ATP7B gene therapy of autologous reprogrammed hepatocytes alleviates copper accumulation in a mouse model of Wilson’s disease
Funding information
The study was supported by National Natural Science Foundation of China No. 81671103 (Xiao-Ping Wang)
Abstract
Background and Aims
Wilson’s disease (WD) is a rare hereditary disorder due to ATP7B gene mutation, causing pathologic copper storage mainly in the liver and neurological systems. Hepatocyte transplantation showed therapeutic potential; however, this strategy is often hindered by a shortage of quality donor cells and by allogeneic immune rejection. In this study, we aimed to evaluate the function and efficacy of autologous reprogrammed, ATP7B gene-restored hepatocytes using a mouse model of WD.
Approach and Results
Sufficient liver progenitor cells (LPCs) were harvested by reprogramming hepatocytes from ATP7B−/− mice with small molecules, which exhibited strong proliferation and hepatic differentiation capacity in vitro. After lentivirus-mediated mini ATP7B gene transfection and redifferentiation, functional LPC-ATP7B-derived hepatocytes (LPC-ATP7B-Heps) were developed. RNA sequencing data showed that, compared with LPC–green fluorescent protein–Heps (LPC-GFP-Heps) with enrichment of genes that were mainly in pathways of oxidative stress and cell apoptosis, in LPC-ATP7B-Heps under high copper stress, copper ion binding and cell proliferation pathways were enriched. LPC-ATP7B-Heps transplantation into ATP7B−/− mice alleviated deposition of excess liver copper with its associated inflammation and fibrosis, comparable with those observed using normal primary hepatocytes at 4 months after transplantation.
Conclusions
We established a system of autologous reprogrammed WD hepatocytes and achieved ATP7B gene therapy in vitro. LPC-ATP7B-Heps transplantation demonstrated therapeutic efficacy on copper homeostasis in a mouse model of WD.
Abbreviations
-
- 5A
-
- 5 additives
-
- AAT
-
- α-antitrypsin
-
- ALB
-
- albumin
-
- ALT
-
- alanine aminotransferase
-
- AST
-
- aspartate aminotransferase
-
- BAX
-
- B cell lymphoma 2–associated X protein
-
- BCL2
-
- B cell lymphoma 2
-
- BCS
-
- bathocuproine sulphonate
-
- CDCFDA
-
- 5(6)-carboxy-2’,7’-dichlorofluorescein diacetate
-
- CK19
-
- cytokeratin 19
-
- COMMD1
-
- copper metabolism domain containing 1
-
- CP
-
- ceruloplasmin
-
- CYP
-
- cytochrome P450
-
- DEG
-
- differentially expressed gene
-
- EpCAM
-
- epithelial cell adhesion molecule
-
- G6PC
-
- glucose-6-phosphatase, catalytic subunit
-
- GFP
-
- green fluorescent protein
-
- GPX
-
- glutathione peroxidase
-
- GSH
-
- glutathione;
-
- HLC
-
- hepatocyte-like cell
-
- HNF
-
- hepatocyte nuclear factor
-
- HREM
-
- hepatocyte reprogramming and expansion medium
-
- KO
-
- knockout
-
- LPC
-
- liver progenitor cell
-
- LPC-ATP7B-Heps
-
- LPC-ATP7B-derived hepatocytes
-
- LV
-
- lentivirus
-
- PDGFα
-
- platelet-derived growth factor α
-
- PH
-
- primary hepatocyte
-
- PXDN
-
- peroxidasin
-
- qPCR
-
- quantitative PCR
-
- SOD
-
- superoxide dismutase
-
- SOX9
-
- SRY (sex-determining region Y)-box 9
-
- WD
-
- Wilson’s disease
-
- WT
-
- wild type
-
- XIAP
-
- X-linked inhibitor of apoptosis protein
INTRODUCTION
Wilson’s disease (WD), also called hepatolenticular degeneration, is an inborn disorder associated with recessive pathogenic variants in ATP7B gene, a copper transporting ATPase expressed mainly in hepatocytes.[1] Impaired copper transport into bile in WD results in the pathologic accumulation of copper in the liver and later in the brain.[2] Early disease detection and identification and treatment prevent the progression of WD. Drug interventions (such as zinc salts and copper chelators) reduce the pathologic copper deposition but are unable to restore normal copper metabolism.[3] Orthotopic liver transplant is an effective treatment for WD, but the shortage of donor organs and need for lifelong immunosuppression limit its application.[3, 4] Therefore, alternative treatments that are focused on restoring copper homeostasis in affected patients with WD are urgently required.
Cell-based therapy has been an alternative approach in rescuing metabolic liver disease.[5] Transplanted hepatocytes can survive and proliferate robustly when the endogenous cells were destroyed, repopulating the liver and performing normal hepatic functions.[6] Primary hepatocytes (PHs) from unaffected individuals were considered the optimal cell type for transplantation, as demonstrated by the effect of the delivery of normal rat PHs into LEC rats, a rodent model of WD eliminates liver copper and reverses the disease.[7, 8] Despite the therapeutic potential, PHs cannot proliferate in vitro and freshly isolated PHs rapidly lose their hepatic gene expression signatures.[9] Given this, several studies were performed focusing on hepatocyte expansion in vitro.[6, 10] Previous reports have also indicated that by small-molecule induction, mouse and human PHs can be converted to duct-like liver progenitor cells (LPCs) that exhibit the potential for cellular proliferation and differentiation into functional hepatocytes that can be tested for their potential to prevent cellular injury.[11, 12] Nevertheless, some drawbacks still exist, including immunological response among allografts and ethical issues. Therefore, gene therapy of autologous reprogrammed hepatocytes from animals or patients with WD becomes a preferable option. Previous data have indicated the feasibility of lentivirus (LV)-mediated ATP7B overexpression in induced pluripotent stem cell (iPSC)-differentiated hepatocyte-like cells (HLCs) from patients with WD or ATP7B gene transfer into bone marrow mesenchymal stem cells from LEC rats. Both were proved to reduce the copper load in cells or rat livers.[13, 14]
To accomplish this, we reprogrammed PHs from ATP7B−/− mice to generate LPCs by modifying earlier established reprogramming protocol.[10] We found that the hepatocyte reprogramming and expansion medium (HREM) significantly promoted the expansion of LPCs to over 40 passages. After ATP7B overexpression, redifferentiated LPC-ATP7B-Heps were capable to repopulate the livers of ATP7B−/− mice with reduced liver copper accumulation. Therefore, LPC-ATP7B-Heps showed therapeutic potential, and the results could contribute to the development of treatment for WD and similar genetic disorders in the liver.
MATERIALS AND METHODS
Reprogramming and redifferentiation of mouse hepatocytes
The purified PHs were seeded on collagen I- or Matrigel-coated six-well or 12-well plates (1% collagen I in PBS, 2% Matrigel in DMEM/F12) at 2 × 104 cells/cm2 and were cultured in HREM. HREM was based on basal medium DMEM/F12 containing 1% fetal bovine serum, insulin-transferrin-selenium-ethanolamine solution (ITS-X, Gibco), 20 ng/ml HGF, and 20 ng/ml EGF (Peprotech), 10 μm Y-27632 (ROCK kinase inhibitor), 1 μm A-8301 (TGF-β signaling inhibitor), 3 μm CHIR99021 (Wnt signaling activator) (MedChem Express), 1 μm sphingosine-1-phosphate, and 5 μm lysophosphatidic acid (yes-associated protein signaling agonists) (Santa Cruz). We named the last five small molecules supplemented in DMEM/F12 as “5 Additives” (5A). About 5–7 days after seeding, cells were dissociated with Accutase (Gibco) and were passaged at a 1:2–1:3 ratio. Approximately after five serial passages of 1:2–1:3 subculture, cells that entered into stable growth and exponential proliferation state were called LPCs (wild-type [WT]-LPCs and knockout [KO]-LPCs). The doubling time was calculated by using the online tool http://www.doubling-time.com/compute.php. [11]
For cell redifferentiation, LPCs (passage 15) were cultured in HREM for 3 days to reach 80% confluence. Then, the medium was changed to hepatocyte maturation medium (HMM) as described previously.[10] HMM contained DMEM/F12, ITS-X, 20 ng/ml oncostatin M (Peprotech), 10 μm N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester and SB431542 (MedChem Express), and 10 μm dexamethasone (Dex) (Sigma-Aldrich). The medium was changed every other day for 3 weeks, and the redifferentiated cells (LPC-Heps) were used in subsequent experiments.
Statistical analysis
Results were presented as mean ± SD or median ± interquartile range (IQR). All data were subjected to the normality test with SPSS 23.0 software (IBM). Comparisons were evaluated by unpaired two-tailed Student t test (2 groups) or one-way ANOVA followed by Student-Newman-Keuls post-test (≥3 groups) with Prism version 7.00 (GraphPad Software). p values < 0.05 were considered statistically significant. Ratios of ATP7B, CD45, sirius red, and alpha-smooth muscle actin (α-SMA) positive areas in liver sections were analyzed using ImageJ version 1.52 (National Institutes of Health) by taking representative pictures from different mice, and 3–5 areas of each mouse were counted.
For experiments involving animals, we assured that all animals received humane care according to the criteria outlined in the NIH Guide for the Care and Use of Laboratory Animals. And all animal studies complied with the guidelines of the Ethics Committee of Shanghai Tong-Ren Hospital and conformed to the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines.
Further methodology may be found in the Supporting Materials.
RESULTS
PH-LPCs exhibit high proliferative capability
First, we isolated PHs from livers of WT mice or ATP7B−/− mice (KO). PHs were both α-antitrypsin (AAT) and albumin (ALB) positive at 4 h after isolation (Figure S1A). Next, we optimized the hepatocyte expansion system by adding combinations of growth factors and 5A (Figure 1A). To explore the optimal plate-coating conditions, cell proliferation on collagen-coated plates was compared with Matrigel-coated plates. We found that PHs exhibited a more rapid expansion on Matrigel than on collagen at 3 and 7 days in HREM (Figure S1B). With Matrigel coating and HREM culture, WT-PHs and KO-PHs could be reprogrammed and serially passaged over 40 times without obvious morphological changes (Figure S1C). Karyotyping of two independently established KO-LPC lines (passage 17) showed that donor 1–derived LPCs exhibited generally normal diploid karyotypes except for aneuploidy of the no. 19 chromosome (dotted box). Donor 2–derived LPCs showed diploid, triploid, and tetraploid changes of karyotypes (Figure S1D). These results were in line with previous reports that aneuploidy was one of the characteristics of hepatocyte proliferation.[15]
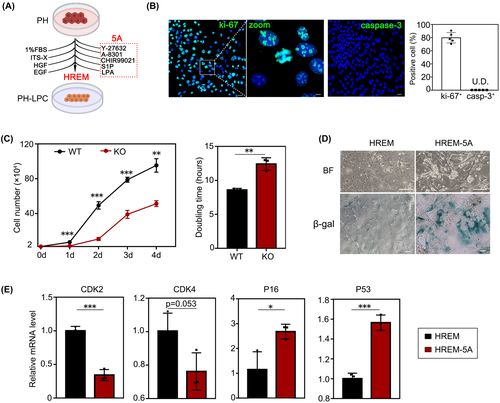
We further evaluated the expression of proliferation-related marker Ki-67 and apoptosis-related marker caspase-3 in KO-LPCs (passage 15). We found that 79.8% ± 7.1% of LPCs were Ki-67+, whereas caspase-3+ cells were not detected, suggesting the extensive proliferation capacity of LPCs (Figure 1B). Meanwhile, we also found that WT-LPCs grew faster than KO-LPCs (passage 15) and KO-LPCs had longer doubling times than WT-LPCs (12.40 h vs. 8.59 h) (Figure 1C). We postulated that this was due to abnormal copper metabolism in KO-LPCs due to loss of ATP7B function. Once withdrawing the 5A, KO-LPCs became senescent and lost their proliferative capability as shown by SA-β-gal staining (Figure 1D). Expression of the cell cycle–related gene CDK2 was reduced, and CDK4 showed a declining tendency, whereas senescence genes like P21 and P53 were up-regulated after the withdrawal of 5A (Figure 1E). These results also implied that the proliferative cells did not undergo oncogenic transformation.
To further clarify the characteristics of KO-LPCs, we assessed the expression of hepatic markers in KO-LPCs. We found that from passage 0 to 14, percentages of ALB and AAT positive hepatocytes gradually decreased, whereas an increased rate of cytokeratin 19 (CK19) positive duct-like liver progenitors was observed (Figure 2A). Consistent with these findings, mRNA analysis indicated that mature hepatocyte-associated genes (ALB, AAT, glucose-6-phosphatase, catalytic subunit [G6PC], and hepatocyte nuclear factor [HNF] 4α) exhibited dramatic down-regulation whereas progenitors-associated genes (CK7, CK19, SRY [sex-determining region Y]-box 9 [SOX9], and epithelial cell adhesion molecule [EpCAM]) were elevated, even in late passages (Figure 2B). Collectively, these data suggested that KO-LPCs acquired stable progenitor features and displayed powerful proliferation potentials when maintained in HREM.

KO-LPCs exhibit the bi-potency of hepatic and biliary differentiation
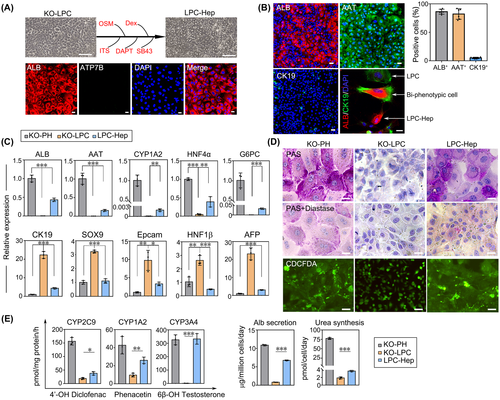
First, we tested whether KO-LPCs could differentiate toward hepatic lineages (KO-LPC-Heps). In line with a previous report,[10] the morphology of KO-LPCs gradually changed from pebbled to polygonal shapes after 3 weeks of differentiation, and ATP7B was not expressed in KO-LPC-Heps (Figure 3A). In addition, increased expression of hepatocyte markers (ALB+, 86.2% ± 4.0% and AAT+, 82.1% ± 8.4%) and decreased expression of progenitor markers (CK19+, 5.0% ± 1.1%) were present in KO-LPC-Heps. It was noted that during the redifferentiation process, LPCs (CK19+), biphenotypic cells (ALB+/CK19+), and LPC-Heps (ALB+) could be observed in the same field (Figure 3B). Consistently, gene expression levels of ALB, AAT, cytochrome P450 (CYP) 1A2, HNF4α, and G6PC were up-regulated whereas CK19, SOX9, EpCAM, HNF1β, and AFP levels were down-regulated after differentiation (Figure 3C). Compared with KO-LPCs, KO-LPC-Heps exhibited typically hepatic glycogen synthesis and 5(6)-carboxy-2’,7’-dichlorofluorescein diacetate (CDCFDA) secretion to bile canaliculi, which was comparable to PHs (Figure 3D). Furthermore, drug metabolism activities of CYP1A2, CYP3A4, and CYP2C9; ALB secretion; and urea synthesis were also elevated in KO-LPC-Heps (Figure 3E). Our results supported the view that relatively mature hepatocytes and liver progenitors could be mutually converted in vitro.[6, 12] Next, we detected the biliary differentiation potential of LPCs. We found the cyst, duct, and sphere formation in three-dimensional cultures (Figure S2A), and cholangiocyte marker CK19 and tight junction marker ZO-1 were coexpressed in the cyst structure (Figure S2B). In addition, rhodamine 123 dye was transported to the luminal space, and verapamil (multidrug resistance protein 1 inhibitor) blocked the efflux process of dye, suggesting that the KO-LPC-Chols were functional (Figure S2C).
Recovery of ATP7B subcellular localization and copper-export function in LPC-ATP7B-Heps
ATP7B gene was overexpressed in KO-LPCs through an LV delivery system. Because the large size of ATP7B genome (5.2 kb) led to unsatisfactorily low-yield virus production, a miniATP7B was constructed based on previous research.[16] To maximize the function of miniATP7B, we kept the species consistency between donor cells and the ATP7B genome. After homology comparison of amino acids between mouse and human ATP7B, we constructed mouse miniATP7B cDNA by deleting the DNA sequences corresponding to amino acids 74–488, and then we packaged this into LV-miniATP7B-green fluorescent protein (GFP; Figure S3A–C). Virus-transfected KO-LPCs expressed green fluorescence, and we estimated that ATP7B mRNA level was 200 times higher than in LPCs with only LV-GFP transfection. After 21 days of differentiation, ATP7B expression in LV-ATP7B-Heps was nearly 600 times higher than in LV-GFP-Heps. Flow cytometry showed that the transduction efficiency was 62.8% ± 0.8% before cell sorting (Figure 4A). Further immunofluorescence suggested that LV-GFP-transfected KO-LPCs expressed GFP but not ATP7B. In contrast, LV-ATP7B-GFP-transfected KO-LPCs expressed both GFP and ATP7B. Moreover, ATP7B was also expressed in redifferentiated binucleated LPC-ATP7B-Heps (Figure 4B). Besides the expression of ATP7B, correct subcellular localization is also a key precondition for copper export. It is known that ATP7B localizes in the Golgi apparatus under conditions with normal intracellular copper. Under high copper conditions, ATP7B will migrate to the lysosomes, exporting excess copper into bile ducts.[17] In KO-LPC-Heps, due to the lack of LV-ATP7B-GFP transfection, no ATP7B signal was found in the Golgi (as detected by Golgin-97+) under conditions with no added CuCl2 and was also not found in lysosomes (as detected by LAMP-1+) under conditions with added 50 μm CuCl2 for 2 h (Figure 4C). By contrast, in LPC-ATP7B-Heps, ATP7B almost completely colocalized with Golgi instead of lysosome under with no CuCl2. Once cells were treated with 50 μm CuCl2 for 2 h, some of the ATP7B migrated to the apical membrane and colocalized with lysosomes (arrows). When cells were treated with the copper chelation bathocuproine sulphonate (BCS), 50 μm, for 1 h, ATP7B was no longer localized in lysosomes but returned to the Golgi. These data suggested that the overexpressed ATP7B was capable of proper subcellular localization in response to changes of cellular copper levels (Figure 4D). Finally, copper levels in the supernatants of LPC-ATP7B-Heps were 3.74, 3.56, and 3.41 times higher than levels in supernatants of LPC-GFP-Heps at 3, 6, and 9 h after copper withdrawal, indicating functional copper export of LPC-ATP7B-Heps (Figure 4E).

RNA sequencing reveals the advantages of KO-LPC-ATP7B-Heps under copper stress
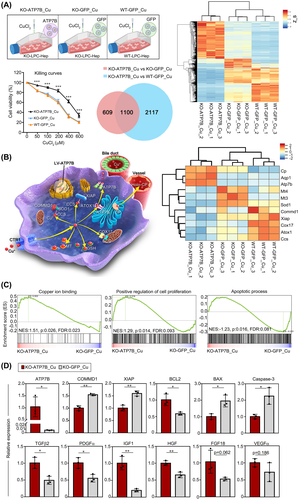
With the increased copper level from 0 to 600 μm in medium, cell viabilities declined much slower in the KO-ATP7B group than those in KO-GFP and WT-GFP groups, suggesting the capacity of copper resistance after ATP7B transduction (Figure 5A). To gain an in-depth understanding of gene expressions in cells under high copper stimulation, transcriptome analyses were performed. There were 1709 differentially expressed genes (DEGs; fold change >2) when comparing KO-ATP7B_Cu with KO-GFP_Cu cells, among which 1100 genes were also differentially expressed in WT-GFP_Cu cells. The heatmap illustrated that KO-GFP_Cu clustered closer to WT-GFP_Cu cells than KO-ATP7B_Cu cells (Figure 5A). The process and pivotal proteins involved in intrahepatic copper trafficking were displayed in Figure 5B. We found that in KO-GFP_Cu and WT-GFP_Cu cells, due to their absent or insufficient expression of ATP7B, non-ATP7B mediated copper metabolic pathways were mobilized, leading to compensatory elevations of metallothionein 3/4, superoxide dismutase (SOD)1, copper metabolism domain containing 1 (COMMD1), X-linked inhibitor of apoptosis protein (XIAP), cytochrome coxidase 17, antioxidant, and copper chaperone for superoxide dismutase (CCS). When ATP7B was restored in KO-ATP7B_Cu cells, all these genes above were down-regulated, accompanied with up-regulated expressions of ATP7B, ceruloplasmin (CP), or even biliary epithelial marker aquaporin 1 (Figure 5B). Furthermore, KO-ATP7B_Cu cells displayed enrichment in genes involved in copper ion binding and the positive regulation of cell proliferation. By contrast, genes involved in the apoptotic process were enriched in KO-GFP_Cu cells (Figure 5C). Further quantitative PCR (qPCR) verified consistent expression of several key genes in pathways of copper metabolism (ATP7B, copper metabolism domain containing 1 [COMMD1], and XIAP), proliferation process (TGFβ2, platelet-derived growth factor α [PDGFα], insulin-like growth factor 1 [IGF1], HGF, FGF18, and VEGFα, which represented common growth factors), and apoptotic process (B cell lymphoma 2 [BCL2], BCL2–associated X protein [BAX], and Caspase-3, which were star molecules in this pathway) (Figure 5D). To further explore if there were unexpected other gene expression profile differences (fold change > 2, p value < 0.05), the Gene Ontology analysis was performed. There were eight DEGs in the copper ion binding pathway (Figure S4A), 56 DEGs in the apoptotic process pathway (Figure S4B), and 62 DEGs in the cell proliferation pathway (Figure S4C).
Oxidative stress was one of the most important events that occurred after cellular copper accumulation.[18] We found that compared with KO-ATP7B_Cu cells, pathways of glutathione (GSH) metabolism and oxidative stress were more enriched in KO-GFP_Cu cells (Figure S5A). And expressions of key genes such as SOD3, ribonucleotide-diphosphate reductase subunit M2 B (RRM2B), glutathione peroxidase (GPX) 1/3, and peroxidasin (PXDN) were also decreased (Figure S5B). Furthermore, lower reactive oxygen species (ROS) levels and higher mitochondrial membrane potentials (5,5,6,6′-tetrachloro-1,1′,3,3′ tetraethylbenzimi-dazoylcarbocyanine iodide red polymers/green monomers) were observed in KO-ATP7B_Cu cells (Figure S5C,D). Besides, both the GSH level and GPX activity were higher in KO-ATP7B_Cu cells than in KO-GFP_Cu cells (Figure S5E), suggesting the pivotal role of ATP7B in relieving cellular oxidative stress.
Next, we compared the transcriptome of KO-ATP7B_Cu versus WT-GFP_Cu under copper stimulation. We found that KO-ATP7B_Cu cells displayed enrichment in genes involved in copper ion binding, zinc ion binding, and cellular defense response, whereas apoptotic process-related genes were enriched in WT-GFP_Cu cells (Figure S6A,B). The qPCR analyses confirmed the expressional differences in pathways of copper ion binding (ATP7B and CP), oxidative stress (SOD2/3, RRM2B, GPX1/3/8, and PXDN), apoptotic process (BCL2, BAX, and their ratio), and cell proliferation (TGFβ2, PDGFα, IGF1, VEGFα, FGF18, EGF, and HGF) (Figure S6C).
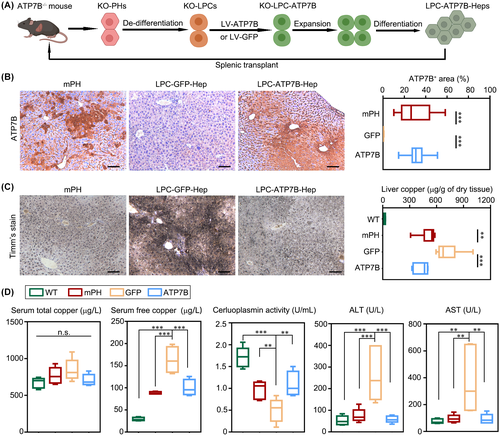
LPC-ATP7B-Heps transplantation reduces copper deposition and improves liver functions
Liver sections from 8-week-old ATP7B−/− mice showed inflammation compared with WT mice, and no ATP7B positive cells were found in ATP7B−/− mice (Figure S8B). Based on the data above, LPC-ATP7B-Heps were then ready to be transplanted into ATP7B−/− mice (Figure 6A). Cell engraftment of LPC-ATP7B-Heps in mice reached an average of 31.5% after 4 months following cell transplantation, which was comparable to mouse healthy PHs transplantation (29.4%). Instead, no ATP7B+ cells were observed in the group of LPC-GFP-Heps (Figure 6B). Consistent with this result, copper staining and liver copper was increased in the LPC-GFP-Heps group compared with the other two groups (Figure 6C). Serum free copper (non-CP bound copper, NCC) was significantly reduced in PHs and LPC-ATP7B-Heps mice, although total copper remained unchanged (possibly due to the offset of NCC). Serum CP activities and biochemical parameters like alanine aminotransferase (ALT) and aspartate aminotransferase (AST) were all improved in LPC-ATP7B-Heps mice compared with LPC-GFP-Heps mice (Figure 6D). Moreover, pathological phenotypes of liver inflammation and fibrosis were also improved in PHs and LPC-ATP7B-Heps groups as evaluated by the presence of CD45+ leukocytes, sirius red+ collagen fibers, and α-SMA+ activated hepatic stellate cells (Figure S7).
DISCUSSION
As a genetic liver disorder, WD often affects the liver early on. Until now, curative treatments were quite limited. Donor shortage and immunosuppression are major obstacles to whole-liver transplantation.[3, 4] Cell-based therapy has shown great potential in liver regenerative medicine. To expand terminally differentiated rodent hepatocytes, various methods of chemically induced hepatic progenitors have been established. Katsuda et al. developed the chemically induced liver progenitors by Y-27632, A-83-01, and CHIR99021 cocktail and verified their regenerative capacity in urokinase-type plasminogen activator-transgenic severe combined immunodeficient mice.[6] Wu et al. reported hepatocytes-derived duct-like progenitors with transition and expansion medium.[10] And Kim et al. established the expandable “chemically derived hepatic progenitors” from hereditary tyrosinemia type 1 mice by a combination of HGF, EGF, A83-01, and CHIR99021.[19] Based on these strategies, in this study, we optimized the reprogramming protocols to generate LPCs from ATP7B−/− mice with HREM culture system. We found that in this medium, LPCs tolerated long-term culture over 40 passages and proliferated extensively with an average doubling time of 8.6 h (WT-LPCs) and 12.4 h (KO-LPCs) when seeding on Matrigel-precoated plates, which were faster than ever before. The reprogrammed LPCs gradually lost their maturation but gained progenitor signatures during passages and could be redifferentiated in the maturation medium. Even in late passages of LPCs, most progenitor markers (CK7, CK19, and SOX9) were still maintained at high transcriptional levels. On withdrawal of the 5A, cell senescence appeared, suggesting such rapid proliferation would not induce tumorigenic transformation.
Our unpublished data suggested that transplantation of patient-iPSCs-derived, gene-restored HLCs into WD mice cannot sufficiently engraft into the liver parenchyma or reduce copper accumulation. This unsatisfying result was also supported by previous studies.[19-23] The low repopulation efficiency for HLCs transplantation was possibly due to the immature state of hepatic differentiation. Other candidates of cell sources like hepatic transdifferentiation from somatic cells and immortalization of hepatocytes show safety issues like oncogenic transformation or other unpredictable risks.[24] Because hepatocytes with high maturation are more desirable to repopulate the liver than unmatured progenitors,[10, 11, 25] differentiated LPC-Heps seem to be an ideal cell source for transplantation.
Whether on immune rejection or cell sources, autologous cells show significant advantages and are the optimal choice of cell therapy. For autologous cells with monogenic mutation, gene replacement or correction are necessary to restore their normal functions. In this study, we adopted the self-inactivating LV-mediated miniATP7B gene overexpression in KO-LPCs. After that, the mRNA level of ATP7B was elevated over 200 times. Moreover, after differentiation, 62.8% of LPC-Heps were ATP7B positive with its expression increasing nearly 600 times, suggesting the high efficiency of gene replenishment. Nevertheless, it must be admitted that LV-mediated gene delivery integrates into the genome semirandomly with a preference for active transcription units.[26] Besides the interference with the expression of endogenous genes, insertional activation of proto-oncogene or disruption of antioncogene adjacent to the insertion locus may increase the insertional oncogenesis.[27, 28] Several factors were reported to affect LV oncogenic risks, including the target cell type and proliferative status, virus design and copy number, the intrinsic genetic mutations in animals and patients, and so on.[29] Compared with γ-retroviral vectors that integrate near transcription start sites, LV exhibited low oncogenic potential.[30] Despite this, oncogenesis should be cautiously evaluated during investigations, and nonviral gene-editing methods should be considered in future research.[19] With the increasing number of clinical trials on LV-based gene therapy, continued follow-up of recipients is essential to know the safety and efficacy of LV vectors.[31]
Correct ATP7B subcellular colocalization is the precondition of normal functions. In our data, we confirmed the copper-responsive organelle localization of ATP7B from Golgi complex to lysosome and further indicated the copper-export capacity of cells in vitro. By subsequent RNA sequencing, we found distinct advantages of LPC-ATP7B-Heps in the high copper niche. In contrast to LPC-GFP-Heps, the expressions of copper binding-related genes, such as ATP7B and holo-CP, and antioxidant-related genes such as SOD3, GPX1/3, and PXDN were increased in LPC-ATP7B-Heps under copper stress. Meanwhile, the GSH degradation process was down-regulated, suggesting that LPC-ATP7B-Heps were responding to lower the production of ROS. Though differentiated from normal LPCs, WT-LPC-GFP-Heps showed much less ATP7B expression level than KO-LPC-ATP7B-Heps, and the latter displayed clear superiority on copper ion binding and defense response.
Our present study demonstrated an average of 31.5% of the engraftment rate in LPC-ATP7B-Heps by measuring ATP7B+ areas in the parenchyma, which was comparable to that in normal PHs. It was reported that 5%–10% of restoration of enzyme function would ameliorate the severe phenotype of WD.[32] In line with this theory, LPC-ATP7B-Heps transplant reduced copper level in the liver and enhanced CP activity in serum. Besides, improved liver functions of ALT and AST as well as alleviated liver inflammation and fibrosis were all indicators of effective therapeutics. Nevertheless, it usually takes several months for transplanted cells to achieve sufficient repopulation. Therefore, increasing the number of initial donor cells and long-term observation to 12 months after transplantation are required. From the clinical perspective, potential candidates including newly diagnosed patients at the relatively early phase, people with intolerance for drug treatment, and those with disease complications such as liver failure will most likely benefit from gene/cell-based therapy.[33]
MiniATP7B retains its biological functions, and its efficacy was further proved in this study. On the one hand, the truncated miniATP7B expressed ATP7B protein in LPC-Heps, which exactly localized at the corresponding organelles with changes of copper levels. Moreover, copper restoration in the miniATP7B group was 3.4–3.75 times higher than that in the GFP group. On the other hand, after LPC-ATP7B-Heps transplantation to WD mice, the copper loading decreased, as was shown by Timm’s staining. Furthermore, the reduced liver copper and transaminase levels confirmed the therapeutic potential of miniATP7B. Thus, miniATP7B could restore functional copper metabolism as expected. However, to guarantee enough LV titer and transduction efficiency, parts of exon 2 and 3 had to be deleted, which made miniATP7B an unsuitable therapeutic way in certain patients with WD (e.g., exon 2 mutation in India).[34] In this case, CRISPR/CRISPR associated protein 9 or prime editing system may be an alternative.
Together, our study provides a promising cell source for WD therapy: expandable and functional LPC-ATP7B-Heps by reprogramming autologous hepatocytes, gene modification, and then redifferentiation in vitro. These preliminary studies on murine model of WD will contribute to future clinical translation.
ACKNOWLEDGMENTS
We thank Prof. Guo-Yuan Yang (Shanghai Jiao Tong University) for his technical support on the copper assay.
CONFLICT OF INTEREST
Nothing to report.
AUTHOR CONTRIBUTIONS
Hongxia Cai: experiments, procedures, and manuscript writing; Xin Cheng: study design, data interpretation; and Xiao-Ping Wang: main idea of the research, and final proof.
Open Research
DATA AVAILABILITY STATEMENT
The raw RNA-seq data have been deposited in Gene Expression Omnibus (GEO) (Accession number: GSE197948).