Prognostic Value of Combined HBV pgRNA and MELD Score for Mortality in HBV-Related Acute-on-Chronic Liver Failure: A Single-Center Retrospective Study
ABSTRACT
Background and Aims
Rapid prognostic assessment of patients with hepatitis B virus-related acute-on-chronic liver failure (HBV-ACLF) is crucial for optimizing clinical interventions. This study aimed to explore the clinical value of combining Hepatitis B Virus (HBV) pregenomic RNA (pgRNA) with the Model for End-stage Liver Disease (MELD) score predicting the short-term outcomes in HBV-ACLF patients.
Methods
This retrospective study enrolled 220 HBV-ACLF patients from Fuqing Hospital in Fujian Province between July 2016 and July 2024. Clinical data collected within 24 h of diagnosis included HBV pgRNA levels, complete blood counts, C-reactive protein, biochemical profiles, coagulation parameters, and MELD scores. Patients were stratified into survival and non-survival groups based on 90-day outcomes. Independent risk factors for mortality were identified through binary logistic regression.
Results
During the 90-day follow-up after hospitalization and discharge, 90 patients died, and 130 survived. Nonsurvivors exhibited significantly higher HBV pgRNA levels (6.07 (5.62–6.71) log10pgRNA [IU/mL] vs. 4.12 [3.81–5.06] log10pgRNA [IU/mL], Z = − 10.14, p < 0.001) and MELD scores (31.88 [27.82–34.66] vs. 21.27 [18.50–23.14], Z = − 12.03, p < 0.001) compared to survivors. Multivariate analysis identified both HBV pgRNA (OR = 12.38, 95% CI = 2.68–57.25) and MELD score (OR = 3.20, 95% CI = 1.33–7.70) as independent predictors of 90-day mortality (both p < 0.01). The combined model demonstrated superior discriminative ability, with an area under the ROC curve (AUC) of 0.988 (95% CI = 0.978–0.998), significantly outperforming HBV pgRNA alone (AUC = 0.902, Z = 4.487, p < 0.001) and MELD score alone (AUC = 0.977, Z = 2.012, p = 0.044). The optimal cutoff for the pgRNA-MELD score (−42.253 + 2.516 × log10pgRNA [IU/mL] + 1.165 × MELD) was 0.302 (Youden index = 0.967). Patients with pgRNA-MELD scores ≥ 0.302 had markedly reduced survival rates (χ² = 253.09, p < 0.01).
Conclusion
The novel pgRNA-MELD score effectively predicts short-term prognosis in HBV-ACLF patients, offering enhanced clinical utility compared to individual models and guiding timely therapeutic decisions in clinical practice.
1 Introduction
Liver failure, characterized by severe impairment of synthetic, detoxification, metabolic, and biotransformation functions, arises from diverse etiologies and is often complicated by multiorgan dysfunction. Among its subtypes, acute-on-chronic liver failure (ACLF), represents a life-threatening syndrome that typically develops in patients with pre-existing chronic liver disease, manifesting as coagulopathy and hyperbilirubinemia [1]. In China, ACLF predominantly occurs secondary tochronichepatitis B infection (HBV-ACLF) [2], a condition marked by rapid clinical deterioration and alarmingly high short-term mortality. Early and accurate prognostication is pivotal for guiding time-sensitive interventions such as liver transplantation and personalized therapeutic regimens. Consequently, multiple scoring systems have been proposed, including the Model for End-Stage Liver Disease (MELD), MELD-Na, integrated MELD (iMELD), Child-Turcotte-Pugh (CTP) score, and Sequential Organ Failure Assessment (SOFA) [3-5]. Hybrid models such as CTP-SOFA and consortium-derived tools—Chronic Liver Failure Consortium-Organ Failure Score (CLIF-COFs), CLIF-SOFA, and CLIF-C ACLF—have also demonstrated utility. Regionally, the Chinese Group on the Study of Severe Hepatitis B-ACLF score (COSSH-ACLFs) was specifically developed for HBV-endemic populations. Serum biomarkers, including interleukin-6 (IL-6) and tumor necrosis factor-alpha (TNF-α), have also been identified as having prognostic value for ACLF, along with estimated liver volume (ELV), histamine levels, and sarcopenia [6-8].
The MELD scoring system, incorporating serum bilirubin, creatinine, and international normalized ratio (INR), has gained widespread acceptance due to its objectivity and ease of application in stratifying liver disease severity and predicting outcomes of patients with end-stage liver disease. Nevertheless, conventional MELD parameters may insufficiently capture the unique pathophysiology and rapid clinical trajectory of HBV-ACLF, limiting its prognostic precision in this population.
Covalently closed circular DNA (cccDNA), which resides persistently within hepatocytes, serves as the fundamental reservoir for hepatitis B virus (HBV) persistence.
While cccDNA detection remains the gold standard for evaluating viral activity, its reliance on invasive liver biopsy hinders routinewidespread clinical application. In contrast, serum RNA (mainly pgRNA), transcribed from cccDNA, reflects the transcriptional activity of cccDNA [9] and can be used to assess the efficacy of different antiviral drugs, predict treatment endpoints, and provide meaningful guidance for clinical antiviral therapy against HBV, appropriate timing for drug withdrawal, and prognosis analysis [10-12]. These attributes position pgRNA as a promising biomarker for prognostic stratification in HBV-ACLF.
Building upon this rationale, the present study proposes a novel prognostic model integrating HBV pgRNA with the Model for End-Stage Liver Disease (MELD) score to predict short-term outcomes in HBV-ACLF patients. This study aims toofferactionable insights for clinical decision-making and future research directions for HBV-ACLF.
2 Patients and Methods
2.1 Patients
The study retrospectively collected data on 319 patients with HBV-ACLF who were hospitalized at Fuqing City Hospital in Fujian Province from July 2016 to July 2024. Inclusion criteria were based on the “Guidelines for the Diagnosis and Treatment of Liver Failure (2018 edition)”: rapid deepening of jaundice on the basis of chronic liver disease, with serum total bilirubin (TBil) ≥ 10 times the upper limit of normal or a daily increase ≥ 17.1 μmol/L, accompanied by bleeding manifestations and prothrombin activity (PTA) ≤ 40% (or international normalized ratio, INR ≥ 1.5). Exclusion criteria included: (1) concurrent malignant tumors, hematological diseases, or other severe underlying diseases that significantly threaten life; (2) autoimmune or cholestatic liver diseases; (3) liver failure caused by alcohol, drugs, or other hepatotoxic substances; (4) pregnant or lactating women; (5) patients on long-term anticoagulant therapy; (6) incomplete patient follow-up information or loss to follow-up. According to these eligibility criteria, a total of 220 patients were included in this study (Figure 1). The study was approved by the Ethics Committee of Fujian Fuqing City Hospital (approval number: K (2022) 43), and all included patients provided informed consent. All patients received comprehensive internal medical treatment, including bed rest, nucleoside analog antiviral drugs, correction of electrolyte disorders, high-carbohydrate, appropriate protein, low-fat diet, liver-protective medication, albumin supplementation, and plasma transfusion to correct coagulation function.
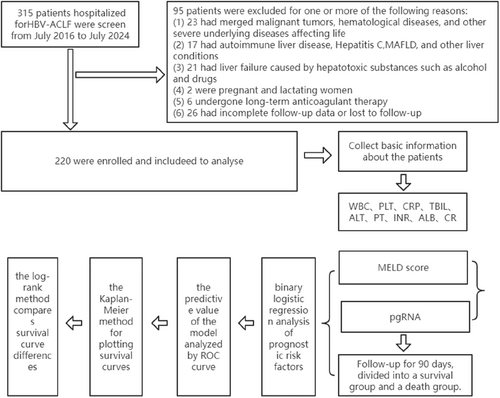
2.2 Data Collection
Basic information and related laboratory results of patients diagnosed with HBV-ACLF were retrospectively collected through the HIS inpatient system. Indicators collected within 24 h after the diagnosis of HBV-ACLF included blood routine, C-reactive protein, biochemical profile, and coagulation function. The MELD score was calculated as follows: 11.2 × ln (INR) + 9.6 × ln[Cr(mg/dL)] + 3.8 × ln[TBil (mg/dL)] + 6.4 × cause (for HBV-ACLF, the cause was assigned a value of 1). Patients were divided into survival and death groups based on the follow-up results within 90 days after the diagnosis of HBV-ACLF.
2.3 Collection and Storage of Peripheral Blood and Serum Specimens
Within 24 h after the diagnosis of HBV-ACLF, 2 mL of venous blood was collected from patients and placed into sterile collection tubes. The samples were centrifuged at 1600 rpm for 5 min at room temperature (not exceeding 4 h) to separate the serum supernatant (at least 0.5 mL), which was then transferred to a 1.5 mL sterile centrifuge tube for storage and frozen at −70°C. Samples were transported using ice flasks with ice or sealed with ice in foam boxes.
2.4 Detection of pgRNA
The detection of serum HBV pgRNA was performed as described in previous studies [13]: (1) Nucleic acid extraction: Nucleic acid was extracted using the nucleic acid extraction or purification kit (S1006) from Hunan Saint Gene Technology Co. Ltd.; (2) DNA digestion: The samples to be tested and negative controls were mixed in proportion and incubated at 37°C for 30 min (quantitative reference samples A ~ D and positive controls did not require DNA digestion); (3) Inactivation of DNA enzyme: The products after DNA digestion were incubated at 75°C for 10 min to inactivate the DNA enzyme; (4) PCR amplification: 10 μL of the product after DNA enzyme inactivation treatment and 10 μL of the nucleic acid extracted positive controls and quantitative reference samples A ~ D were added to a 40 μL prepared PCR-mix and detected on a fluorescent PCR instrument; (5) Fluorescence detection channel selection: (i) The FAM detection channel was selected to detect the HBV RNA target; (ii) The HEX/VIC channel was selected to detect the internal standard of HBV RNA; (iii) The reference fluorescence was set to ROX. The unit of HBV pgRNA is IU/mL.
2.5 Statistical Analysis
Statistical analysis was performed using SPSS Statistics25.0 software, Graphpad Prism10.3.1 software, and MedCalc18.2.1software. Normally distributed measurement data were expressed as mean ± standard deviation, and the independent samples t-test was used for comparison between two groups with normal distribution and equal variance; skewed distributed measurement data were expressed as M (QR), and the Mann-Whitney U test was used for group comparison. Categorical data were expressed as numbers and percentages, and the chi-square test was used for group comparison. Binary logistic regression analysis was used to analyze the risk factors for the prognosis of HBV-ACLF patients. The predictive value of the HBV pgRNA, MELD, and combined HBV pgRNA-MELD score models was analyzed using the ROC curve. The Kaplan-Meier method was used to draw survival curves, and the log-rank test was used to compare the differences in survival curves (survival rates) between groups. A p < 0.05 was considered statistically significant.
3 3 Results
3.1 Demographic Data and Baseline Situation
Among the 220 patients, 90 (death group) died after treatment, with an average age of 60 (50, 71) years, and a mortality rate of 40.91%; 130 (survival group) survived after treatment, with an average age of 59 (51.75,67) years. There was no significant difference in age between the two groups (Z = −0.371, p = 0.71). In the death group, there were 59 males (65.6%) and 31 females (34.4%); in the survival group, there were 81 males (62.3%) and 49 females (37.7%). The difference in gender between the two groups was not statistically significant (χ² = 0.242, p = 0.62). In the death group, 39 patients (43.3%) had a history of antiviral treatment; in the survival group, 67 patients (51.5%) had a history of antiviral treatment. The difference in antiviral treatment history between the two groups was not statistically significant (χ² = 1.434, p = 0.23). In the death group, there were 81 cases (90%) of liver cirrhosis, 78 cases (86.7%) of ascites, 73 cases (81.1%) of bacterial or fungal infections, 26 cases (28.9%) of gastrointestinal bleeding, and 38 cases (42.2%) of hepatic encephalopathy; in the survival group, there were 87 cases (66.9%) of liver cirrhosis, 75 cases (57.7%) of ascites, 71 cases (54.6%) of bacterial or fungal infections, 9 cases (6.9%) of gastrointestinal bleeding, and 9 cases (6.9%) of hepatic encephalopathy; the differences between the two groups were all statistically significant (χ2 = 15.69, 21.08, 16.51, 19.18, 39.45, all p < 0.001), Shown as Table 1. The HBV pgRNA, white blood cells, total bilirubin, prothrombin time, INR, MELD score, creatinine, and so forth, were statistically significantbetween the death and survival groups (all p < 0.001). Among the 220 patients, there were 106 cases (48.2%) with a history of antiviral treatment at baseline and 114 cases (51.8%) without a history of antiviral treatment. The HBV pgRNA levels in the group with a history of antiviral treatment were lower than those in the group without antiviral treatment [4.91 (4.84, 5.34) lg IU/mL compared to 5.3300 (4.98, 5.49) lg IU/mL], but the difference was not statistically significant (Z = −0.796, p = 0.43). For the detailed results, please refer to Table 1 below and continuous variables in the Table are presented as medians (25%, 75%) due to non-normal distribution.
| Variable | Survival (n = 130) | Nonsurvival (n = 90) | χ2/Z | p |
|---|---|---|---|---|
| Sex (male) | 81 | 59 | 0.242 | 0.62 |
| Age (years) | 59 (51.75, 67) | 60 (50, 71) | −0.371 | 0.71 |
| Cirrhosis | 87 | 81 | 15.691 | < 0.001 |
| Ascites | 75 | 78 | 21.080 | < 0.001 |
| Infection | 71 | 73 | 16.511 | < 0.001 |
| GIB | 9 | 26 | 19.181 | < 0.001 |
| HE | 9 | 38 | 39.445 | < 0.001 |
| History of antiviral treatment | 67 | 39 | 1.434 | 0.23 |
| HBV RNA (lgIU/mL) | 4.12 (3.81, 5.06) | 6.07 (5.62, 6.71) | −10.144 | < 0.001 |
| WBC (× 10 ^ 9/L) | 6.36 (4.32, 9.71) | 9.64 (5.63, 12.63) | −3.454 | 0.001 |
| PLT (× 10 ^ 9/L) | 60.5 (29.24, 93.75) | 65.0 (37.47, 107.25) | −1.255 | 0.21 |
| CRP (mg/L) | 17.66 (9.95, 36.86) | 15.25 (7.59, 33.77) | −1.001 | 0.32 |
| TBIL (umol/L) | 225.3 (187.73, 316.9) | 251.55 (206.25, 363.53) | −2.636 | 0.008 |
| ALT (U/L) | 97.05 (33.75, 378.45) | 66 (30.53, 326.5) | −0.776 | 0.44 |
| PT (s) | 20.7 (15.68, 24.73) | 29.0 (22.68, 39.13) | −7.232 | < 0.001 |
| INR | 2.02 (1.76, 2.40) | 2.72 (2.36, 3.54) | −8.207 | < 0.001 |
| MELD score | 21.27 (18.50, 23.14) | 31.88 (27.82, 34.66) | −12.033 | < 0.001 |
| ALB (g/L) | 29.05 (26.68, 31.65) | 28.7 (25.85, 31.83) | −1.209 | 0.23 |
| CR (umol/L) | 62 (51, 75) | 116.5 (89.75, 169.0) | −9.960 | < 0.001 |
3.2 Univariate and Multivariate Analysis of Short-Term Prognosis in HBV-ACLF Patients
Univariate logistic analysis of factors that may affect the prognosis of HBV-ACLF patients showed that HBV pgRNA, white blood cells, total bilirubin, prothrombin time, INR, MELD score, creatinine, and the presence of liver cirrhosis, bacterial or fungal infections, ascites, gastrointestinal bleeding, and hepatic encephalopathy in patients with HBV-ACLF were related to the prognosis (all p < 0.01). Further multivariate logistic regression analysis of variables that may have an impact showed that the level of HBV pgRNA and MELD score were related factors affecting patient prognosis (Table 2). Based on this, our new predictive model formula is: −42.253 + 2.516 × lg (pgRNA) + 1.165 × MELD, which we name the pgRNA-MELD.
| Variable | Univariate logistic | Multivariate logistic | ||
|---|---|---|---|---|
| OR (95% CI) | p | OR (95% CI) | p | |
| RNA | 5.573 (3.615, 8.591) | 0.000 | 12.378 (2.676, 57.246) | 0.001 |
| WBC | 1.063 (1.018, 1.111) | 0.006 | ||
| PLT | 1.001 (0.997, 1.005) | 0.518 | ||
| CRP | 0.999 (0.990, 1.008) | 0.866 | ||
| TBIL | 1.003 (1.001, 1.005) | 0.005 | ||
| ALT | 1.000 (1.000, 1.000) | 0.964 | ||
| PT | 1.149 (1.099, 1.201) | 0.000 | ||
| INR | 10.797 (5.155, 22.614) | 0.000 | ||
| MELD score | 2.037 (1.658, 2.503) | 0.000 | 3.205 (1.334,7.702) | 0.009 |
| ALB | 0.949 (0.896, 1.005) | 0.074 | ||
| CR | 1.057 (1.040, 1.075) | 0.000 | ||
| Cirrhosis | 4.448 (2.040, 9.7) | 0.000 | ||
| Ascites | 4.767 (2.366, 9.602) | 0.000 | ||
| Infection | 3.568 (1.899, 6.705) | 0.000 | ||
| GIB | 5.462 (2.414, 12.355) | 0.000 | ||
| HE | 9.825 (4.432, 21.777) | 0.000 | ||
| History of antiviral treatment | 0.719 (0.419, 1.235) | 0.232 | ||
3.3 The Prognostic Value of HBV pgRNA, MELD Score, and Combined HBV pgRNA-MELD Score Models for Short-Term Outcomes in HBV-ACLF Patients
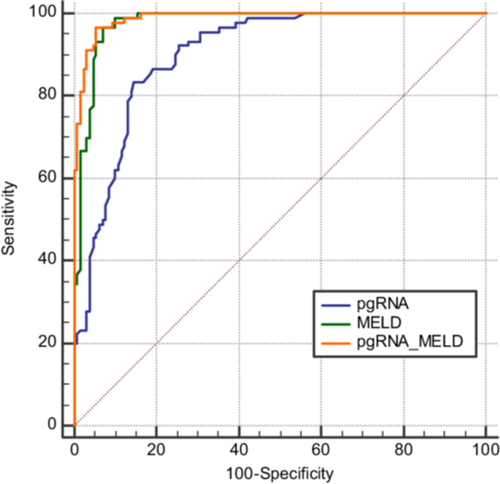
The areas under the receiver operating characteristic curves for HBV pgRNA, MELD score, and the combined HBV pgRNA-MELD score models were 0.902, 0.977, and 0.988, respectively, with 95% confidence intervals of 0.863 to 0.941, 0.960 to 0.995, and 0.978 to 0.998, respectively (Figure 2). This indicates that all models have good predictive capabilities. The area under the curve for the combined HBV pgRNA-MELD score model was significantly larger than that for the HBV pgRNA and MELD score models alone (Z = 4.487, p < 0.0001; Z = 2.012, p = 0.0442). The area under the curve for the MELD score model was also significantly larger than that for the HBV pgRNA model (Z = 3.433, p = 0.0006). The Youden index for HBV pgRNA was 0.687, with a sensitivity of 0.833 and a specificity of 0.854, and the optimal cutoff value was 5.45. For the MELD score, the Youden index was 0.967, with a sensitivity of 0.898 and a specificity of 0.931, and the optimal cutoff value was 25.0973. For the combined HBV pgRNA-MELD score, the Youden index was 0.967, with a sensitivity of 0.913 and a specificity of 0.946, and the optimal cutoff value was 0.301679.
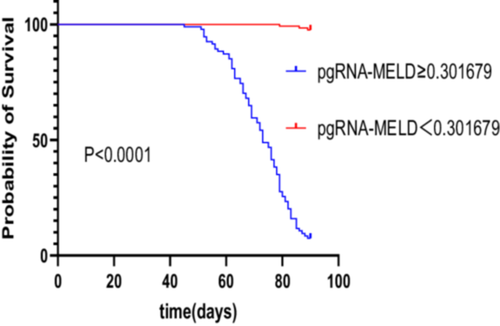
3.4 Analysis of Survival Curves for Patients With Different pgRNA-MELD Scores
Patients were categorized based on thepgRNA-MELD scores (calculated using the formula: − 42.253 + 2.516 × lg pgRNA + 1.165 × MELD), with scores of ≥ 0.301679 and < 0.301679. The 90-day survival curves for these groups were plotted, revealing that patients with scores of ≥ 0.301679 had a significantly lower cumulative survival rate compared to those with scores < 0.301679 (χ² = 234.3, p < 0.0001, Figure 3).
4 Discussion
Despite advancements incomprehensive medical therapies, liver failure remains a highly lethal condition, particularly in its acute-on-chronic form (ACLF) [14]. Prognostic stratification of ACLF—categorized into early, intermediate, and late stages based on clinical severity—is critical for timely interventions such as transplantation.
Consequently, prognostic assessment for liver failure should be an integral part of the entire diagnostic and therapeutic process, with a particular emphasis on early-stage evaluation.
Currently, univariate indicators hold certain value in the prognostic assessment of liver failure, and numerous multifactorial models such as MELD, MELD-Na, iMELD, HINT, CLIF-COFs, CLIF-C ACLF, COSSH-ACLFs, and COSSH-ACLF IIs have been identified as potential prognostic indicators for HBV-ACLF patients [3-5, 15], offering clinical utility but with inherent limitations. Models like CLIF-C OFs and CLIF-C ACLFs were established in Western countries, where the predominant causes of liver disease are hepatitis C virus and alcohol, not HBV. Theyalso necessitate complex organ function assessments. Although COSSH-ACLFs is an HBV-specific prognostic scoring model involving multiple parameters, it requires intricate organ function evaluation. Daxianet al. [16] identified fibrinogen as a potential prognostic biomarker for HBV-ACLF through quantitative proteomics using tandem mass tag (TMT) labeling. A subsequent prospective cohort study of 207 cases over a 30-day survival prognosis follow-up revealed significantly lower fibrinogen levels in non-survivors at admission compared to survivors (p < 0.001). They constructed the P5 scoring model, which demonstrated an AUC of 0.907 for predicting 30-day mortality, outperformingthe Child-Pugh, MELD, and HINT scores (AUCs of 0.718, 0.680, and 0.856, respectively, all p < 0.05), as well as outperforming CLIF-C ACLF and COSSH scores (AUCs = 0.857 and 0.868, respectively; p = 0.057 and p = 0.069). Recently, two studies have been published. The COSSH-onset-ACLF score [17] based on four predictive factors (0.101 × ln (alanine aminotransferase) + 0.819 × ln (total bilirubin) + 2.820 × ln (INR) + 0.016 × ln (ferritin)), can accurately predict the onset of ACLF in patients with HBV-related chronic liver disease within 7, 14, and 28 days, but its predictive power for 90-day survival remains unknown. Additionally, the COSSH-ACLF IIs score [4], based on six predictive factors (1.649 × ln (international normalized ratio) + 0.457 × hepatic encephalopathy score + 0.425 × ln (neutrophils) + 0.396 × ln (total bilirubin) + 0.576 × ln (serum urea nitrogen) + 0.033 × age), is significantly correlated with 28-day mortality rates and demonstrates superior predictive power compared to other models (COSSH-ACLF, CLIF-C ACLF, MELD, MELD-Na). However, the assessment of hepatic encephalopathy in the P5, HINT, and COSSH-ACLF IIs models still has certain limitations. For instance, distinguishing between grade 0 (possible minimal hepatic encephalopathy, MHE) and grade 1 is highly subjective and may vary among different physicians. Diagnosing MHE is challenging due to its lack of overt clinical symptoms, necessitating specialized neuropsychological tests or brain function imaging for identification. Test results may be influenced by various factors, including the patient's age, education level, cooperation, and learning outcomes, potentially affecting the accuracy and repeatability of the tests. Therefore, there is an urgent need to develop an accurate and convenient HBV-ACLF prognostic scoring system, which is vital for providing effective treatment and reducing mortality rates.
HBV covalently closed circular DNA (cccDNA) serves as the most critical intracellular replication intermediate, and its presence and transcriptional activity are the most direct and effective indicators for evaluating the efficacy of antiviral therapy. However, detecting cccDNA requires liver biopsy, which is not widely applicable in clinical practice. HBV pgRNA, as a direct transcription product of cccDNA, reflects cccDNA's transcriptional activity and has been widely studied and applied in hepatitis B. Numerous studies have shown that even when HBV DNA is below the detection limit or after HBV surface antigen (HBsAg) becomes negative, HBV pgRNA is still present in the serum of HBV-infected individuals [18-20]. Therefore, compared to other indicators such as HBV DNA, HBV pgRNA holds higher clinical value in assessing the efficacy of antiviral therapy against HBV, determining the appropriate time to discontinue medication, and predicting the risk of relapse after discontinuation [21, 22].
Regarding the treatment of HBV-ACLF, the current emphasis is on a comprehensive treatment model that includes internal medicine, artificial liver support, and liver transplantation [14]. Oral nucleos(t)ide analogs have a proven efficacy against chronic hepatitis B, rapidly controlling viral replication, reducing viral load, minimizing viral spread between hepatocytes, decreasing cytotoxic T-lymphocyte (CTL) damage to liver cells, alleviating excessive immune responses in the body, and thus achieving rapid disease control and preventing relapse [23]. Administering nucleos(t)ide analogs early in the treatment of HBV-ACLF can effectively improve liver function, increase patient survival rates, and enhance prognosis [24]. Similarly, assessing the efficacy of antiviral drugs in liver failure, in conjunction with the determination of serum HBV pgRNA, provides better guidance. The proportion of patients who had not previously received regular antiviral treatment in the death groupwas high; however, this difference may not be statistically significant due to the small number of patients. All HBV-ACLF patients included in this study were promptly started on antiviral treatment upon admission to our hospital to suppress viral replication and improve prognosis.
Our study demonstrates that integrating HBV pgRNA with the MELD score significantly enhances short-term prognostic accuracy in HBV-ACLF. This finding corroborates prior evidence from Keli et al. [25] where HBV RNA outperformed MELD in prognostic discrimination (AUC = 0.745 vs. 0.66). Discrepancies in MELD performance across studies [26-28] may stem from heterogeneous cohorts, follow-up durations, or variations in baseline data collection. The pgRNA-MELD model's simplicity (incorporating pgRNA, INR, creatinine, and bilirubin) and high sensitivity (Youden Index = 0.967 at cutoff ≥ 0.302) facilitate rapid risk stratification, enabling clinicians to prioritize interventions like artificial liver support or transplantation.
5 Limitations
Nevertheless, our study has limitations. First, it omits parameters reflecting systemic inflammation or complications—factors implicated in ACLF progression. Second, pgRNA assay variability across laboratories and limited accessibility may hinder widespread adoption. Third, the retrospective design and single-center cohort necessitate validation in prospective, multicenter studies. Future research should explore integrating novel biomarkers (e.g., interleukin-6, histamine) and machine learning approaches to refine prognostication.
6 Conclusion
In conclusion, the pgRNA-MELD score represents a pragmatic, high-performance tool for short-term prognosis in HBV-ACLF. By synthesizing viral activity and hepatic dysfunction metrics, this model addresses critical gaps in existing scoring systems and holds immediate translational potential for guiding HBV-ACLF therapeutic decisions.
Author Contributions
Fenlan Chen, Xiongle Zhang, Wen Lin and Shizhong Lin collected and analyzed the data. Fenlan Chen, Jun Huang and Liping Chen drafted the manuscript. Fenlan Chen, Jun Huang, Liping Chen, Chaohui Liu and Qinqi Wang did critical revision of manuscript. All authors have read and approved the final version of the manuscript. Fenlan Chen, Jun Huang and Liping Chen had full access to all of the data in this study and takes complete responsibility for the integrity of the data and the accuracy of the data analysis.
Acknowledgments
We also express our gratitude for the support and assistance provided by some teachers at Beijing You An Hospital and Fuqing City Hospital. The study was supported by a grant from the Startup Fund for Scientific Research Fujian Medical University (2022QH1351).
Ethics Statement
The current retrospective study was approved by the Ethics Committee of Fujian Fuqing City Hospital (approval number: K (2022) 43).
Conflicts of Interest
The authors declare no conflicts of interest.
Transparency Statement
The lead author Jun Huang, Liping Chen affirms that this manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned (and, if relevant, registered) have been explained.
Open Research
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. Fenlan Chen, Jun Huang and Liping Chen had full access to all of the data in this study.