WARS1 and SARS1: Two tRNA synthetases implicated in autosomal recessive microcephaly
Abstract
Aminoacylation of transfer RNA (tRNA) is a key step in protein biosynthesis, carried out by highly specific aminoacyl-tRNA synthetases (ARSs). ARSs have been implicated in autosomal dominant and autosomal recessive human disorders. Autosomal dominant variants in tryptophanyl-tRNA synthetase 1 (WARS1) are known to cause distal hereditary motor neuropathy and Charcot-Marie-Tooth disease, but a recessively inherited phenotype is yet to be clearly defined. Seryl-tRNA synthetase 1 (SARS1) has rarely been implicated in an autosomal recessive developmental disorder. Here, we report five individuals with biallelic missense variants in WARS1 or SARS1, who presented with an overlapping phenotype of microcephaly, developmental delay, intellectual disability, and brain anomalies. Structural mapping showed that the SARS1 variant is located directly within the enzyme's active site, most likely diminishing activity, while the WARS1 variant is located in the N-terminal domain. We further characterize the identified WARS1 variant by showing that it negatively impacts protein abundance and is unable to rescue the phenotype of a CRISPR/Cas9 wars1 knockout zebrafish model. In summary, we describe two overlapping autosomal recessive syndromes caused by variants in WARS1 and SARS1, present functional insights into the pathogenesis of the WARS1-related syndrome and define an emerging disease spectrum: ARS-related developmental disorders with or without microcephaly.
1 INTRODUCTION
Ribosomal protein biosynthesis requires translation of the information encoded in messenger RNA (mRNA) into a chain of amino acids. At the ribosome, amino acids are introduced into the elongating chain via their specific transfer RNA (tRNA). The transfer of each amino acid to its cognate tRNA (aminoacylation) is carried out by enzymes called aminoacyl-tRNA synthetases (ARSs) (Rubio Gomez & Ibba, 2020). These evolutionary conserved enzymes are of vital importance, since they ensure the fidelity of the translational process (Swanson et al., 1988). In humans, ARSs are denominated using the one-letter code of the amino acid substrate (e.g., LARS for leucyl-tRNA synthetase) and an added number indicates the cytosolic (1) and the mitochondrial (2) homologues (Rubio Gomez & Ibba, 2020). In higher eukaryotes, ARSs have acquired functions that surpass the original catalytic activity, for example, they are able to function as amino acid sensors (Yu et al., 2021) or to form multi-enzyme complexes involved in various signal transduction pathways (Lee et al., 2004). While pathogenic variants in the mitochondrial ARSs have been associated with a variety of mitochondriopathies (Fine et al., 2019), variants in the cytosolic ARSs have been associated with different autosomal dominant and autosomal recessive phenotypes. Most recessively inherited, ARS-related phenotypes have largely overlapping features including a variable combination of microcephaly, growth retardation, developmental delay, intellectual disability, brain anomalies, seizures, ataxia, and additional features.
We describe five individuals, from three unrelated families, with severe microcephaly, variable growth retardation, brain anomalies, severe developmental delay, and intellectual disability, in whom we identified a homozygous variant in the genes WARS1 (MIM# 191050), encoding the cytosolic tryptophanyl-tRNA synthetase (WARS1), or compound heterozygous variants SARS1 (MIM# 607529), encoding the cytosolic seryl-tRNA synthetase (SARS1). Structural mapping of the identified variants in both WARS1 and SARS1 showed that they most likely disturb enzymatic function or protein integrity. To examine the functional effects of the WARS1 variant in vivo, we used a zebrafish model to determine the pathogenicity of the variant and studied the variant's effect on patient fibroblasts. We discuss our findings in the context of the existing knowledge of ARSs in human disease and suggest a systematic nomenclature for ARS-related autosomal recessive disorders, introducing the term “aminoacyl-tRNA synthetase-related developmental disorder with or without microcephaly” (ARS-DDM).
2 METHODS
2.1 Editorial policies and ethical considerations
All studies were performed in accordance with the Declaration of Helsinki protocols. The studies were reviewed and approved by the local institutional ethics board (University Medical Center Göttingen). Written informed consent was obtained from all affected subjects, parents, or legal representatives participating in this study.
2.2 Exome sequencing
DNA isolation from EDTA blood was carried out following standard protocols for all participants. In family 1, we performed exome sequencing (ES) for individual 1 and both her parents. Sequencing libraries were prepared using the Twist enrichment workflow (Twist Bioscience) and the Twist Core exome kit with spiked-in Twist RefSeq probes as well as a custom-design spike-in to enrich mitochondrial DNA. Library preparation and capture were performed according to the manufacturer's instructions and paired-end sequencing was performed on a NovaSeq. 6000 instrument (Illumina). Sequence data were aligned to GRCh37 (GCA_000001405.14) with bowtie2 (v2.4.3.1) (Langmead & Salzberg, 2012). Aligned data were further processed with GATK4 (Poplin et al., 2017) and joint calling was applied to generate vcf-files. Finally, resulting data were processed with custom scripts and uploaded for analysis with VarFish (Holtgrewe et al., 2020). In family 2, we performed ES of the index and both affected brothers. Exome capture was carried out with BGI Exome kit capture (59M) and the library was then sequenced on a BGISEQ-500, paired-end 100bp, at BGI laboratory in Shenzhen, China. Analysis of the raw data was performed using the software Varfeed (Limbus; Rostock) and the variants were annotated and prioritized using the software Varvis (Limbus; Rostock). In family 3, we performed ES for individual 5 and both her parents as previously described (Altmüller et al., 2016). ES data analysis and filtering of variants were carried out using the exome and genome analysis pipeline varbank (https://varbank.ccg.uni-koeln.de) of the Cologne Center for Genomics (University of Cologne, Cologne, Germany). We filtered for variants with a min. coverage of 10 reads, a minimum quality score of 10, and a MAF <1%. We have prioritized all potential protein-influencing variants with regard to their pathogenicity and clinical relevance according to all possible inheritance modes.
2.3 In silico prediction tools
- −
PolyPhen2 (http://genetics.bwh.harvard.edu/pph2/), HumVar and HumDiv models (Adzhubei et al., 2010);
- −
PROVEAN (http://provean.jcvi.org/index.php) (Choi & Chan, 2015);
- −
SIFT (http://sift.jcvi.org/) (Ng & Henikoff, 2003);
- −
MutationTaster (http://www.mutationtaster.org/) (Schwarz et al., 2014).
2.4 Zebrafish model
Zebrafish (Danio rerio) were raised and maintained in an AALAC-approved facility at the Oklahoma Medical Research Foundation (OMRF) under standard conditions, and all experiments were performed as per protocol 20-07 approved by the Institutional Animal Care Committee of OMRF (IACUC). All zebrafish work was carried out in wild-type strain NHGRI-1 (LaFave et al., 2014). We designed four single guide RNAs (sgRNAs), targeting functional domains of wars1, and synthesized by in vitro transcription, from templates generated by annealing two oligos as described earlier (Varshney et al., 2016). We mixed the four sgRNAs with Cas9 protein (University of Berkley, CA, USA) to form the ribonucleoprotein complexes (RNPs) and injected into one-cell stage zebrafish eggs. Knock-down efficiency was confirmed by Quantitative reverse transcription polymerase chain reaction (RT-qPCR) (Supporting Information: Figure 1). Phenotyping was performed as described in previous methods (Lin et al., 2021). For size measurements, the mean values of head size and eye size of uninjected embryos were set as 100% and then comparative percentages were calculated for each other group (wars1 F0, F0 + WT, and F0 + Arg133Cys). Statistical analysis was performed using the one-way analysis of variance (ANOVA) with Tukey's multiple comparisons test. Variant-specific complementary DNA (cDNA) was generated from a wild-type WARS1 cDNA cloned into pCS2+ vector as a template using quick change site-directed mutagenesis kit (Agilent Inc.). mRNA was synthesized from a linearized template using SP6 mMessage kit (Thermo Fisher, Inc.), and purified by RNA clean & concentrator kit (Zymo Inc.).
2.5 RNA extraction and RT-qPCR from zebrafish
Total RNA was extracted from uninjected control, Cas9 protein injected, and wars1 F0 mutant embryos at 3 dpf using the TRIzol Reagent (Thermo Fischer Scientific) and purified by miRNeasy Mini kit (Qiagen), following the manufacturer's instructions. The mRNA was reverse-transcribed into cDNA using iScript cDNA-synthesis kit (Bio-Rad) according to the manufacturer's instructions. The cDNA was used as a template for RT-qPCR reactions using SYBR Green Supermix according to the manufacturer's instructions (Thermo Fisher Scientific) and the Light Cycler® 96 System (Roche). All relative quantifications were done using three independent biological replicates and three technical replicates, the values were normalized to the housekeeping gene 18S. The primer sequences for RT-qPCR are wars1: Forward: 5′-GCGAGTCCTGCCTTCTACAG-3′, Reverse: 5′-TAGCACACGGAATGAGGCAC-3′, 18S: Forward: 5′-TCGCTAGTTGGCATCGTTTATG-3′, and Reverse: 5′-CGGAGGTTCGAAGACGATCA-3′. The quantification was performed using the method and corresponding age-matched control samples as calibrators.
2.6 Structural mapping of disease-related variants in WARS1 and SARS1
Structure figures were prepared with PyMol 2.5 (Schrödinger LLC).
2.7 Cell culture
Fibroblasts were grown at 37°C in a 5% CO2 humidified atmosphere in Dulbecco's modified Eagle's medium containing 4.5 g/L glucose, l-glutamine, sodium pyruvate, and 3.7 g/L NaHCO3 (DMEM; PAN-Biotech GmbH), additionally supplemented with 10% fetal bovine serum (Merck KGaA; Darmstadt), penicillin-streptomycin (10,000 U/ml penicillin, 10 mg/ml streptomycin; PAN-Biotech GmbH), and 0.7 µg/ml amphotericin B (Merck KGaA).
2.8 Western blot
Fibroblasts were grown in 100 × 20 mm cell culture dishes (Greiner Bio-One GmbH) to a confluence of approximately 90%, before they were harvested by trypsinization and pelleted. Pellets were then incubated with 100 μl of protein lysis buffer (20 mM Tris/HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% w/v NP-40), and 1× Halt™ Protease Inhibitor Cocktail (Thermo Scientific) for 10 min on ice. After centrifugation at 4°C and 19,000g for 10 min, supernatants were collected, protein concentrations were determined using Pierce™ BCA Protein Assay Kit (Thermo Scientific) and 40 μg (33 µg for replicate 2) of protein was loaded onto a 4%–20% gradient precast Mini-PROTEAN TGX Stain-Free Gel (Bio-Rad Laboratories, Inc.). Following electrophoresis, the gel was blotted onto an Immun-Blot® PVDF membrane (Bio-Rad Laboratories, Inc.) using a Trans-Blot® Turbo™ Transfer System (Bio-Rad Laboratories, Inc.). Immunoblots were then blocked for 1 h at room temperature in 5% milk powder (blotting grade; Carl Roth Gmbh & Co. Kg) in tris-buffered saline with 0.1% TWEEN 20 (TBS-T) and subsequently incubated overnight at 4°C with horseradish peroxidase (HRP)-conjugated primary anti-WARS1 antibody (0.5 µg/ml, sc-374401 HRP; Santa Cruz Biotechnology, Inc.) in 2.5% milk/TBS-T. WesternBright Chemilumineszenz Substrat (Biozym Scientific GmbH) was used as HRP substrate for enhanced chemiluminescence detection of bands. Chemiluminescence was recorded using a ChemiDoc™ Touch Imaging System with Image Lab™ Touch Software (version 1.2.0.12; Bio-Rad Laboratories, Inc.). Membranes were then reprobed with rabbit anti-glyceraldehyde 3-phosphate dehydrogenase (α-GAPDH; 1:5000) monoclonal antibody (#2118; Cell Signaling Technology) followed by secondary mouse anti-rabbit IgG-HRP antibody (1:10000, sc-2357; Santa Cruz Biotechnology, Inc.) as loading control.
2.9 Quantification and statistical analysis of Western blot data
Densitometric quantification of western blot bands on raw 16-bit images was performed using Gel Analyzer within Fiji/ImageJ 2.1.0/1.53c (Schindelin et al., 2012). Results are presented as boxplots (center line corresponds to median, upper and lower hinge correspond to 25th and 75th percentiles, upper and lower whisker extend to largest/smallest value within 1.5× inter-quartile range of the respective hinge, outliers beyond the whiskers are presented individually). For statistical analyses, ANOVA was performed (significance level α = 0.05), followed by post hoc Tukey analysis. Results are presented as compact letter display of all pair-wise comparisons in increasing order.
2.10 Aminoacylation assay
Aminoacylation was assessed by measuring WARS1 activity in cultured fibroblasts. Fibroblast lysates (cytosolic fraction) were incubated in triplicate at 37°C for 10 min in a reaction buffer containing 50 mmol/L Tris buffer pH 7.5, 12 mmol/L MgCl2, 25 mmol/L KCl, 1 mg/ml bovine serum albumin, 0.5 mmol/L spermine, 1 mmol/L ATP, 0.2 mmol/L yeast total tRNA, 1 mmol/L dithiothreitol, 0.3 mmol/L [13C4, 15N]-tryptophan, [D5]-phenylalanine, and [D2]-tyrosine. The reaction was terminated using trichloroacetic acid. After sample washing with trichloroacetic acid, ammonia was added to release the labeled amino acids from the tRNAs. [13C]-phenylalanine and [13C9]-tyrosine were added as internal standards and the labeled amino acids were quantified by liquid chromatography with tandem mass spectrometry (LC-MS/MS). Phenylalanyl-tRNA synthetase and tyrosyl-tRNA synthetase activities were simultaneously detected as control enzymes.
2.11 Cycloheximide chase assay
Patient and control fibroblasts were seeded into six-well plates (CytoOne®; Starlab) at 2.2 × 105 cells/well and grown to a confluence of approximately 90%, before 10 mg/ml cycloheximide (≥94% TLC-grade; Merck) in H2O at a final concentration of 250 μg/ml was added to the medium. Cells were treated for 8, 4, 2, or 0 h and all fibroblasts were harvested at the same time by trypsinization. Proteins were lysed using 25 µl of protein lysis buffer and protein levels were analyzed by immunoblot densitometry as described before, again with GAPDH as loading control. Protein levels were normalized to their respective 0 h treatment.
3 RESULTS
3.1 Clinical reports
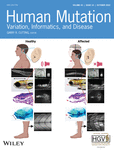
Individual 1 is the index patient of family 1 (Figure 1a). She is the second child of a healthy consanguineous couple from Syria. The parents are first cousins and they also have a healthy son (Figure 2a). The girl was presented to the genetics clinic at the age of 5 years for microcephaly, autism, and intellectual disability. Pregnancy and birth were reported as uncomplicated, but there are no detailed records. Her birth measurements were reported as follows: birthweight 2200 g (1st percentile, −2.5 SD) and head circumference 33 cm (11th percentile, −1.2 SD). Developmental delay was noticed at 9 months when she failed to reach motor milestones. She learned to walk at 26 months. The parents reported that she had learned a few words of Arabian, but stopped speaking altogether at the age of 3 years, either as a symptom of early childhood autism or of developmental regression. At age 5 years she is not toilet trained. She received the diagnoses of autism, speech-language disorder and, tentatively, mild intellectual disability. Physical examination revealed a height of 100 cm (1st percentile, −2.4 SD), a weight of 15 kg (3rd percentile, −1.8 SD), and a head circumference of 46 cm (<1st percentile, −4.2 SD) (Table 1). Besides the severe microcephaly, she showed no striking dysmorphic features. Her gait was ataxic, but otherwise she showed no obvious neurological abnormalities, especially no signs of neuropathy. Apart from secondary loss of active speech, she showed no signs of a progressive neurodegenerative disorder at the age of 5 years. Cranial MRI showed a normal configuration of the cerebral cortex, the inner and outer cerebrospinal fluid spaces, the basal ganglia, and the pituitary gland. The corpus callosum was described as thin and periventricular white matter hyperintensities were reported (images not shown).

| Family | 1 | 2 | 2 | 2 | 3 | Musante et al. | Ravel et al. |
|---|---|---|---|---|---|---|---|
| Individual | 1 | 2 | 3 | 4 | 5 | 4 individuals | 4 individuals |
| Genotype | WARS1 | WARS1 | WARS1 | WARS1 | SARS1 | SARS1 | SARS1 |
| c.397C>T; | c.397C>T; | c.397C>T; | c.397C>T; | c.904C>T; | c.514G>A; | c.638G>T; | |
| p.(R133C) | p.(R133C) | p.(R133C) | p.(R133C) | p.(R302C)/c.1168C>T; p.(R390C) | p.(D172N) | p.(R213L) | |
| Genomic location | chr14:100826916G>A | chr14:100826916G>A | chr14:100826916G>A | chr14:100826916G>A | chr1:109777988C>T/ | chr1:109773566G>A | chr1:109774299G>T |
| chr1:109779081C>T | |||||||
| Gender | Female | Male | Male | Male | Female | M (2)/f (2) | M (2)/f (2) |
| Consanguinity | Yes | Yes | Yes | Yes | No | Yes | Yes |
| Origin | Syria | Iraq | Iraq | Iraq | Germany | Iran | Turkey |
| Birth | SVD | SVD | SVD | SVD | SVD | ND | ND |
| At term | At term | At term | At term | 38 + 4 weeks | At term | ND | |
| Weight (g) | 2220 (−2.5 SD) | ND | ND | ND | 3360 (+0.1 SD) | ND | −2.2 to +0.03 |
| Height (cm) | ND | 52 (+0.4 SD) | −1.0 to +0.3 | ||||
| OFC (cm) | 33 (−1.2 SD) | 33 (−1.2 SD) | −1.8 to −0.2 | ||||
| Age (y) | 5 | 13 | 8 | 7 | 11 | 11-30 | 2–9 |
| Weight (kg) | 15 (−1.8 SD) | 24.8 (−3.9 SD) | 18.9 (−2.9 SD) | 18 (2.4 SD) | 17 (−5 SD) | Thin body (2/4) | −1.77 to +0.2 |
| Height (cm) | 100 (−2.4 SD) | 144 (−2.2 SD) | 121 (−1.8 SD) | 113 (−2.3 SD) | 129 (−2.5 SD) | −0 to −3 SD | −0.6 to +1.3 |
| OFC (cm) | 46 (−4.2 SD) | 49 (−4.3 SD) | 48 (−3.8 SD) | 48 (−3.5 SD) | 46,5 (−6.2 SD) | −4 to −5 SD | −2.2 to −1.5 |
| Facial features | − | + | + | ND | + | - | + |
| Development | Delayed | Delayed | Delayed | Delayed | Delayed | Delayed (4) | Delayed (4) |
| Ataxia | + | Unsteady gait | − | − | − | + (4) | − (4) |
| Spasticity | − | − | − | − | + | ND | ND |
| Muscle weakness | − | Hypotonia | Hypotonia | − | − | + (4) | + (4) |
| Seizures | − | − | − | − | + | + (4) | + (2) |
| Behavior | Autism | Autistic feat. | − | − | Restlessness | Aggressiveness | Aggressiveness (2) |
| Speech delay | + | + | + | + | + | + (4) | + (3) |
| ID | Mild | Severe | Severe | Severe | Severe | + (4) | ND |
| Brain anomaly | CCD, WMH | RBV, CCD, LE | Unknown | RBV, CCD, WMH | RBV, CCD, RCV, WMH, MTS | ND | DM, CCD, RCV |
| Other anomaly | − | Melanoma | − | Mov. disord. | PVS, esotropia | Pes planus (2/4) | ED, CM, deafness |
- Abbreviations: C, Cys; CCD, corpus callosum dysgenesis; CM, cardiomyopathy; D, Asp; DM, delayed myelination; ED, early death; ID, intellectual disability; L, Leu; LE, leukencephalopathy; Mov. disord., movement disorder; MTS, mesial temporal sclerosis; N, Asn; ND, not documented; OFC, head circumference; PVS, pulmonary valve stenosis; R, Arg; RBV, reduced brain volume; RCV, reduced cerebellar volume; SD, standard deviation; SVD, spontaneous vaginal delivery; WMH, white matter hyperintensities.
Individual 2 is the index patient of family 2 (Figure 1b). He is one of eight children of a consanguineous couple from Iraq. He has two similarly affected younger brothers (individuals 3 and 4), and a healthy brother and sister (Figure 2b). Three siblings were already deceased. One sister passed away at age 13 years, she reportedly had severe intellectual disability and was nonverbal. She likely suffered from the same condition as her affected brothers. Another sister and another brother passed away in their second year of life and were reported to have had poor growth. It is unclear, whether the latter two children had the same condition as their affected siblings. Individual 2 was 13 years and 6 months old at last presentation. He has global developmental delay and intellectual disability. He learned to sit at 2.5 years, learned to crawl in the second year of life, and learned to walk with assistance at 3 years, but he never learned to walk independently. He is not toilet-trained and he is nonverbal. He cannot eat independently but is orally fed. Physical examination revealed a height of 144 cm (1st percentile, −2.2 SD), a weight of 24.8 kg (<1st percentile, −3.9 SD), and a head circumference of 49 cm (<1st percentile, −4.3 SD) (Table 1). Cardio-pulmonary status was normal. He showed muscular hypotonia, his gait was unsteady, and walking only possible with assistance. He did not react to his own name or show any interest in his siblings. He showed the following dysmorphic features: a microcephalic aspect with a sloping forehead, a large nose with a high nasal bridge, synophrys, long palpebral fissures, and large ear lobes (Figure 1b). Cranial MRI revealed reduced cerebral volume with enlarged internal and external cerebrospinal fluid spaces, especially supratentorial, a thin corpus callosum, and supratentorial leukoencephalopathy (Figure 1d). Individual 2 has a history of childhood malignant melanoma of the occiput with multiple pulmonary, lymphatic, and osseous metastases. Individual 3 is the younger brother of individual 2. He was 8 years and 2 months old at last presentation. He has a history of developmental delay. He learned to sit at approximately 1 year of age, crawled at about 2 years of age, and walked at 3 years. He shows a steady but broad-based gait. He was toilet trained at 4 years but needs assistance in all aspects of his daily life. He interacts with other children but is unable to communicate verbally. He has a height of 121 cm (4th percentile, −1.8 SD), a weight of 18.9 kg (<1st Percentile, −2.9 SD), and a head circumference of 48 cm (<1st percentile, −3.8 SD). He showed dysmorphic features, similar to his elder brother (Table 1). Individual 4 is the youngest affected child from family 2. He was 7 years and 2 months old at last presentation. He has a history of global developmental delay similar to his elder brothers. He is unable to stand or walk unassisted. He does not speak, except for one word, and communicates with noises. He is unable to feed himself independently. Neurological examination revealed a movement disorder that was classified as mainly dystonic. He makes eye contact, but does not follow any instructions. Cardio-pulmonary status, and kidney and liver function were normal. Audiological and ophthalmological examinations were normal. EEG showed no signs of epilepsy. Cranial MRI revealed slightly enlarged subarachnoid spaces compared to healthy peers indicative of reduced cerebral volume, white matter T2-hyperintensities, and a thin corpus callosum (Figure 1e). He has a height of 113 cm (1st percentile, −2.3 SD), a weight of 18 kg (1st percentile, −2.4 SD), and a head circumference of 48 cm (<1st percentile, −3.5 SD) (Table 1). In none of the affected children from family 2 there was evidence of degenerative neurological disease.
Individual 5 is the index patient of family 3 (Figure 1c). She is the second child of healthy, unrelated, German parents (Figure 2c). She was born via spontaneous vaginal delivery at 38+4 weeks of gestation with a birth weight of 3360 g (54th percentile, +0.1 SD), a length of 52 cm (65th percentile, +0.4 SD), and a head circumference of 33 cm (11th percentile, −1.2 SD). Postnatal adaptation was good, APGAR was 9/10/10 at 1/5/10 min and the arterial cord blood pH was normal with 7.28. Early routine examinations were unremarkable, however at 2 months of age she was hospitalized for bronchopneumonia and at this time point microcephaly was noted, as well as pulmonary valve stenosis and esotropia. Cerebral ultrasound revealed pachygyria and an enlargement of the inner and outer cerebrospinal fluid spaces. TORCH serology was negative. She received physiotherapy for hypotonia. At the age of 7 months she reportedly had a first febrile seizure and she continued to have focal seizures with secondary generalization regularly and was hospitalized several times for status epilepticus. Her development is severely delayed, she neither walks nor speaks. She has a history of eating her own hair. Post-ictal EEGs revealed sharp-slow-wave complexes temporally on both sides, and a right temporal focal point was identified after status epilepticus. At 11 years of age, cMRI showed reduced cerebral volume compatible with the severe microcephaly, likely secondary dysgenesis of the corpus callosum, reduced cerebellar volume, mesial temporal sclerosis on the left side and parahippocampal white matter intensities. Physical examination at 11 years revealed increased tendon reflexes and spasticity of all four limbs, most prominently of the legs. The patient was friendly, but quite restless and tried to put objects in her mouth and to bite her caregivers, out of curiosity rather than aggressiveness. Her height was 129 cm (1st percentile, −2.5 SD), weight 17 kg (<1st percentile, −5.0 SD), and her head circumference was 46.5 cm (<1st percentile, −6.2 SD) (Table 1). She exhibited the following dysmorphic features: a microcephalic aspect with a sloping forehead, deep-set eyes, a long nose with a high nasal bridge, long palpebral fissures, and low-set ears (Figure 1c). Before ES, she received a diagnostic next-generation sequencing panel for epilepsy-related genes, as well as chromosome analysis and array-CGH, all with normal results.
3.2 Molecular findings
We performed trio ES on DNA of individual 1 and her parents. The data set was initially analyzed for variants in known genes for microcephaly. As this analysis did not reveal a pathogenic variant, we then analyzed the data for homozygous variants, due to the consanguineous background, and identified the homozygous missense variant c.397C>T, p.(Arg133Cys) (chr14:100826916G>A; GRCh37/hg19) in the WARS1 gene (NM_004184.4; Figure 2a). We also checked for relevant de novo variants but found none. The variant c.397C>T is present in one heterozygous allele in the gnomAD browser. It affects a highly conserved residue within the N-terminal domain and is predicted to be damaging by the prediction programs SIFT (Score: 0.000), PROVEAN (Score: −7.12), MutationTaster (Score: 180), and PolyPhen2 (HumDiv Score: 1.000; HumVar Score: 0.999). Both parents were shown to be heterozygous carriers of the variant c.397C>T (Figure 2a).
In family 2, we performed ES for three affected brothers and identified the identical homozygous variant c.397C>T, p.(Arg133Cys) in WARS1 that had also been identified in individual 1, in all three of them (Figure 2b). Targeted Sanger sequencing in the two healthy siblings showed that they did not carry the variant in the homozygous state (data not shown). There was no parental DNA available for segregation analysis.
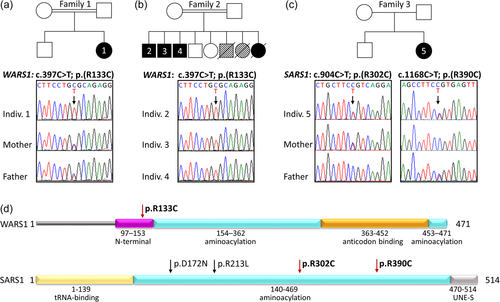
The identification of the identical missense variant in the WARS1 gene in two families prompted us to ask whether there might be a shared haplotype. We, therefore, compared rare and common SNVs at the WARS1 locus and in a 1-Mb region flanking WARS1 (Figure 3). We found that the homozygous haplotypes of individual 1 and individuals 2–4 shared no rare variants with a MAF <1% except WARS1 c.397C>T in the analyzed region. Identical homozygous variants in individuals 1 and 2–4 all had a MAF >50%. This analysis likely excludes a common ancestral allele in families 1 and 2. The fact that the same mutation arose independently in two patients with very similar phenotypes lends weight to the supposed pathogenicity of the variant.
Family 3 was analyzed via patient-parent trio ES. We identified the compound-heterozygous variants c.904C>T, p.(Arg302Cys) (chr1:109777988C>T; GRCh37/hg19), and c.1168C>T, p.(Arg390Cys) (chr1:109779081C>T; GRCh37/hg19) in the SARS1 gene (NM_006513.4). The variant c.904C>T was inherited from the healthy mother and the variant c.1168C>T was inherited from the healthy father (Figure 2c). The variant c.904C>T, p.(Arg302Cys) is absent from the gnomAD browser, while c.1168C>T, p.(Arg390Cys) is present in two heterozygous alleles. Both variants are predicted to be damaging by the prediction programs SIFT (Scores: 0.007 and 0.000, respectively), PROVEAN (Scores: −7.53 and −7.91, resp.), MutationTaster (Scores: 180, resp.), and PolyPhen2 (HumDiv Scores: 1.000, resp.; HumVar Scores: 1.000, resp.) and both variants affect highly conserved residues within the catalytic domain of SARS1 (Figure 2d).
3.3 Zebrafish studies
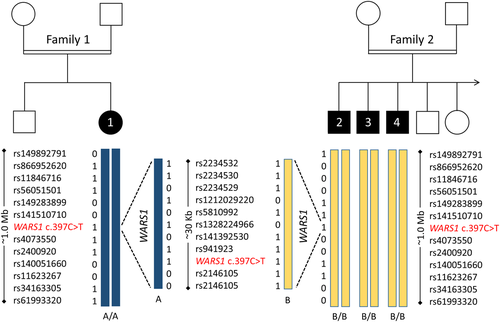
Zebrafish have a single ortholog of human WARS1. First, we investigated the effect of loss of wars1 function on animal development by generating a wars1 knockout using CRISPR/Cas9-mediated targeted mutagenesis and analyzed phenotypes in the F0 (founding generation) generation. Injected embryos were raised, and the phenotypes scored within the first 5 days of post-fertilization (dpf) (F0 generation). Wars1 F0 mutant embryos did not show any obvious phenotypes until 2 dpf. At 3 dpf, mutants show a smaller head and eyes, as well as heart edema compared to control animals (Figure 4). The smaller head phenotype is similar to the microcephaly observed in individuals affected by the homozygous WARS1 variant. To confirm whether the phenotypes in the zebrafish model arise from the loss of wars1 function, we used an in vitro synthetic mRNA rescue method by using wild-type (WT) human WARS1 mRNA, co-injected with wars1 sgRNA/Cas9 RNP. mRNA rescue restored phenotypes shown by F0 mutants, confirming the specificity of the phenotypes (Figure 4). To test the pathogenicity of the p.Arg133Cys variant in WARS1, we compared the efficiency of variant WARS1 mRNA to rescue the F0 mutant phenotypes to WT for both head and eye size. mRNA carrying the variant failed to rescue the eye and head size phenotypes (Figure 4), suggesting the variant significantly impacts protein function.
3.4 Structural mapping
To gain further insight into the potential functional consequences of the variants identified, we mapped them onto previously determined structures of human WARS1 (Shen et al., 2006) and SARS1 (Xu et al., 2013) (Figure 5a).
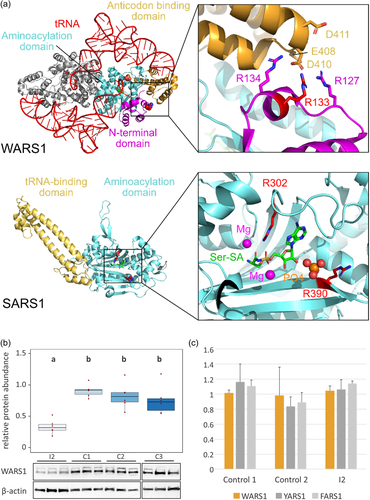
In the case of WARS1, we used the structure of the enzyme in complex with its target (Shen et al., 2006). WARS1 functions as a homodimer, and the anticodon binding domain of one monomer interacts with the anticodon loop of the tRNA to position the substrate in the active site of the other monomer (Shen et al., 2006). Structural mapping shows that Arg133 is not in vicinity of the active site and does not appear to be directly involved in interactions with the tRNA substrate (Figure 5a). Instead, it is part of a basic patch in the N-terminal domain (comprised of Arg127, Arg133, and Arg134), which forms a contact interface with an acidic patch (comprised of Glu408, Asp410, Asp411) in the anticodon-binding domain. Arg133 may form a salt bridge with Asp410, and its substitution with cysteine would abolish this salt bridge.
For structural mapping of the two SARS1 variants, we used the structure of the enzyme in complex with the substrate analog Ser-SA (Xu et al., 2013). This reveals that both Arg302 and Arg390 are located within the active site of the enzyme (Figure 5a). Arg302 is highly conserved among seryl-tRNA synthetases and forms direct contacts with the substrate Ser-SA (Xu et al., 2013). Thus, substitution of this residue with cysteine is likely to affect substrate binding by the enzyme. Arg390 is located at the edge of the active site and may thus be involved in tRNA binding. This residue is conserved as a basic residue (arginine or lysine) from human to Escherichia coli (C. Wang et al., 2015), suggesting that its substitution to cysteine would likely affect proper function of the enzyme.
3.5 Functional analyses in WARS1 mutant patient fibroblasts
Western blot analysis showed that the abundance of WARS1 was significantly reduced in WARS1 mutant patient fibroblasts from individual 2 compared with three controls. Protein abundance was determined as pixel intensity of WARS1 immunoblot band, relative to β-actin loading control, on six replicates. ANOVA was performed (significance level α = 0.05, [F(3,20) = 13.49, p = 4.85e−05]), followed by post hoc Tukey analysis (Figure 5b). A cycloheximide chase assay did not show significantly faster protein degradation in patient fibroblasts than in controls (Supporting Information: Figure 2). Aminoacylation activity was normal in patient fibroblasts when compared with controls and endogenous YARS1 and FARS1 activity (Figure 5c).
4 DISCUSSION
Here we describe the identification of a novel homozygous missense variant in the WARS1 gene as well as compound heterozygous variants in the SARS1 gene in five individuals presenting an overlapping developmental disorder with microcephaly. WARS1 and SARS1 are members of the family of human ARSs, which are responsible for the attachment of an amino acid to its cognate tRNA. For each of the 20 canonical amino acids, there is one specific ARS, except for glutamic acid and proline, which, in humans, interact with the same multifunctional ARS (EPRS1). Apart from their recognized enzymatic function, ARSs also act beyond aminoacylation (Fukui et al., 2009; Guo et al., 2010; Xu et al., 2018).
WARS1 encodes the cytosolic tryptophanyl-tRNA synthetase (WARS1). In addition to its enzymatic function of charging tryptophan to the complementary tRNA, this protein has been reported to play a role in angiogenesis regulation and immune response (Ahn et al., 2017; Xu et al., 2018). The variant c.397C>T in WARS1, identified in families 1 and 2 of this study, leads to the substitution of a highly conserved arginine residue with cysteine at position 133 within the N-terminal domain (Shen et al., 2006). The variant c.397C>T is annotated in the gnomAD database in a single allele, which is compatible with a rare recessive disease allele. Of note, populations from the Middle East are commonly underrepresented in the available genome databases. However, given the high number of alleles (251.256) annotated in the gnomAD database from a variety of population backgrounds, we believe this to be a strong argument in favor of pathogenicity. The variant is predicted as damaging by several in silico prediction programs (MutationTaster, SIFT, PROVEAN, and PolyPhen2). Based on these results, we assumed this variant to be likely causative.
Since we found the variant c.397C>T, p.(Arg133Cys) in two unrelated families originating from neighboring Arabian countries, we hypothesized that there might be a common ancestral allele. However, comparison of rare and common SNPs within a 1-Mb region around the WARS1 locus in the exome data set of both families likely excluded a shared haplotype (Figure 3). This result might indicate that the mutated cytosine at position 397, as part of a CpG dinucleotide, could be prone to mutation to thymine through methylation and spontaneous deamination. The fact that the same variant arose on different haplotypes in two individuals with a very similar clinical phenotype strongly argues for pathogenicity of the variant.
To characterize the mutational effect of the WARS1 variant c.397C>T, p.(Arg133Cys), we tested the variant in a CRISPR/Cas9 wars1 knockout zebrafish model. The wars1 knockout zebrafish shows a significant reduction in head size and ocular diameter, which indicates a defect in the development of the central nervous system (Figure 4). While the wars1 knockout phenotypes could be rescued with wild-type human mRNA, the p.Arg133Cys variant mRNA was unable to rescue either phenotype (Figure 4). We, therefore, concluded that the variant c.397C>T, p.(Arg133Cys) in WARS1 leads to a, possibly incomplete, loss of function. This observation is in line with other reports in the literature, which have shown that biallelic mutations in ARS genes confer loss-of-function effects (Botta et al., 2021; Friedman et al., 2019; Helman et al., 2021; Mendes et al., 2020; Ravel et al., 2021).
To further explore the WARS1 variant c.397C>T, p.(Arg133Cys), we performed structural mapping, which indicated that arginine 133 is part of an intramolecular domain interface, where it may form electrostatic interactions with residues in the anticodon-binding domain (Figure 5a). We, therefore, hypothesized that the substitution of arginine 133 by cysteine could destabilize the interaction between the N-terminal and the anticodon-binding domains, which could, in turn, affect the structural integrity of the enzyme or the correct binding and positioning of the tRNA substrate. To test our hypotheses, we first performed western blot analyses of WARS1 in patient fibroblasts from individual 2 and observed a significant reduction in protein abundance (Figure 5b). A cycloheximide chase assay showed no significant difference in protein degradation between mutant and control fibroblasts. However, protein levels of WARS1 were so low in patient fibroblasts that the method might be unable to discern differences with densitometric quantification (Supporting Information: Figure 2). We therefore cannot fully exclude protein instability. We then applied an LC-MS/MS-based enzyme assay to test the aminoacylation activity of the mutant WARS1 protein. We observed that under Michaelis–Merten conditions, there was no significant difference in aminoacylation activity between mutant and control fibroblasts (Figure 5c). This result is unexpected; however, normal enzymatic activity in this in vitro assay does not necessarily reflect aminoacylation in vivo or in other cell types. A similar example of a complex mutational mechanism with normal enzymatic function, in a patient with a severe neurological phenotype due to biallelic mutations in RARS1, has been described before (Mendes et al., 2020). Based on the results from structural mapping, Western blot analyses, and the cycloheximide chase assay, we hypothesize that the mutation c.397C>T; p.(Arg133Cys) might lead to inefficient protein synthesis or misfolding of the WARS1 protein, resulting in early degradation and inefficient delivery of aminoacylated tRNA to the ribosome. Given the genetic and functional evidence, we classified this variant as likely pathogenic according to ACMG (PS3, PM2, PP3, PP4).
Heterozygous, dominant variants in the WARS1 gene are known to cause distal hereditary motor neuropathy type 9 (HMN9; MIM# 617721) (Li et al., 2019; Tsai et al., 2017; B. Wang et al., 2019) and have been linked to Charcot-Marie-Tooth disease (LaFave et al., 2014). Dominant and recessive variants in the same ARS gene, underlying two distinct phenotypes, have previously been documented (Table 2). For example, dominant variants in alanyl-tRNA synthetase AARS1 can cause Charcot-Marie-Tooth disease type 2N (CMT2N; MIM# 613287), while recessively inherited variants cause developmental and epileptic encephalopathy type 29 (DEE29; MIM# 616339), and biallelic loss-of-function variants in HARS1 have recently been described as the cause of a multisystem neurodevelopmental disorder with microcephaly, ataxia, and additional neurological features, such as spasticity and athetosis (Galatolo et al., 2020), whereas dominant variants in the HARS1 gene have been associated with CMT type 2W (CMT2W; MIM# 616625). It has been suggested that, in contrast to biallelic variants, autosomal dominant variants in ARS genes exert a dominant-negative effect (Tsai et al., 2017; Wallen & Antonellis, 2013). It is therefore inconspicuous that our patients showed no signs of a distal neuropathy. Since we investigate young individuals, we cannot exclude that some signs of neuropathy may develop later in life. However, the healthy carrier parents did not report such symptoms either. The differential phenotypes caused by dominant or recessive variants in some of the ARS genes indicate variant-specific effects. Interestingly, HGMD-annotated variants for the WARS1-associated autosomal dominant neuropathy (CM1915624, CM175385, CM199660, and CM2028011) are distributed between amino acid residues 138 and 410 within the N-terminal, the aminoacylation of the anticodon binding domains. Thus, the specific phenotypic effects apparently do not depend on the affected protein domain, but rather on the individual amino acid substitution.
| Amino acid | Short | ARS | Phenotype dominant | Phenotype recessive | ARS-DDM |
|---|---|---|---|---|---|
| Alanine | A | AARS1 | CMT2N | Developmental and epileptic encephalopathy/TTD (Botta et al., 2021) | ARS-DDM-A |
| Cysteine | C | CARS1 | NR | Microcephaly, developmental delay, and brittle hair syndrome (Kuo et al., 2019) | ARS-DDM-C |
| Aspartic acid | D | DARS1 | NR | Hypomyelination with brainstem and spinal cord involvement and leg spasticity (Taft et al., 2013) | ARS-DDM-D |
| Glutamic acid + Proline | E P | EPRS1 | NR | Leukodystrophy, hypomyelinating (Mendes et al., 2018) | ARS-DDM-E |
| Phenylalanine | F | FARSA/FARSB | NR | NDD with brain, liver, and lung abnormalities (Krenke et al., 2019) | ARS-DDM-F |
| Glycine | G | GARS1 | CMT2D/dHMN type VA/SMAJI | Multisystem developmental syndrome with microcephaly (Oprescu et al., 2017) | ARS-DDM-G |
| Histidine | H | HARS1 | CMT2W | NDD with microcephaly and ataxia (Galatolo et al., 2020) | ARS-DDM-H |
| Isoleucine | I | IARS1 | NR | Growth retardation, impaired intellectual development, hypotonia, and hepatopathy (Kopajtich et al., 2016) | ARS-DDM-I |
| Lysine | K | KARS1 | NR | CMTRIB/deafness/leukencephalopathy (McMillan et al., 2015) | ARS-DDM-K |
| Leucine | L | LARS1 | NR | Infantile liver failure syndrome 1 (Casey et al., 2012) | ARS-DDM-L |
| Methionine | M | MARS1 | CMT2U | Interstitial lung and liver disease/TTD/NDD with microcephaly (Botta et al., 2021) | ARS-DDM-M |
| Asparagine | N | NARS1 | NDD with microcephaly | NDD with microcephaly (Manole et al., 2020) | ARS-DDM-N |
| Glutamine | Q | QARS1 | NR | Microcephaly, progressive, seizures, and cerebral and cerebellar atrophy (Zhang et al., 2014) | ARS-DDM-Q |
| Arginine | R | RARS1 | NR | Leukodystrophy, hypomyelinating (Wolf et al., 2014) | ARS-DDM-R |
| Serine | S | SARS1 | NR | Evidence reported in this study | ARS-DDM-S |
| Selenocysteine | U | (SARS1) | - | - | - |
| Threonine | T | TARS1 | NR | TTD (Theil et al., 2019) | ARS-DDM-T |
| Valine | V | VARS1 | NR | NDD with microcephaly, seizures, and cortical atrophy (Okur et al., 2018) | ARS-DDM-V |
| Tryptophan | W | WARS1 | dHMN type IX/CMT | Reported in this study | ARS-DDM-W |
| Tyrosine | Y | YARS1 | CMTDIC | NDD with microcephaly, brain anomalies, lung, and liver disease (Williams et al., 2019) | ARS-DDM-Y |
- Abbreviations: ARS, aminoacyl-tRNA synthetase; ARS-DDM, aminoacyl-tRNA synthetase-associated developmental disorder with or without microcephaly; CMT, Charcot-Marie-Tooth; dHMN, distal hereditary motor neuropathy; NDD, neurodevelopmental disorder; NR, not reported; SMA, spinal muscular atrophy; s.a.; see above; TTD, Trichothiodystrophy.
Very recently, while this manuscript was in revision, Okamoto et al. described the compound heterozygous missense variants c.1342C>T; p.(Arg448Trp) and c.997G>A; p.(Ala333Thr) in WARS1 in a girl with developmental delay, profound intellectual disability, seizures, microcephaly, and hypomyelination of the brain. Peripheral neuropathy was excluded in this patient as well as in her parents, who were heterozygous carriers of one variant each (Okamoto et al., 2022). This report supports the findings of this study as well as the notion that symptoms of peripheral neuropathy are unlikely to occur in patients suffering from ARS-related developmental disorders or in their parents.
SARS1 encodes the cytosolic seryl-tRNA synthetase (SARS1), which is responsible for charging of tRNA with serine. The variants c.904C>T and c.1168C>T in SARS1 each lead to the substitution of a highly conserved arginine residue with cysteine within the aminoacylation domain of SARS1. Both variants were predicted as damaging by several in silico prediction programs and structural mapping indicated that the substitution of Arg302 with cysteine is likely to impair substrate binding and/or catalysis, while the substitution of Arg390, located at the edge of the active site, might interfere with tRNA binding and/or enzymatic activity (Figure 5a). Thus, these variants likely affect proper function of SARS1. Given the phenotypic overlap with previously reported patients, together with the evidence described above, we classified both variants in SARS1 as likely pathogenic, according to ACMG criteria (PM1, PM2, PP3, and PP4). The phenotype observed in individual 5 closely resembles that described by Musante et al., who found the homozygous variant c.514G>A, p.(Asp172Asn) in four individuals from a consanguineous Iranian family who suffered from a severe neurodevelopmental disorder with microcephaly and epilepsy (Table 2) (Musante et al., 2017). Epileptic encephalopathy and neurodevelopmental abnormalities have also been described for four individuals from a consanguineous family in whom the homozygous variant c.638G>T; p.(Arg213Leu) in SARS1 was identified (Ravel et al., 2021; Thevenon et al., 2016). Microcephaly was not a feature in this family. It is noteworthy that SARS1 not only conducts the synthesis of seryl-tRNA but also catalyzes the first step of selenocysteine (Sec) synthesis, namely the serylation of Sec-specific tRNA (tRNA(Sec)) (Amberg et al., 1996; C. Wang et al., 2015). Selenocysteine is required at the catalytic center of selenoproteins, which play an important role in neurogenesis (Pitts et al., 2014). Thus, pathogenic variants in SARS1 likely impair nervous system development by reduced availability of seryl-tRNA as well as tRNA(Sec). It is therefore not surprising that our patient's phenotype resembles cerebellocerebral atrophy, a syndrome with microcephaly, intellectual disability, and spastic quadriplegia caused by biallelic variants in the O-phosphoserine-tRNA-selenocysteine-tRNA synthase gene (SEPSECS; MIM# 613811), which is another important enzyme in the selenocysteine synthesis pathway (Ben-Zeev et al., 2003). Apart from aminoacylation activity, SARS1 has also been implicated in vascular development (Fukui et al., 2009) and there is one report implicating the de novo missense variant c.971T>C (p.Ile324Thr) in SARS1 in nonsyndromic congenital brain arteriovenous malformation. However, this disease association needs to be verified through identification of additional patients (K. Wang et al., 2018).
The individuals described herein show an overlapping phenotype with secondary microcephaly, developmental delay, intellectual disability, and brain anomalies including cerebral and cerebellar atrophy, white matter hyperintensities, and a thin corpus callosum. A closer look at recessive phenotypes previously described for the cytosolic ARS genes revealed that they are almost invariably associated with developmental delay and intellectual disability. Microcephaly is a feature of most of these phenotypes, and many are associated with seizures or ataxia, or both. Hypomyelination, cerebral and cerebellar atrophy are also common characteristics (Table 2). These phenotypes have each been described in a small number of individuals and a range of disease names have been defined for them, based on their most prominent features. However, these classifications fail to pinpoint the features that are common to the ARS-related phenotypes and, most importantly, they do not follow a systematic approach. For example, the recessive phenotype for AARS1 (a combination of microcephaly, short stature, failure to thrive, skeletal abnormalities, nystagmus, and blepharospasm, cerebral atrophy and hypomyelination, cognitive impairment, spasticity, dystonia, and chorea, in addition to epilepsy) is termed developmental and epileptic encephalopathy 29 (DEE29; MIM# 616339) (Simons et al., 2015). Meanwhile, although phenotypically largely overlapping, the recessive phenotype for EPRS1 (microcephaly, short stature, failure to thrive, hearing loss, nystagmus and optic atrophy, cerebellar atrophy and hypomyelination, cognitive impairment, spasticity, dystonia, ataxia, athetosis, and seizures), is termed hypomyelinating leukodystrophy 15 (HLD15; MIM# 617951). To unify the recessive ARS-related disorders, we propose to reclassify them as a group or spectrum of disorders and propose the name “aminoacyl-tRNA synthetase-related developmental disorder with or without microcephaly” (ARS-DDM). Each subtype should be denominated by addition of the one-letter abbreviation of the amino acid corresponding to the ARS in question, that is, ARS-DDM-S for SARS1 (Table 2). This new disease nomenclature for ARS-related developmental disorders is in line with the dyadic approach to the delineation of disease entities as proposed by Biesecker et al. (2021). We modified the suggested approach by using the common denominator “ARS” to include a group of related gene names into one disease group, instead of using the single-gene name for each disorder, as suggested for single-gene disorders.
We are aware that in addition to the common neurodevelopmental phenotype, some ARSs are associated with specific organ impairment. Some of these more specific phenotypes indicate a high demand for certain amino acids in certain tissues, such as lung, liver, or hair. For instance, a developmental phenotype with brittle hair is caused by biallelic variants in CARS1 (Kuo et al., 2019) or TARS1 (Theil et al., 2019). The TARS1-related phenotype has previously been classified as nonphotosensitive trichothiodystrophy (TTD7; MIM# 618546), which is not incorrect, because the typical tiger band pattern has been observed upon polarized light microscopy. However, at least one of the individuals also had a neurodevelopmental phenotype and almost no clinical data were available for the second individual described. Very recently, recessive variants in AARS1, as well as MARS1, were identified in individuals with clinical signs of nonphotosensitive trichothiodystrophy in addition to a severe neurodevelopmental disorder with microcephaly (Botta et al., 2021). These findings strongly indicate that brittle hair and (follicular) keratosis belong to the plethora of ARS-DDM-associated abnormalities. A systematic approach to the ARS-related developmental disorders is especially warranted since it has recently been shown that patients with certain ARS-DDM profit from substitution with the corresponding amino acid (Kok et al., 2021). Therefore, stronger efforts to identify, correctly diagnose, and treat these patients should be undertaken. We believe that a structured nomenclature system, systematically capturing all ARS-related developmental disorders, might aid in achieving this goal.
In summary, we add a new member to the growing number of ARS-related phenotypes, the WARS1-related autosomal recessive developmental disorder, and extend the knowledge of the rarely described SARS1-related autosomal recessive disorder. We provide potential functional explanations for the severe effects of the observed variants and propose a systematic nomenclature for the emerging phenotypic spectrum of ARS-related developmental disorders with or without microcephaly (ARS-DDM).
AUTHOR CONTRIBUTIONS
Nina Bögershausen, Gökhan Yigit, and Yun Li analyzed the genetic data of individuals 1 and 5. Hannah E. Krawczyk performed cell culture and cellular functional experiments. Saskia Biskup performed ES for individual 1. Arne Zibat processed ES data for individual 1. Rami A. Jamra, Johannes R. Lemke, Konrad Platzer, and Johannes R. Lemke performed and analyzed ES for individuals 2–4. Konrad Platzer, Johannes R. Lemke, Rami A. Jamra, and Martin Zenker performed exome analysis and analyzed the exome data of individuals 2–4. Janine Altmüller and Peter Nürnberg performed ES for individual 5. Anna M. Polo acquired and reviewed the clinical data of individual 1. Christian P. Kratz and Beate B. Dörgeloh acquired and reviewed the clinical data of individuals 2–4. Sheng-Jia Lin, Kevin Huang, Barbara Vona, and Gaurav K. Varshney coordinated and performed the zebrafish work and created Figure 4. Marisa I. Mendes, Desiree E. C. Smith, and Gajja S. Salomons performed the LC-MS aminoacylation assay. Eva Bültmann performed and analyzed MRIs of individuals 2 and 4. Irina Hüning, Malte Spielmann, and Andreas Busche acquired and reviewed the clinical data of individual 5. Julia Schmidt helped with data analysis and clinical evaluations. Hauke S. Hillen performed structural mapping. Nina Bögershausen and Bernd Wollnik wrote the paper. Bernd Wollnik coordinated the study.
ACKNOWLEDGMENTS
We thank the families for their participation in this study and Karin Boss for careful proofreading. This study was funded by the Heidenreich von Siebold program 2016 grant (University Medical Center Göttingen, Germany) to NB, the DFG (Deutsche Forschungsgemeinschaft) grant EXC 2067/1-390729940 and DZHK (German Centre for Cardiovascular Research; partner site Göttingen) grant 81Z0300112 to BW, DFG grants FOR2848, SFB1190, and EXC 2067/1-390729940 to HSH, DFG VO 2138/7-1 grant 469177153 to B.V., the Oklahoma Medical Research Foundation, and Presbyterian Health Foundation (PHF-4411-07-04-0) (GKV), as well as funding through the DFG Collaborative Research Center 889. Open Access funding enabled and organized by Projekt DEAL.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
WEB RESOURCES
The following databases were used for this study: Ensembl: https://www.ensembl.org/index.html; HGMD: http://www.hgmd.cf.ac.uk/ac/index.php; gnomAD: https://gnomad.broadinstitute.org/; The UCSC browser: http://genome.ucsc.edu/; The human protein reference database: http://www.hprd.org/; Uniprot: https://www.uniprot.org/; PubMed: http://www.ncbi.nlm.nih.gov/pubmed/; RSCB Protein Data Bank: https://www.rcsb.org/
Open Research
DATA AVAILABILITY STATEMENT
Variants have been submitted to the database ClinVar: https://www.ncbi.nlm.nih.gov/clinvar/?term=WARS1 [gene]; https://www.ncbi.nlm.nih.gov/clinvar/?term=SARS1 [gene]. Due to ethical considerations (protection of personal data), we cannot share our patients' original exome data in public databases. Accession numbers: WARS1: NM_004184.4, UniProt ID P23381. SARS1: NM_006513.4, UniProt ID P49591.