CAPN3 c.1746-20C>G variant is hypomorphic for LGMD R1 calpain 3-related
Magdalena Mroczek and Inna Inashkina have equal contribution.
Abstract
The investigated intronic CAPN3 variant NM_000070.3:c.1746-20C>G occurs in the Central and Eastern Europe with a frequency of >1% and there are conflicting interpretations on its pathogenicity. We collected data on 14 patients carrying the CAPN3 c.1746-20C>G variant in trans position with another CAPN3 pathogenic/likely pathogenic variant. The patients compound heterozygous for the CAPN3 c.1746-20C>G variant presented a phenotype consistent with calpainopathy of mild/medium severity. This variant is most frequent in the North/West regions of Russia and may originate from that area. Molecular studies revealed that different splicing isoforms are produced in the muscle. We hypothesize that c.1746-20C>G is a hypomorphic variant with a reduction of RNA and protein expression and only individuals having a higher ratio of abnormal isoforms are affected. Reclassification of the CAPN3 variant c.1746-20C>G from variant with a conflicting interpretation of pathogenicity to hypomorphic variant explains many unidentified cases of limb girdle muscular dystrophy R1 calpain 3-related in Eastern and Central Europe.
Limb girdle muscular dystrophy (LGMD) R1 calpain 3-related (MIM# 253600) is the most common autosomal recessive form of LGMD constituting approximately 30% of all LGMD cases (Lasa-Elgarresta et al., 2019). LGMD R1 calpain 3-related is caused by biallelic inactivating variants in the CAPN3 gene. It has been proposed that some variants at intron-exon boundaries of CAPN3 may modify precursor messenger RNA (mRNA) and thus create multiple new transcripts (Nascimbeni et al., 2010). The clinical significance of nucleotide changes in intronic regions is very difficult to predict in silico and this makes distinguishing between benign and pathogenic variants a challenge. Identifying CAPN3 variants that affect splicing may have therapeutic consequences, because preliminary studies suggested the possibility of restoring the normal splicing with antisense oligonucleotides (Blázquez et al., 2013). There is conflicting information about the pathogenicity of the CAPN3 variant c.1746-20C>G (NM_000070.3:c.1746-20C>G, dbSNP: rs201892814). The conflict arises from its higher allelic frequency (minor allele frequency [MAF] >1%) than it is commonly accepted for the pathogenic variants (Karczewski et al., 2020). In LGMD cohorts this variant has been identified several times and classified as pathogenic/likely pathogenic on the clinical level (Table S1). The compound heterozygous patients with the c.1746-20C>G variant in a trans position with a second pathogenic calpain three variant showed a significantly decreased calpain level or a complete loss of calpain on Western blots (Macias et al., 2021; Nascimbeni et al., 2010; Schutz et al., 2017).
Furthermore, there are controversial experimental data published regarding the variants’ effect on RNA splicing. Krahn et al. (2007) reported no deviation from the norm in the complementary DNA (cDNA) analysis of exon 14, in a patient with severely reduced calpain on Western blot and symptoms of calpainopathy. Interestingly, the second variant for this patient (NM_000070.3:c.1745+4_1745+7del) was located in the same intron, and cDNA analysis showed abnormal exon 13 splicing and reported an activated cryptic splice site within it (Krahn et al., 2007). However, Nascimbeni et al. (2010) reported a generation of four aberrant transcripts, present exclusively in the muscles of patients with the c.1746-20C>G variant (Nascimbeni et al., 2010).
Although most of the families with calpainopathy follow a recessive inheritance pattern (Angelini, 2005), inheritance is also described as dominant several times (MIM# 618129). Two in-frame deletions in the CAPN3 (NM_000070.3:c.643_663del and NM_000070.3:c.598_612del) and more recently, two substitutions (NM_000070.3:c.1333G>A and NM_000070.3:c.1715G>C) have been linked to autosomal dominant (AD) forms of calpainopathy (Cerino et al., 2020; González-Mera et al., 2021; Vissing et al., 2016, 2020). In the cases of dominantly inherited calpainopathy, the phenotype was comparable with a mild LGMD. However, it varied substantially between individuals, and some patients presented only with a late-onset mild muscle weakness, creatine kinase (CK - creatine phosphokinase) elevation and muscle pain. At the same time, a few family members were affected by a severely disabling neuromuscular disease (Vissing et al., 2016). Establishing the pathogenicity and models of inheritance in calpainopathies may greatly influence genetic counselling.
Our study aimed to characterize the pathogenicity of the CAPN3 c.1746-20C>G variant on the clinical and molecular levels, its population frequency and relation to the LGMD R1 calpain 3-related.
We gathered clinical, molecular and genotype data from 14 patients: three of Latvian origin, two of Lithuanian origin, six from the Czech Republic, one from Slovenia and two from the Netherlands (Table S1). Materials and methods are in Supporting Information.
Segregation analysis was performed in all cases where parental DNA was available. The variant c.1746-20C>G was found six times together with the deletion common in Central and Eastern Europe (c.598_612del) and three times with the variant NM_000070.3:c.643T>C (p.Ser215Pro).
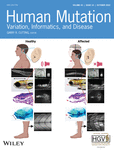
The disease presentation in patients varied from childhood to late onset with mild to moderate proximal weakness. Mean age at the last examination was 49.7 years, SD = 11.9, median was 48 years. CK ranged from 306 U/L in Patient number 14 to >4000 U/L in Patient number 5. (Table S1). All patients, apart from Patient number 14, were ambulatory at the time of the last examination. Although muscular hypertrophy is somewhat unusual for calpainopathy, six of the patients showed pseudohypertrophy of quadriceps muscles or mild hypertrophy of both calves. During the disease course, some muscle groups tended to become atrophic after being hypertrophic at the beginning of the disease (Figure 1). Electromyography demonstrated myopathic changes in five patients of those included in this study. Two patients (Patient number 1 and Patient number 14) had mild cardiac left ventricle hypertrophy and one had a mild facial involvement (Patient number 5). None of the patients had contractures or respiratory insufficiency. Muscle magnetic resonance imaging (MRI) demonstrated fatty transformation predominantly of the posterior thigh muscles (M. adductor magnus, semimembranosus, semitendinosus, biceps) with relative sparing of M. sartorius and M. rectus femoris (Figure S1). In Patient number 1 additionally M. vastus, M. gastrocnemius medialis (asymmetrical involvement) and Mm. glutei were affected. The muscle biopsy in Patient number 1 revealed mild myopathic changes with variation in fiber size, internal nuclei, fiber splitting and lipomatosis. Fiber atrophy and Type 2 fiber hypertrophy were noted. In Patient number 12 myopathic-neurogenic changes, variability of muscle fibers and numerous internal nuclei has been reported. The muscle biopsy of Patient number 14 showed a severe muscular dystrophy.
There are several publications in which authors considered the CAPN3 gene variant c.1746-20C>G, together with another pathogenic/likely pathogenic CAPN3 variant, to be causal for calpainopathy. Clinically the patients presented with the late disease onset: in the adolescence or adulthood. The ambulation was relatively well preserved. Cardiac involvement was diagnosed in four cases (Table S1). There was no respiratory involvement described. CK levels were variable from normal to elevated more than 10 times. The severe reduction or a total loss of calpain 3 expression in Western blot was described in all cases, where the test was performed (Macias et al., 2021; Nascimbeni et al., 2010; Schutz et al., 2017; Ten Dam et al., 2019).
Interestingly, several patients in our cohort presented with an accentuated muscular hypertrophy as a prominent clinical feature and only a minor component of muscle weakness. Muscular hypertrophy is a rare clinical feature of calpainopathy, and even if calf hypertrophy occurs, it is discrete. Only a few reports highlight the hypertrophy in LGMD R1 calpain 3-related. Rekik et al. (2019) reported a patient presenting with a calf hypertrophy and carrying a homozygous (c.1681T>C) variant affecting exon 13. Hypertrophy associated with the CAPN3 variant c.1746-20C>G was previously described in six cases (Krahn et al., 2007; Macias et al., 2021; Schutz et al., 2017) (Table S1), primarily as calf hypertrophy.
In the study we identified two unaffected individual homozygous for c.1746-20C>G within the self-reported healthy cohort from the Latvian biobank and the Estonian cohort. Both individuals had normal CK. The individual from Estonia was clinically investigated at 48 years of age and had no signs of neuromuscular weakness (Table S2). An extensive literature search identified further patients homozygous for the calpain variant: one affected in The Netherland LGMD database (Ten Dam et al., 2019) and one affected individual from the Turkish population (Balci et al., 2006) (Table S2). The disease onset for the patient from the Netherlands was at 46 years of age with the first clinical diagnosis at the age of 61 years. The course of the disease was mild with a mildly elevated CK—up to 480 U/L (Ten Dam et al., 2019). The patient is ambulatory until now, but requires walking sticks. Neurological investigation revealed mild facial diplegia, neck flex and extension according to MRC scale: 4/5, shoulders muscle strength 2–4/5, proximal arms 4/5, distal arms 5-/5 proximal legs 2–3/5 and distal legs 5-/5. Shoulder muscles and thigh muscles were atrophic. No hypertrophy has been observed. Histology reported severe muscular dystrophy. The patient from Turkey showed an intermediate phenotype with the onset at the age of 17 years. There was no information about calpain reduction in Western blot or particular changes in MRI, therefore diagnosis is based on clinical symptoms of LGMD and molecular findings. Unfortunately, an affected patient from Turkey was not available for the follow-up, however given an early disease onset and the fact that large-throughput sequencing method diagnostic has not been performed, he might have been affected with the other genetic disease.
Through literature search of the PubMed and ClinVar databases we identified 34 reported patients with c.1746-20C>G and other pathogenic/likely pathogenic CAPN3 variant. The phenotype reflected mild LGMD R1 calpain 3-related (Table S1). Interestingly, the c.1746-20C>G variants occurred with deletions c.643_663del and c.598-612del, both being described as dominant (Cerino et al., 2020; Vissing et al., 2016).
Recently, a growing number of variants in the CAPN3 gene, found in isolated populations, were identified as dominant. However, some of the individuals carrying the heterozygous mutation did not present with symptoms despite advanced age or showed only with unspecific myalgias related to exercise and intermittently elevated CK (Cerino et al., 2020; González-Mera et al., 2021; Vissing et al., 2016, 2020). Of particular interest is the deletion c.598_612del, rare (MAF 0.00005) in Eastern and Central Europe, while absent in other populations (Karczewski et al., 2020). The deletion was suggested to be dominant (Cerino et al., 2020; Nallamilli et al., 2018), although it was described several times as being recessive (Dorobek et al., 2015; Stehlíková et al., 2007). Our study identified the deletion c.598_612del together with the c.1746-20C>G variant in six individuals. The deep intronic splicing variants, such as c.1746-20C>G, may often remain undiagnosed, because most laboratories are investigating only a 10 bp flanking sequence (Nallamilli et al., 2018). In the dominant calpainopathies calpain was suggested to act in a dominant-negative mechanism. Since calpain 3 is a homodimer, wild type calpain could polymerase with an altered protein and inactivate the whole complex (Vissing et al., 2016). On the western blot calpain expression was reduced to less than 15% of normal and according to the dominant negative mechanism a reduction to 25% activity or less is expected (Vissing et al., 2016). We could only speculate that altered binding properties of calpain will result in dominant phenotype as for the patient described here no quantitative assay was available. The molecular mechanism underlying different calpain variants certainly requires further investigation.
Another explanation for the variants reported as dominant could be that there is a second hit on the same allele and that in fact a pseudodominance occurs. Especially for a high frequency variant, this inheritance model could be considered. Pseudo-dominant inheritance pattern has been described in the GNE myopathy (Alrohaif et al., 2018; Chamova et al., 2015) and in the LGMD R8 TRIM32-related, in the Hutterite population (Frosk et al., 2005). In those cases, the pseudodominance was caused either by high consanguinity or the high disease-causing allele frequency in the isolated population. The pseudodominance has also been discussed in case of titinopathies. Expression data analysis identified that in the French family where a dominant inheritance model was reported, in fact both mother and son were carrying the second titin variant. (Evilä et al., 2017). Recently a pseudo-dominant inheritance of heterozygous c.598_612del and c.1746-20C>G variants in trans position in the CAPN3 gene has been reported in a Russian family (Sharkova et al., 2021). Both mother and daughter carried both variants, whereas other family members carried only one of the variants and were asymptomatic. This is a very rare example of the pseudo-dominant inheritance in LGMD, where one of the variants (c.1746-20C>G) is common (MAF>1%) not only in an isolated population, but in a large geographical region. As the variant frequency in some regions is high, other cases of pseudodominance are likely to be reported. Attentively, in the case of calpainopathy variants should not be reported being dominant without fully comprehensive molecular genomic DNA and mRNA transcript assessments.
The frequency of the c.1746-20C>G (rs201892814) in the healthy population varies significantly between different geographic regions according to the gnomAD database. We found the MAF of the c.1746-20C>G in healthy individuals from the Latvian National Genome database as 0.0237. Additionally, the frequency of the variant of interest was determined in four sets of ethnic Russians from geographically different regions of European Russia (Arkhangelsk region [Mezen], Tver region [Staritsa], Kursk region [Pryamitsino], and Smolensk region [Sychevka]). The corresponding MAF in Polish population was obtained from publicly available database and reaches 0.0114 (Table S3). The variant c.1746-20C>G has a frequency of 0.0237 in the Latvian general population. Notably, the high frequency was also identified in the ethnic Russian populations from the Tver and Arkhangelsk regions. Genetically, Russians from central-south regions of European Russia (Tver, Smolensk, Kursk etc.) are closer to populations Eastern and Central European populations (Khrunin et al., 2013). In contrast, northern Russians (Arkhangelsk region) are differed from them by preservation of substantial portion of Finno-Ugric genetic ancestry (Khrunin et al., 2013). However, it seems not to be the case in the context of the c.1746-20C>G variant that could be arisen in the territories to the south/southeast of the Baltic Sea around the time of split of Balto-Slavic community (Kushniarevich et al., 2015) and spread later to the North-East with Russian migrants (Khrunin et al., 2013). We cannot also exclude that high c.1746-20C>G variant frequency in Latvia is due to the population admixture with ethnic Russians from neighboring regions of the European Russia (Khrunin et al., 2013) (Figure S2).
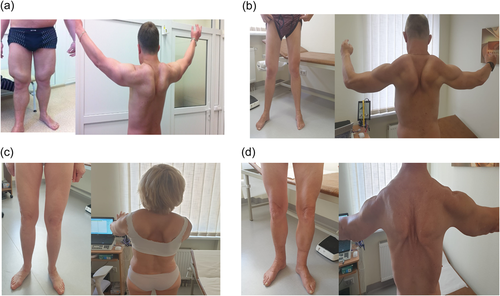
The analysis of the transcriptional profile of CAPN3 revealed an increased number of abnormal reads, mapping to the intronic region between exons 13 and 14 of the CAPN3 gene and crossing the exon-intron border in samples of patient with the c.1746-20C>G variant. Additionally, our results show low level of alternative transcripts, present between exons 12 and 15 that are not present in control sample. The major muscle isoform ENST00000397163.7 of CAPN3 includes exon 13 and exon 14, without alternative events in the region of interest (GTExPortal, 2021). See Figure 2. To verify our findings on RNAseq, we amplified the region of interest using primers located in both exons and the intron-exon boundaries in various combinations. Direct sequencing of these products revealed that at least five splicing variants were present—86 bp, 122 bp, 305 bp, 466+ bp fragments of intron 13 from the side of exon 14, and one that includes the entire 13th–14th intron (Figure 2). Three of these were previously described by Nascimbeni et al. (2010) in a patient harboring this variant. In addition, we found a transcript variant that includes the entire 13th intron, also previously described by Nascimbeni et al. (2010) and proposed to be a naturally occurring benign transcript (Nascimbeni et al., 2010). Apparently, the primary transcript form exists without alterations, while aberrant isoforms are present in a low percentage in comparison. We could speculate that their low level is caused by nonsense mediated decay, as all aberrant isoforms form a pseudo-exon, followed by a nonsense variant and premature protein termination.
The variant c.1746-20C>G has a MAF >1%, reaching even more than 2% in some regions. It is not uncommon that variants, classified as pathogenic, are frequently occurring in the general healthy population. Analyzing data from the 1000 Genomes Project, Cassa et al. (2013) found that 3.744 variants with MAF ≥0.01 (4.6%) and 2.837 variants with MAF ≥0.05 (3.5%) are considered to be pathogenic. Some of these variants may be erroneous findings or have lower penetrance than previously expected, or possibly other inheritance mechanisms should be considered. Although an allele frequency of >1% is a strong evidence that the variant is benign (BS1), stand-alone benign (BA1) evidence should be applied to variants with an allele frequency of ≥5%, according to the ACMG variant classification guidelines (Gudmundsson et al., 2022). For example, CLCN1 gene variant NM_000083.3:c.352G>T with a proven pathogenicity in the protein studies, occurs with the frequency of 0.0196 in the all Finnish population (gnomAD; Raheem et al., 2012). The pathogenicity criteria for some variants above the threshold of 1% or 5% in the future could be revised to comply with current scientific data. We hypothesize that c.1746-20C>G is a hypomorphic variant. By definition the recessive hypomorphic variant reduces gene function, but only causes a biochemical abnormality—or disease—if it is in trans to a loss of function allele. Hypomorphic variants may often have MAF between 0.01 and 0.05 (Houge et al., 2021). The variant c.1746-20C>G is associated with the certain reduction of RNA and protein expression, but not severely affected functional performance. Therefore, a person homozygous for the c.1746-20C>G variant may never present with clinical symptoms, or the onset of the disease may be late. As a result, many were considered healthy and participating in the nation-wide genome databases. The proportion of CAPN3 isoforms may also cause interindividual variability, with affected individuals having a higher ratio of abnormal isoforms.
Although the variant c.1746-20C>G at the population level does not fulfill the criteria of a pathogenic variant, it acts like a hypomorphic allele, associated with LGMD R1 calpain 3-related on a clinical level. Particular care should be given to attributing the pathogenicity of variants based only on the population frequency criteria.
ACKNOWLEDGEMENTS
We thank Leroy Ten Dam and Pervin Dincer for the information regarding the homozygous patients from their cohorts. This work was supported by European Regional Development Fund (No. 1.1.1.1/18/A/096 “The determination of rare inherited diseases' causative mechanisms using whole genome sequencing approach”). We acknowledge support from the Estonian Research Council grants PRG471 and MOBTP175, the Russian Foundation for Basic Research (Grant No.: 20-04-00824) to Andrey Khrunin, and the Ministry of Health of the Czech Republic (FNB RVO 65269705).
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.