause autosomal dominant deafness type 8/12 (DFNA8/12, MIM #601543), which is characterized by nonsyndromic moderate to severe hearing loss affecting mid to high frequencies with onset typically from early childhood to adolescence. Biallelic loss-of-function (LOF) variants in TECTA cause autosomal recessive deafness type 21 (DFNB21, MIM #603629), which is characterized by moderate to profound prelingual hearing loss, often with a distinctive U-shaped audiogram (Alloisio et al., 1999; Balciuniene et al., 1999; Moreno-Pelayo et al., 2001; Yamamoto et al., 2017). Most TECTA variant entries in the Human Gene Mutation Database (HGMD, 72%) and those classified as “pathogenic” or “likely pathogenic” in ClinVar are missense or result in LOF due to frameshift, nonsense, or splicing changes. Synonymous TECTA variants are found in only two HGMD entries but are reported in >128 ClinVar entries. About 75% of the synonymous variants in ClinVar are classified as likely benign by at least one clinical laboratory, while 27% have uncertain clinical significance. These synonymous variants may be important for establishing a genetic diagnosis of hearing loss.
One uncharacterized variant is NM_005422.4(TECTA):c.327C>T, p.(Gly109=), a synonymous change with multiple VUS entries in ClinVar (Variation ID: 179381). The variant is computationally predicted to moderately impact splicing, but a functional study has not been performed to validate this prediction. Given multiple occurrences of this variant in hearing loss cases, we were motivated to identify and describe a case series of patients with this variant and to further evaluate its clinical significance through functional and population genetic assessments.
We performed a retrospective review of the genetic testing results of individuals with hearing loss in three diagnostics laboratories offering next-generation sequencing (NGS) based panels for hearing loss, namely the Children's Hospital of Philadelphia (CHOP, Philadelphia, PA, USA), the Laboratory for Molecular Medicine (LMM, Cambridge, MA, USA), and GeneDx (Gaithersburg, MD, USA). We identified seven individuals with the c.327C>T, p.(Gly109=) alteration from six unrelated non-consanguineous families. Four of these individuals carry the variant in the homozygous state, while the other three are compound heterozygous for this variant in addition to another variant in TECTA in trans, including two likely pathogenic variants, [c.5754_5755delAA, p.(Ile1919*)] and [c.6162+1G>A, p.?], and a rare VUS [c.6115C>T, p.(His2039Tyr)] (Table 1). Detailed clinical and genetic information available for these individuals indicated all of them had congenital or prelingual onset, moderate to severe bilateral sensorineural hearing loss, and four of them were reported to have a U-shaped configuration which was stable through their most recent auditory brainstem response testing (Figure 1a,b, Table 1). No other clinical features were observed. NGS panel analysis and/or research-based exome analysis revealed no additional variants related to hearing loss in these patients.
| Family-Individuala | Hearing loss featuresb | Genotypec | Ethnicity | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Onset | Severity | Frequency affected | Shape | Progressive | c. synonymous | p. synonymous | c.[701A>G;3097C>T] | ||
| Pt1-1 | Congenital | moderate-severe | 1500−8000 Hz | Shallow U-shaped | Stable | c.[327C>T];[327C>T] | p.[(Gly109=)];[(Gly109=)] | Homozygous | Latino/Puerto Rico |
| Pt1-2 | Prelingual | moderate-severe | 1500−8000 Hz | Shallow U-shaped | Stable | c.[327C>T];[327C>T] | p.[(Gly109=)];[(Gly109=)] | Homozygous | Latino/Puerto Rico |
| Pt2-1 | Prelingual | moderate-severe | NA | NA | NA | c.[327C>T];[327C>T] | p.[(Gly109=)];[(Gly109=)] | Homozygous | NA |
| Pt3-1 | Prelingual | moderate-severe | 500−8000 Hz | Shallow U-shaped | NA | c.[327C>T];[5754_5755delAA] | p.[(Gly109=)];[(Ile1919*)] | Heterozygous | Latino/Puerto Rico |
| Pt4-1d | Prelingual | moderate-severe | NA | NA | NA | c.[327C>T];[6115C>T] | p.[(Gly109=)];[(His2039Tyr)] | Heterozygous | Latino/Puerto Rico |
| Pt5-1 | Prelingual | moderately severe to severe | 500−8000 Hz | NA | Stable | c.[327C>T];[6162+1G>A] | p.[(Gly109=)];[?] | Heterozygous | Caucasian + Latino |
| Pt6-1 | Congenital | Moderate-severe | NA | Shallow U-shaped | Stable | c.[327C>T];[327C>T] | p.[(Gly109=)];[(Gly109=)] | Homozygous | Latino/Puerto Rico |
- a The patients were tested at different clinical laboratories (CHOP: 1−2; LMM: 3−5; GeneDX: 6−7).
- b All patients were affected with bilateral nonsyndromic sensorineural hearing loss.
- c The phase of the TECTA variants was determined by parental studies. The variants, c.6162+1G>A and p.(Ile1919*), are likely pathogenic, whereas the clinical significance of c.6115C>T remains uncertain. Thess variants were reported in relation to the TECTA transcript NM_005422.4.
- d The synonymous variant did not segregate in this individual's similarly affected sibling.
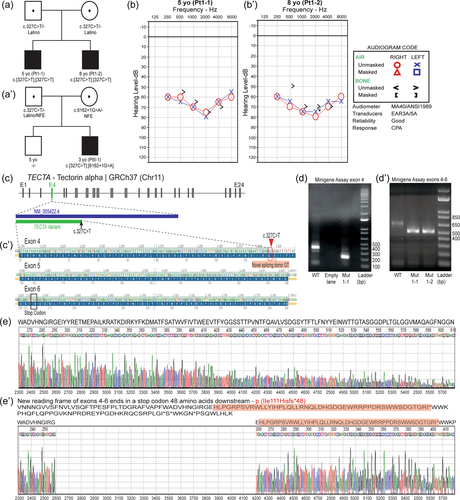
The c.327C>T variant is located 160 base pairs (bp) upstream from the canonical donor splice site of intron 4 of TECTA (c.486+1,2) (Figure 1c). Conservation analysis indicates that the nucleotide position at c.327 is not conserved according to the default thresholds of three algorithms [GERP: −1.61 (>4), PhastCons: 0.038 (>0.8), and PhyloP: −0.58 (>2)]; however, five popular splice site predictor algorithms predict a possible gain of a splice donor site 2 bp upstream at c.325 [SpliceAI: 0.21−0.51 (≥0.5), SSF: 77.72−87.25 (≥70), MaxEnt: 7.66 (≥0), NNSPLICE: 0.97 (≥0.4), GENESPLICER: 4.55(≥0)] (Supporting Information: Figure S1). The activation of this cryptic splice site may supplant the natural donor splice site and cause alternative splicing, resulting in the deletion of 161 nucleotides in exon 4, as well as the canonical donor splice site in intron 4, leading to a loss of 54 amino acids and leaving the remainder of the transcript out of frame (Figure 1c’).
According to GTEX, the TECTAgene is very weakly expressed in blood, EBV-transformed lymphocytes (EBV-LCLs), or cultured fibroblasts with a low number of transcripts per million (TPM); hence, a direct assessment of the effect of a variant on splicing in these tissues may not be possible (Bournazos et al., 2022; Collin et al., 2008). Therefore, we established an in vitro assay to investigate the effect of this synonymous change on mRNA splicing. To do this, we cloned both the wild-type and the variant sequences of exon 4, along with its flanking introns, into the pET01 Exontrap vector, which has previously been used to characterize RNA splicing (details in supplementary methods, Supporting Information: Table S1). When we transfected a vector containing the wild-type TECTAsequence into HEK293T cells, polymerase chain reaction (PCR) amplification of cDNA yielded the expected 374 bp product containing the entirety of TECTAexon 4 (288 bp) and portions of the native 5′ and 3′ exons of pET01. However, HEK293T cells transfected with the variant vector yielded a shorter 213 bp product containing a truncated TECTA exon 4 (127 bp) (Figure 1d). The splicing occurred 2 bp before c.327C>T, supporting the creation of a cryptic splice donor site (GT) by this variant. We also confirmed that the exon 4 truncation results in a frameshift by repeating the in vitro splicing assay using a TECTA sequence extended to the 3′ intronic region flanking exon 6 (Figure 1d’). The new reading frame results in a premature stop codon downstream, c.326_486+2del, p.(Ile111Hisfs*48) (Figures 1e and e’), which is predicted to result in a truncated protein impacting the NIDO domain and all other downstream protein domains (VWFC, VWFD1-4, TIL1-3, and ZP, Uniprot O75443). Given the location of this variant, this mutated transcript is almost certainly subjected to nonsense-mediated mRNA decay, leading to the absence of protein from this allele. Importantly, we did not detect expression of expected-length (wild-type) transcripts with either variant vector, suggesting leaky expression of the wild-type transcript does not occur with this variant.
We next investigated the population distribution of c.327C>T, p.(Gly109=). This variant was observed in a heterozygous state in four individuals using exome sequencing in the Genome Aggregation Database (gnomAD) v2.1.1 with a global minor allele frequency (MAF) of 0.0016% (4/251,416 alleles, 0 homozygotes), including one occurrence in the non-Finnish European (NFE) subpopulation and three in the “other” subpopulation (unknown ethnic background). In this version of gnomAD, the variant was absent from the 34,586 alleles of Latino/Admixed Americans; however, using genome sequencing in gnomAD v3.1.2, this variant (chr11:121109339C>T, hg38) is significantly enriched in Latino/Admixed Americans with a frequency of 0.2161% (33/15,272 alleles, 0 homozygotes). The local ancestry inference (LAI) data recently released for this subpopulation disclosed that all carriers of this variant are Latinos of African descent (2.26%, 33/1460 alleles). The variant is absent or extremely rare in other Latino and non-Latino ancestries. With two NFE individuals and one African American, its global MAF is estimated at 0.024% (36/152,108 alleles).
Six patients in this study self-reported as Puerto Ricans with Latino ancestry (Table 1). In addition, this variant was observed in a heterozygous state in three other individuals with or without NSHL tested at CHOP (Murrell et al., 2022), all of whom are also Puerto Rican. Although these ancestry reports were not verified through genetic analysis, our observation of two additional TECTA variants shared by all these cases led us to an ancestry-specific assessment of the gnomAD. All the patients in this study and the three additional heterozygous carriers of c.327C>T from CHOP also carried two additional TECTA variants, chr11: 120984338A>G (hg19), c.701A>G, p.(Gln234Arg), and chr11: 121008285C>T (hg19), c.3097C>T, p.(Arg1033Trp) (Murrell et al., 2022). These two additional variants are relatively common in the African/African American population (MAF 0.45% and 0.35%, respectively) but are rare among Latino/Admixed Americans (0.05% and 0.02%, respectively) in gnomAD v2.1.1 (Supporting Information: Figure S2). The available parental studies from four individuals demonstrated that all three TECTA variants (c.327C>T, c.701A>G, and c.3097C>T) are inherited together from one parent, indicating that they are in cis. Using the gnomAD co-occurrence tool, we determined that the synonymous variant (c.327C>T) and either of the two common variants (c.701A>G and c.3097C>T) are most likely present on the same haplotype in the individuals from the “other” subpopulation, but on different haplotypes in the NFE individuals. Interestingly, these two common variants are also prevalent in Latinos (MAF ~0.42% for both) but mainly in those of African descent according to the LAI in gnomAD v3.1.2 (Supporting Information: Figure S2). Homozygotes of c.327C>T are absent in either version of gnomAD.
The original diagnostic reports of the three laboratories in this study classified this variant as of uncertain significance (VUS). LMM and GeneDX, along with three other laboratories, submitted this variant to ClinVar as a VUS between 2017 and 2022, with LMM providing additional genetic data on their patients, deeming this a VUS in favor of being likely pathogenic (ClinVar variation ID: 179381). Besides these data, no information was available for c.327C>T, p.(Gly109=) in the medical literature. Using the new information in this study, we reassessed this variant for classification according to the ACMG/AMP and ClinGen Hearing Loss ACMG specifications v1 guidelines (Oza et al., 2018; Richards et al., 2015). The variant is observed in the compound heterozygous state with a likely pathogenic variant in two individuals and in the homozygous state in four patients. The variant was also observed in the compound heterozygous state with a VUS in patient #4 (Table 1); however, because the two variants did not segregate in this individual's similarly affected sibling (who is not included in this study), the TECTA variants are unlikely to explain the hearing loss in this patient. Overall, the available information provides evidence for a classification of at least the PM3_strong category. The variant segregates with the disease in two affected relatives (PP1_supporting). Furthermore, in vitro minigene analysis suggests that this variant likely creates a cryptic donor site, leading to an out-of-frame transcript (PS3_supporting). Although this variant is prevalent in Latinos of African descent, it occurs very rarely in other subpopulations (MAF ~0% in non-Latinos and 0.02% global), strongly suggesting that the synonymous TECTA variant could be a founder variant in Latinos of African origin. To be conservative, the frequency ACMG criteria (PM2 or BS2) were not used in the classification. According to the applied points (PM3_strong, PS3_supporting, PP1_supporting), this synonymous variant can be reclassified as likely pathogenic for autosomal recessive deafness type 21 (DFNB21).
Synonymous variants are routinely considered to be nondeleterious in genetic testing since they do not alter the amino acid sequences. They are only prioritized for analysis and reporting when previously reported in affected individuals or when strong computational or functional data indicates an effect on splicing. For TECTA, the clinical significance of many synonymous variants detected in hearing loss patients remains uncertain. A different synonymous variant in this gene, NM_005422.4(TECTA):c.5331G>A, p.(Leu1777=), has previously been shown to result in an in-frame deletion of a partial exon due to loss of an exonic splice enhancer (ESE) predicted by ESE finder, causing autosomal dominant deafness type 8/12 (DFNA8/12) (Collin et al., 2008). Our study, therefore, presents an additional disease-causing synonymous variant in TECTAwith functional evidence of gene disruption through a different splicing mechanism.
In the absence of functional studies, clinical laboratories utilize bioinformatic strategies to filter out nondeleterious synonymous variants at an early stage in the analysis to minimize the burden of variant analysis and reporting. While many computational tools are available for use in the clinical setting, they may generate conflicting or inaccurate predictions that can result in overlooking the true cause of a genetic disorder, especially when a synonymous variant is relatively common in controls, as seen with the c.327C>T, p.(Gly109=) variant. The synonymous variant in this study is predicted to introduce a cryptic splice donor site 2 bp upstream of the variant; however, the predicted score provided by an individual bioinformatics tool is still insufficient to satisfy the PP3 criteria (computational evidence of pathogenicity), even for the SpliceAI tool, which outperforms other tools in sensitivity and specificity (Bournazos et al., 2022; Jaganathan et al., 2019; Wai et al., 2020). Of note, SpliceAI predicts no impact of c.5331G>A, p.(Leu1777=) on splicing (Collin et al., 2008). The two synonymous variants would have remained unreported had multiple tools not been used or had they not been previously submitted to ClinVar as a VUS. The subsequent collaboration between the laboratories involved in this study led to the identification of multiple patients carrying this variant, along with additional genotyping information, which was a key step in further studying this variant. This example highlights the importance of data sharing by clinical laboratories.
RNA sequencing (RNA-seq) is emerging as a promising assay for detecting splicing abnormalities in constitutional disorders. Its clinical utility, however, remains limited due to the inaccessibility of certain cell types (Bournazos et al., 2022). The TECTA gene has high expression in the cochlea but not in peripheral blood cells, EBV-LCLs, or fibroblasts, the most accessible sample types for clinical laboratories. One study showed that only genes expressed at TPM >0.5 levels are feasible for analysis by reverse transcription-polymerase chain reaction (RT-PCR), but the median TPM value of TECTA for blood, ETV-LCLs, and fibroblasts is all well below this threshold, and RT-PCR is generally more sensitive than RNA-seq for splicing studies (Bournazos et al., 2022; Collin et al., 2008). For this reason, we used an in vitro minigene assay to characterize the effect of this synonymous variant on splicing. Although there is an inherent limitation to the minigene-based analysis of splicing (Lin et al., 2021), our experiment successfully demonstrates the activation of a cryptic splice site with no leaky expression of the wild-type transcript. This was used as supportive evidence in clarifying the clinical significance of this TECTA synonymous variant.
Population analysis and internal database queries suggest this synonymous variant has arisen in two different haplotype blocks, one in Latino ancestry in cis with the two common TECTA variants, and the other in NFE ethnicity with the two common variants more likely in trans with this synonymous variant. According to the LAI data, this variant and the two other variants appear with a high frequency specifically in Latinos of African ancestry but are rare in other subpopulations. Consistent with this data, all of the heterozygous carriers identified at CHOP and most of the affected subjects included in this study have Puerto Rican ancestry; although the Puerto Rican population is highly diverse, at least 10% self-identify as both African and Latino, and the modern Latino population is known to be formed via genetic admixture among ancestral source populations from Africa, the Americas and Europe (Norris et al., 2018). Our analysis thus strongly suggests this variant arose as a founder variant in Latinos of African ancestry, which explains why this variant is rare or absent in other subpopulations of gnomAD and our internal databases. Disease-relevant variants occurring at elevated frequencies due to bottleneck events in a population's history are expected to occur in the LAI data, and several studies have identified founder variants in genes associated with other diseases in the Latino ancestry (Gonzaga-Jauregui et al., 2015). Our study further demonstrates the utility of LAI in evaluating allele frequencies of rare and common variants during variant classification.
In summary, we describe a rare synonymous variant in TECTA that results in a splicing defect. The variant has been identified in the biallelic state in multiple individuals with bilateral NSHL, is likely pathogenic for TECTA-related autosomal recessive NSHL, and likely arose as a founder variant in Latinos of African origin.
ACKNOWLEDGMENTS
We would also like to thank all of the patients and their families for participating in this study. This research was partially supported by funds from the Division of Genomic Diagnosis at CHOP.
CONFLICT OF INTEREST
Erin Torti, Gabriele Richard, and Renkui Bai, are employees of GeneDx, LLC. The remaining authors declare no conflict of interest.




