Excretion of infectious hepatitis E virus into milk in cows imposes high risks of zoonosis
Potential conflict of interest: Nothing to report.
This work was financially Supported by the National Natural Science Foundation of China (31360619), the Natural Science Foundation of Yunnan pProvince (2013FB032), and the China Postdoctoral Science Foundation (2014M562672 and 2015T81138).
Abstract
Hepatitis E virus (HEV) represents the main cause of acute hepatitis worldwide. HEV infection in immunocompromised patients involves a high risk for the development of chronic hepatitis. Because HEV is recognized as a zoonotic pathogen, it is currently believed that swine is the primary reservoir. However, this is not sufficient to justify the strikingly high seroprevalence of HEV in both developing and Western countries. Thus, this study aimed to identify new zoonotic sources that bear a high risk of transmission to humans. We collected fecal, blood, and milk samples of cows in a typical rural region of Yunnan Province in southwest China, where mixed farming of domestic animals is a common practice. HEV RNA was quantified by quantitative real-time polymerase chain reaction, and the whole genome was sequenced. HEV infectivity was assessed in rhesus macaques. We found a high prevalence of active HEV infection in cows as determined by viral RNA positivity in fecal samples. Surprisingly, we discovered that HEV is excreted into milk that is produced by infected cows. Phylogenetic analysis revealed that all HEV isolates from cow/milk belong to genotype 4 and subtype 4h. Gavage with HEV-contaminated raw and even pasteurized milk resulted in active infection in rhesus macaques. Importantly, a short period of boiling, but not pasteurization, could completely inactivate HEV. Conclusion: Infectious HEV-contaminated cow milk is recognized as a new zoonotic source that bears a high risk of transmission to humans; these results call attention to understanding and establishing proper measurement and control of HEV zoonotic transmission, particularly in the setting of mixed farming of domestic animals. (Hepatology 2016;64:350-359)
Abbreviations
-
- cDNA
-
- complementary DNA
-
- HEV
-
- hepatitis E virus
-
- IgG/IgM
-
- immunoglobulins G and M
-
- ORF
-
- open reading frame
-
- PCR
-
- polymerase chain reaction
-
- qRT-PCR
-
- quantitative real-time PCR
Hepatitis E virus (HEV) is a positive single-stranded RNA virus with four defined genotypes and other newly discovered strains that have not been assigned to these known genotypes.1 It is the most common cause of acute hepatitis globally.2 In the Western world, chronic hepatitis has been frequently described in immunocompromised patients.3 Thus, HEV infection has emerged as a global public health issue with a particularly high mortality rate in pregnant women.4 Seroprevalence is rather high in the developing world, ranging 30%-80%. Strikingly, it is also very high in Western countries. In the United States, population-based surveys have indicated a seroprevalence of 21% from 1988 to 1994 and 6% from 2009 to 2010.5 An overall seroprevalence of 22.4% and 27% was found in French6 and Dutch7 blood donors, respectively.
In the developing world, epidemics of hepatitis E occur periodically and are mainly attributed to genotypes 1 and 2. They account for annually 20 million infections, over 3 million cases with symptomatic diseases, and 70,000 deaths.8 Fecal contamination of drinking water is a major route of transmission of these two genotypes.8
In contrast, in developed countries, HEV genotype 3 is predominant and spread by zoonotic transmission. It is believed to be mainly transmitted through consumption of uncooked or undercooked pig meat.9 Genotype 4 is also a zoonotic strain, mainly found in China10-12 and Japan13, 14 as well as some sporadic cases reported in Europe.15-17 Thus, zoonotic strains are circulating in both developing and developed countries. The initial concept of zoonotic transmission was based on the identification of HEV stains in various animals, including swine, boars, deer, and rabbits.18-21 Recent studies have confirmed the genetic similarity between strains circulating in pigs and in indigenous human cases as well as cross-species infection,18, 22 indicating that swine is the most important reservoir of HEV zoonotic transmission.
Given the factor of cross-species infection, traditional mixed farming of different types of domestic animals may potentially expand the zoonotic sources that mediate the transmission of HEV to humans. Thus, besides direct contact with infected pigs or consumption of uncooked pork contaminated with HEV, other unidentified zoonotic sources may also contribute to the high prevalence of HEV. In this study, we unexpectedly found a high prevalence of HEV infection in cows in a rural area of Yunnan Province, southwest China. We discovered that infected cows excreted HEV into milk. Furthermore, we demonstrated that HEV from raw and even pasteurized fresh milk are infectious in rhesus macaques yet can be inactivated by boiling.
Materials and Methods
SAMPLE COLLECTION
Fresh stool, serum, and milk samples of Holstein cows were collected from Dali, Yunnan, China, during September to December 2015. The region where samples were collected has traditional mixed farming of different types of domestic animals. Typically, each household owns one to three cows, whereas a few families own larger numbers of cows. The samples were stored at -80°C until use. Details are described in Supporting Table S1.
DETECTION OF HEV RNA
Stool specimens were suspended at 10% w/v in phosphate-buffered saline (pH 7.4), containing 0.01% diethyl pyrocarbonate, and centrifuged at 12,000g for 10 minutes. Total RNA was extracted from the supernatant of stool, milk, or serum using the QIAamp Viral RNA mini-kit (Qiagen, Germany) according to the manufacturer's instructions. Reverse-transcription was performed using a reverse transcriptase kit (AMV XL for real-time polymerase chain reaction [RT-PCR]; Takara, Japan) according to the manufacturer's directions. A 348-nucleotide amplicon from HEV open reading frame 2 (ORF2) was amplified by nested RT-PCR as described (Supporting Table S2).23
GENOME SEQUENCING
Three full-length nucleotide sequences of HEV were obtained by amplification of five overlapping fragments covering the complete genome (Supporting Table S2). The extreme 5′ and 3′ ends of the viral genome were amplified using the rapid amplification of complementary DNA (cDNA) ends technique as in our previous study.24 The complete nucleotide sequence was assembled and analyzed using the MegAlign software. All sequences have been submitted to the GenBank database.
VIRAL TITER QUANTIFICATION BY QUANTITATIVE RT-PCR
The viral titer of HEV in serum, feces, or milk was quantified using SYBR green-based quantitative RT-PCR (qRT-PCR) with HEV-specific primers as described (Supporting Table S2).23 In brief, 200 μL of milk or serum or 0.4 g of fecal samples was subjected to RNA isolation. Isolated RNA was used to synthesize the first-strand cDNA, and cDNA was added as a template for qRT-PCR. qRT-PCR was performed under the following conditions: 95°C for 30 seconds, followed by 39 cycles of 95°C for 5 seconds and 60°C for 31 seconds. It was performed using an ABI PRISM 7300 Real-Time PCR System.
VIRAL TITER CALCULATION
A standard plasmid was constructed by cloning the RT-PCR-amplified ORF2 partial gene (348 bp) into the pMD 18-T clone vector. The copy number of the recombinant plasmid was calculated using the following formula: (DNA concentration [ng/μL] × 10−9 × 6.0233 × 1023 copies/mol)/(DNA size [bp] × 660). The standard curve was drawn based on the copy number and the cycle threshold value of qRT-PCR. The accuracy of HEV RNA quantification by qRT-PCR was validated with a 10-fold serial dilution of this standard plasmid. We first performed dilutions from 1 × 101 copies/mL to 1 × 107 copies/mL to generate a linear range. However, there was no clear distinction of cycle threshold values between 1 × 101 copies/mL and 1 × 102 copies/mL, suggesting that this assay is not sensitive to quantify at a low copy number of 1 × 101 copies/mL. Therefore, a linear range was generated from 1 × 102 copies/mL to 1 × 107 copies/mL with R2 = 0.9966 (Supporting Fig. S1).
PHYLOGENETIC ANALYSIS
The nucleotide sequences of the amplified PCR products and of prototypes of different genotypes of HEV strains were aligned using MEGA 4.1 software (version 4.10, http://www.megasoftware.net). The reference sequences of prototype HEV strains were obtained from the GenBank database. The standard classification of HEV genotypes and subtypes was according to previous studies.25, 26
Phylogenetic trees were generated by the minimum evolution and interior branch methods. Bootstrapping with 1,000 resamplings of the data was performed to calculate branch percentages. The identity between nucleotide sequences was calculated using the MegAlign program (DNAstar package, version 5.03).
HEV INFECTIVITY IN RHESUS MACAQUES
Healthy male rhesus macaques (n = 7), 2-3 years old, negative for HEV RNA and anti-HEV immunoglobulin G (IgG) and immunoglobulin M (IgM) antibodies, were used. The protocol of animal experimentation was approved by the committee of Laboratory Animal Welfare and Ethics of the Kunming University of Science and Technology. Monkeys were numbered randomly and housed in individual cages. HEV-contaminated raw or pasteurized milk and a swine HEV strain (KM01, 2 × 104 copies/mL) that we have demonstrated has robust infectivity in rhesus macaques27 were used. The first monkey was inoculated with gavage of 5 mL supernatant of KM01 (viral titer 2 × 104) pretreated at 62°C for 30 minutes. The second monkey was inoculated with gavage of 5 mL supernatant of KM01 pretreated at 72°C for 30 seconds. The third monkey was inoculated with gavage of 5 mL supernatant of KM01 pretreated at 100°C for 3 minutes. The fourth monkey was gavaged with 5 mL of pasteurized (the exact pasteurization condition was unclear) commercial milk positive for HEV RNA (shelf life 24 hours, viral titer 3 × 104 copies/mL). The fifth monkey was gavaged with 5 mL of raw milk from the cow that was HEV-positive in both feces and milk (viral titer in milk 2 × 104 copies/mL). The sixth monkey was gavaged with 5 mL of phosphate-buffered saline. The seventh monkey was gavaged with 5 mL of milk negative for HEV RNA. Heating treatment of the indicated samples was performed using a PCR machine. The effectiveness of our pasteurization protocols was validated on Escherichia coli (Supporting Fig. S2). Fecal samples were collected from each monkey twice per week for HEV RNA detection/quantification. Sera were collected every week for anti-HEV IgG and IgM testing, HEV RNA quantification, or liver enzymes analysis. All samples were tested immediately or stored at -80°C until use.
DETECTION OF ANTI-HEV IgG AND IgM ANTIBODIES
Serum samples were tested for the presence of anti-HEV IgG and IgM antibodies using commercial enzyme-linked immunosorbent assay kits (KHB, China) containing recombinant ORF2 peptides from the HEV genome as well as both positive and negative controls. Samples were tested in duplicate according to the manufacturer's instructions, with cutoff values for IgG and IgM assays set at 0.22 and 0.26, respectively, which were determined based on the mean optical density 450 values from the negative controls (± standard deviation).
SERUM LIVER CHEMISTRY PROFILE
The activities of alanine aminotransferase and aspartate aminotransferase in serum were measured with an automated biochemistry analyzer (Olympus 2700, Japan).
Results
HIGH PREVALENCE OF ACTIVE HEV INFECTION IN COWS
To investigate the prevalence of active HEV infection in cows, we collected stool samples from 140 individual Holstein cows from Dali, Yunnan Province, southwest China, from September to December 2015. Livestock are important components of the agricultural economy in that region, and many of the households typically own one to three cows, as well as other types of domestic animals (Supporting Table S1). The presence of HEV genomic RNA (a hallmark of active infection) in stool samples was examined by nested RT-PCR as described.23 We found that 52 out of 140 samples were positive (37.14%), in line with previous reports of HEV RNA positivity in cows (8.79%, 8/91) on dairy farms in Xinjiang Province (northwest China)28 and in swine (7.8%, 20/256) in Kunming, the capital of Yunnan Province.29
Complete genomic nucleotides of three strains of cow HEV were sequenced by amplification of five overlapping fragments covering the complete genome from fecal samples (Supporting Table S2). The extreme 5′ and 3′ ends of the viral genome were amplified using the rapid amplification of cDNA ends technique, similar to our previous study.24 The complete genomic sequence was assembled and analyzed using the MegAlign software. Sequences were submitted to the GenBank database (KU356187, KU356188, and KU356189 for HEV isolated from fecal samples of cows and KU974927, KU974928, KU974934, KU974936, KU974938, KU974946, KU974947, KU974948, KU974949, KU974950, and KU974952 for partial HEV ORF2 sequences isolated from raw milk in Dali, Yunnan Province, China) (Supporting Table S3).
EXCRETION OF HEV INTO COW MILK
A recent case report showed the presence of HEV in breast milk of a woman with acute hepatitis E.30 We thus investigated whether HEV is excreted in the milk of infected cows. Paired serum, feces, and milk samples were collected from 6 out of the 52 HEV-infected cows that we have identified. As shown by qRT-PCR analysis, all samples were positive, and the titers in milk were considerably high (Fig. 1A; Supporting Table S4).
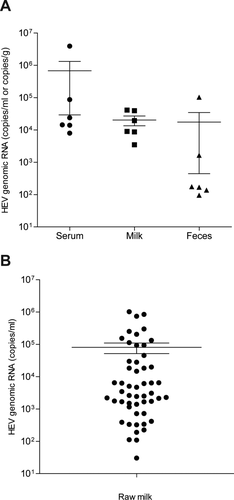
We further extended the collection of milk samples from all of the identified HEV-positive cows. The presence of HEV RNA (100%, 52/52) was first confirmed by RT-PCR, and the genomic copy number was subsequently quantified by qRT-PCR (Fig. 1B; Supporting Table S4). With respect to the milk samples derived from cows for which the complete HEV genome was sequenced, the amplified ORF1 and ORF2 (partial) sequences were also determined. As expected, these ORF1 and ORF2 sequences were perfectly matched with their parental genome (Supporting Fig. S3), providing further confirmation that HEV in milk was excreted by the infected cows.
HIGH HOMOLOGY OF COW/MILK HEV TO HUMAN AND SWINE STRAINS
To find the homology relationship, phylogenetic analysis was performed based on the complete genomic sequences of our identified cow HEV strains and ORF2 (partial) sequences from milk. A phylogenetic tree clearly illustrated that all HEV isolates from cow/milk belong to genotype 4 (Fig. 2A), subtype 4h (Fig. 2B). More importantly, these HEV strains were closely clustered to human and swine HEV isolated in Kunming City, the capital of Yunnan Province.
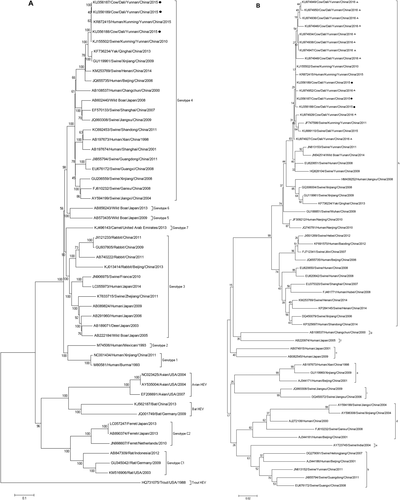
Homology analysis based on the complete sequence of HEV indicated that these cow HEVs shared 99.2%-99.4% similarity to human HEV isolated from a pregnant woman in Kunming City, in early 2015,31 and shared 99.5%-99.8% identity with swine HEV isolated in Kunming City in 2010.32 These results strongly suggest that these HEV isolates prevalent in human, swine, and cows in Yunnan Province probably originated from the same source.
HEV FROM RAW OR PASTEURIZED MILK IS INFECTIOUS IN RHESUS MACAQUES
Cow milk produced in this area is mainly for household consumption and local market supplies. To address the key issue of whether HEV-contaminated milk can be a mediator of zoonosis, we next evaluated whether it is infectious in rhesus macaques. We employed this sophisticated animal model for HEV infection that had been previously used by our group27 and others.33, 34 We selected a representative milk sample with intermediate HEV viral titers (approximately 2 × 104 copies/mL of genomic RNA) (Fig. 1B).
Experimental rhesus macaques were prescreened and confirmed as being HEV-negative. Oral gavage of raw milk containing HEV resulted in active infection in a monkey showing an elevated viral titer from 7-10 days postinoculation in feces (Fig. 3A) and a high titer at 4 weeks postinoculation in serum (Fig. 3B). In contrast, HEV was not detectable in the monkey gavaged with HEV-negative raw milk.
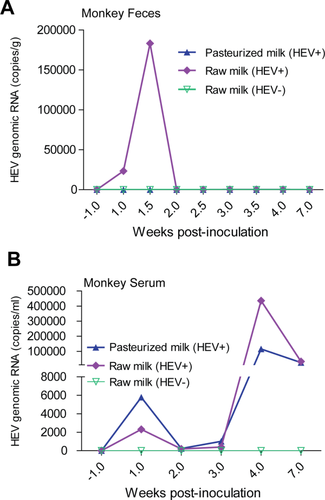
Fresh pasteurized milk is readily available at local markets supplied by the farmers. As expected, we identified HEV-contaminated packages but were curious to know whether they remained infectious. Surprisingly, oral gavage of pasteurized milk in a rhesus macaque resulted in a substantially high level of HEV viral load (viral genomic RNA approximately 1.1 × 105 copies/mL) in serum at 4 weeks postinoculation (Fig. 3B), although viral RNA was hardly quantifiable in feces (Fig. 3A). Thus, pasteurization seems insufficient to completely inactivate HEV.
COMPLETE INACTIVATION OF HEV BY BOILING
To more clearly demonstrate the effects of pasteurization on HEV infectivity, we mimicked the commonly used protocols of pasteurizing raw milk by preheating at 62°C for 30 minutes or at 72°C for 30 seconds using a PCR machine. We used a swine genotype 4 strain that we have previously demonstrated to produce robust infection in monkeys.27 We found that pretreatment with our pasteurization protocols is not sufficient to completely inactivate the HEV infectivity in monkeys as shown by quantified viral load in both feces and serum (Fig. 4).
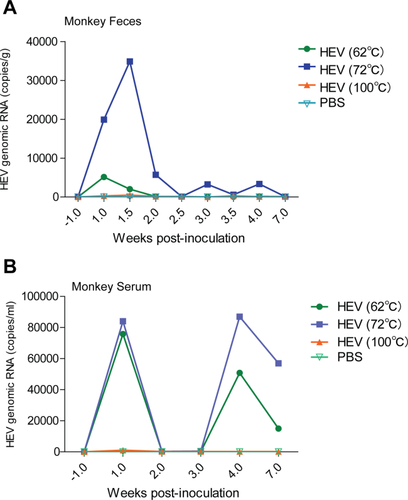
We next explored whether boiling of HEV at 100°C for 3 minutes is sufficient for complete inactivation. Indeed, similar to the phosphate-buffered saline-treated negative control monkey, HEV viral RNA was not detectable in both feces and serum of the monkey inoculated with the boiled HEV sample (Fig. 4). Thus, these results alert us to the potential zoonotic risk of both raw and pasteurized cow milk. Fortunately, a short period of boiling is already sufficient to completely inactivate the virus, providing a cost-effective approach that can be easily implemented even in resource-limited regions.
Discussion
Shortly after the discovery of the HEV genome, which was attributed to the cause of non-A, non-B hepatitis in patients,35 the identification of HEV in pigs36 has raised the assumption of a potential zoonotic feature of this virus. The cross-species infectivity of swine HEV demonstrated in nonhuman primates37 and of human HEV in pigs,22 as well as the strong association of the consumption of undercooked animal meat with HEV infection in humans,38 have now firmly established its zoonotic nature. The evidence of HEV infection in animals is rapidly expanding.1, 39 Besides in swine, genotype 3 and 4 strains have also been identified in other hosts, including deer, wild boar, mongoose, macaque, sheep, yak, and cattle. Many new strains that have not been assigned to any genotype were recently discovered from camel, birds, rabbits, bats, rats, ferrets, and fish.1, 39
Because many of these newly identified animal hosts are not closely associated with human life, we currently only recognize swine as the primary source of zoonotic transmission. However, the recent discovery of HEV in other common domestic animals may revise our classical concept. In cattle, a seroprevalence of 15% was described in the United States40 and an HEV RNA positivity of 8% was reported in cows on dairy farms in Xinjiang Province, China.28 Seroprevalence of HEV in cows has been described in various regions in China.41-43 In this study, we described a 37% positivity of HEV RNA in cows surveyed in a rural area of Yunnan Province, China. In this area, cows are regarded as an important component of the agricultural economy and each household typically owns one to three cows. Mixed farming in this region is a common practice. Family-based, small-size farms host diverse domestic animals including buffalo, cows, goats, sheep, pigs, chicken, and ducks. In fact, a mixed farming system is one of the oldest and most traditional farming methods practiced all over the world and remains the backbone of agriculture in Asia and many developing countries.44 However, an increased risk of zoonosis has been raised as an important concern in this system.45 Domestic animals are reared primarily on grass and naturally grown crops, while composted animal wastes are often used to fertilize the soil for growing crops. When livestock are reared outdoors, this increases the potential for contact with disease vectors including rodents, wild birds, and insects, as well as interaction among different types of domestic animals. Given the ability of cross-species infection, we were not astonished by a high rate of HEV infection in cows found in our study with a high homology to human and swine HEV strains as a high prevalence of HEV has been reported in both swine and patients in Yunnan Province.11, 46 Importantly, especially with respect to mixed farming systems, we call attention to the proper measurement and control of HEV circulating among different types of domestic animals, to eventually prevent transmission to humans.
We were indeed concerned by the discovery of HEV in milk that was produced by the infected cows. The most common virus contaminating cow milk is bovine leukemia virus. Bovine leukemia virus-infected lymphocytes circulate through the blood of infected cattle and can also infect the mammary epithelial cells of cows. Infected cells could be disseminated into milk. Exposure to bovine leukemia virus through consumption of cow milk has been associated with breast cancer in women.47 Although there is no direct evidence yet to illustrate milk-mediated transmission of HEV, a recent study has demonstrated isolation of the virus from breast milk of an acute hepatitis E patient, suggesting that breast-feeding could be a potential route of HEV transmission from mother to child.30 Another case of liver transplantation with chronic hepatitis E infected with a camel strain was related to regular consumption of camel meat and milk.48 Because camel meat is often well cooked but the milk is often consumed fresh without processing, this might indicate that camel milk is a possible source of transmission.
To concretely prove whether milk represents a valid source for HEV transmission, we assessed the infectivity of HEV-contaminated cow milk in rhesus macaques in this study. The rhesus macaque is one of the most robust animal models for HEV infection,27, 34, 37 although it is not commonly accessible for most researchers. We successfully demonstrated that gavage of raw milk resulted in active infection, as shown by the quantification of HEV RNA in feces and blood. Surprisingly, inoculation of pasteurized milk also led to active infection, although it appeared less robust. Consistently, mimicking the commonly used protocols of pasteurization by preheating at 62°C for 30 minutes or at 72°C for 30 seconds still resulted in active infection in monkeys. In contrast, boiling of HEV at 100°C for 3 minutes is sufficient for complete inactivation. Rhesus macaques are in general tolerant of HEV infection, often with weak or no clear humoral immune response, as we have described.27 Except for one monkey that was inoculated with HEV preheated at 62°C for 30 minutes and showed a dramatic elevation of alanine transaminase 2 weeks postinoculation (Supporting Fig. S4), a strong indication of liver injury, all of the others had normal alanine aminotransferase and aspartate aminotransferase levels (Supporting Figs. S4 and S5). Most of these monkeys had no clear humoral immune responses (Supporting Figs. S6 and S7), except one that was inoculated with HEV preheated at 72°C for 30 seconds and had a high level of anti-HEV IgM at 3 weeks postinoculation (Supporting Fig. S7).
In fact, several factors have been proposed to affect the thermal stability of HEV. An early study reported that an HEV isolate from Guangzhou, China, could be inactivated by heating at 56°C for 30 minutes.49 There are probably also genotype-dependent differences and variations between different isolates. The genotype 1 strain (Akluj) could be completely inactivated at 56°C for 1 hour. Another genotype 1 strain (Sar55) was more heat-resistant and was not inactivated at 56°C, but ∼80% of the viruses were inactivated at 60°C for 1 hour. Approximately 50% of the viruses of a genotype 2 strain (Mex 14) could be inactivated at 56°C, and 96% of the viruses were inactivated by incubation at 60°C for 1 hour.50 Heat treatment for 1 minute up to 75°C was not able to completely inactivate a genotype 3 strain (47832c). Complete inactivation was achieved at 80°C or higher temperatures. These data have been used to calculate predictive heat inactivation models for HEV.51 Thus, we speculate that the tradition of consuming raw pork and raw milk in regions where HEV is highly prevalent could be the key route of transmitting HEV to humans.
In summary, we found a high prevalence of active HEV infection in cows in a typical rural region of Yunnan Province in China, where mixed farming of domestic animals is a common practice. Furthermore, we discovered that infectious HEV is excreted into the milk that is produced by the infected cows, although the mechanism of action remains to be elucidated. Importantly, we demonstrated that a short period of boiling, but not pasteurization, could completely inactivate HEV. This provided a cost-effective approach that can be easily implemented even in resource-limited regions to eliminate the risk of milk-mediated HEV transmission to humans. Finally, we call attention to understanding and establishing proper measurement and control of HEV zoonotic transmission, in particular in the setting of mixed farming of domestic animals.
Acknowledgment
We thank Chunxiang Shi, Huchun Peng, and his colleagues for their valuable assistance in sample collection and Dr. M.S. Hakim (Erasmus University Medical Center, Rotterdam, The Netherlands) for critical editing of the manuscript.
REFERENCES
Author names in bold designate shared co-first authorship.




