Questioning the challenging role of epithelial-to-mesenchymal transition in liver injury†
Potential conflict of interest: Nothing to report.
Osterreicher CH, Penz-Österreicher M, Grivennikov SI, Guma M, Koltsova EK, Datz C, et al. Fibroblast-specific protein 1 identifies an inflammatory subpopulation of macrophages in the liver. Proc Natl Acad Sci U S A 2011;108:308-313. (Reprinted with permission.)
Abstract
Cirrhosis is the end result of chronic liver disease. Hepatic stellate cells (HSC) are believed to be the major source of collagen-producing myofibroblasts in cirrhotic livers. Portal fibroblasts, bone marrow-derived cells, and the epithelial-to-mesenchymal transition (EMT) might also contribute to the myofibroblast population in damaged livers. Fibroblast-specific protein 1 (FSP1, also called S100A4) is considered a marker of fibroblasts in different organs undergoing tissue remodeling and is used to identify fibroblasts derived from EMT in several organs, including the liver. The aim of this study was to characterize FSP1-positive cells in human and experimental liver disease. FSP1-positive cells were increased in human and mouse experimental liver injury including liver cancer. However, FSP1 was not expressed by HSC or type I collagen-producing fibroblasts. Likewise, FSP1-positive cells did not express classical myofibroblast markers, including alpha smooth muscle actin (α-SMA) and desmin, and were not myofibroblast precursors in injured livers as evaluated by genetic lineage tracing experiments. Surprisingly, FSP1-positive cells expressed F4/80 and other markers of the myeloid-monocytic lineage as evaluated by double immunofluorescence staining, cell fate tracking, flow cytometry, and transcriptional profiling. Similar results were obtained for bone marrow-derived and peritoneal macrophages. FSP1-positive cells were characterized by increased expression of COX2, osteopontin, inflammatory cytokines, and chemokines but reduced expression of MMP3 and TIMP3 compared with Kupffer cells/macrophages. These findings suggest that FSP1 is a marker of a specific subset of inflammatory macrophages in liver injury, fibrosis, and cancer.
Comments
Liver fibrosis and cirrhosis are the outcome of many chronic liver diseases that leads to extracellular matrix (ECM) production and collagen deposition.1 Hepatic stellate cells (HSCs) are considered the major but not the only source of ECM-producing myofibroblasts in the injured liver. Myofibroblasts originate from portal fibroblasts, interphase (septal) myofibroblasts, and, to a smaller extent, bone marrow-derived mesenchymal cells as well. Epithelial-to-mesenchymal transition (EMT), where epithelial cells acquire features of mesenchymal cells, is claimed to be another significant source of myofibroblasts in several organs and in liver.2
Fibroblast-specific protein 1 (FSP1), also known as S100 calcium binding protein A4 (S100A4) or calcium placental protein (CAPL), was identified as relatively gene-specific expressed in fibroblasts but not in epithelial cells, mesangial cells, or embryonic endoderm.3 Therefore, it was suggested to facilitate the mapping of cell fate among fibroblasts.3 Because FSP1 is further expressed in fibroblasts in different organs that undergo tissue remodeling including kidney, lung, and heart FSP1 is commonly used as a marker to identify epithelial cells undergoing EMT during tissue fibrogenesis.2 Therefore, FSP1 was used as proof of EMT by hepatocytes and cholangiocytes in several studies. However, the role of FSP1-positive cells in ECM deposition in liver injury was never shown.
In the current study it was demonstrated that FSP1-positive (FSP1+) cells are increased in injured human and murine livers.4 These cells locate along fibrotic septa and show the same spatial distribution as ECM-producing myofibroblasts. As well as in fibrotic injury, FSP1+ cells could be shown in a tumor microenvironment of a murine model of hepatocellular carcinoma (HCC) in mice that were challenged with diethylnitrosamine (DEN).
Most interestingly, the authors demonstrate by a plenitude of excellent in vitro and in vivo experiments that hepatic fibroblasts do not express FSP1 (Fig. 1). In one set of experiments the previously described Col-GFP mouse in which the collagen α1(I) promoter/enhancer drives green fluorescent protein (GFP) expression was utilized. These animals were subjected either to carbon tetrachloride (CCl4)- or bile duct ligation (BDL)-induced liver injury and colocalization for GFP and FSP1 was analyzed by immunofluorescence microscopy. In a total of 6,185 GFP+ cells no colocalization was observed. Also, primary HSC were isolated and culture-activated by culture on a plastic surface representing a well-defined in vitro model of HSC transdifferentiation.5 In contrast to mouse skin fibroblasts, isolated HSC again lacked expression of FSP1. Next FSP1-GFP mice, in which the FSP1 promoter drives GFP expression, were subjected to CCl4- or BDL-induced liver injury. Again, costaining with alpha smooth muscle actin (α-SMA) and desmin was performed but no colocalization could be detected. To determine if FSP1 is a marker for precursors of myofibroblasts and disappears before the upregulation of typical myofibroblast markers like α-SMA or desmin, the authors used lineage tracing techniques crossing FSP1-Cre mice to ROSA26 reporter mice. This genetic labeling technique allows the identification of cells that have expressed FSP1 at some time during differentiation but lack FSP1 expression at the time of analysis. Again, a total of 1,563 GFP-positive cells (indicating FSP1 driven Cre-LoxP recombinase activity) were analyzed but no colocalization with α-SMA or desmin was observed. The same results were obtained with isolated primary HSCs.
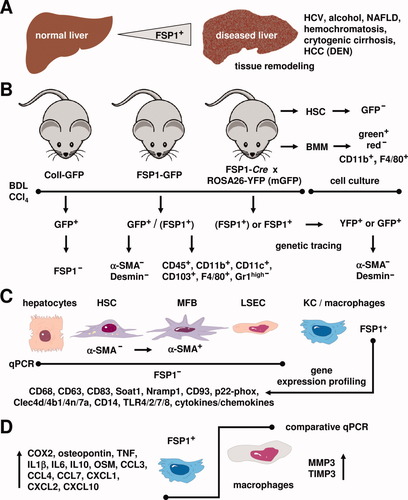
Expression of FSP1 in inflammatory liver macrophages. (A) In the liver, the number of FSP1+ cells increases during liver disease including chronic hepatitis C virus (HCV) infection, alcoholic liver disease, nonalcoholic fatty liver disease (NAFLD), hereditary hemochromatosis, cryptogenic cirrhosis, and DEN-induced HCC. (B) Transgenic mouse models that contain a ColI-GFP or a FSP1-GFP transgene were subjected to BDL or challenged with CCl4. GFP+ cells from respective animals were isolated and stained for FSP1, α-SMA, and desmin. GFP+ cells from transgenic FSP1-GFP animals were positive for CD45, CD11b, CD11c, CD103, F4/80 and negative for Gr1high. Using genetic tracing techniques in animals that were obtained by crossing of FSP1-Cre mice with animals that contain either the ROSA26-YFP or ROSA26-membrane targeted GFP transgenic cassette allowing the identification of cells that actually express or have expressed FSP1 during differentiation revealed that respective YFP+ or mGFP+ cells were negative for α-SMA and desmin. In addition, HSC or BMM were isolated from untreated animals and stained for GFP, CD11b, and F4/80. (C) Although primary hepatocytes, quiescent HSC (α-SMA−), MFB (α-SMA+), and liver sinusoidal endothelial cells (LSEC) were negative for FSP1 in quantitative polymerase chain reaction (PCR), Kupffer cells (KC)/macrophages were positive for FSP1. In addition, gene expression profiling revealed that FSP1-positive liver cells are positive for CD68, CD63, CD83, Soat1, Nramp1, CD93, p22-phox, Clec4d, Clec4b1, Clec4n, Clec7a, CD14, TLR4, TLR2, TLR7, TLR8 and several cytokines and chemokines involved in phagocytosis. (D) Comparative quantitative PCR revealed that the expression of COX2, osteopontin, TNF, IL1β, IL6, IL10, oncostatin, CCL3, CCL4, CCL7, CXCL1, CXCL2, and CXCL10 is higher, whereas the expression of MMP3 and TIMP3 is lower in FSP1+ liver cells than in blood macrophages. Please note that immunoreactivity for a specific marker is marked by a plus sign and the absence of respective marker protein is indicated by a minus sign.
To identify the cellular lineage of FSP1+ cells, the authors performed gene expressing profiling on FSP1+ cells isolated from CCl4-treated FSP1-GFP mice. Surprisingly, the analysis of the gene expression profile showed FSP1+ cells closest to peritoneal macrophages stimulated with zymosan. FSP1+ cells expressed genes typical for macrophages and cells of dendritic lineage like CD68, Nramp1, Soat1, CD63, CD83, CD93, Clec4d, Clec4b1, Clec4n, Clec7a, and p22-phox. Also, these cells expressed genes involved in innate immunity like CD14, TLR4, TLR2, TLR7, and TLR8. These profiles suggest that FSP1+ cells in liver injury belong to myeloid-monocytic lineage. Analyzing purified hepatocytes, HSCs, and Kupffer cells for FSP1 expression revealed FSP1 messenger RNA (mRNA) expression primarily in Kupffer cells. Immunofluorescence staining of mice genetically labeled for FSP1 with the macrophage marker F4/80 showed significant colocalization (46.7% ± 6.3%). In fluorescence-activated cell sorting (FACS) analysis, FSP1+ cells expressed CD45, indicating these cells as bone marrow-derived. Furthermore, FSP1+ cells expressed CD11b, CD11c, and F4/80 but lacked expression of the granulocyte marker Gr1high. A significant amount of FSP1+ cells also expressed CD103, a marker for resident dendritic cells of the intestine and the skin. To further confirm these results, bone marrow-derived macrophages (BMM) from FSP1-Cre × ROSA26-reporter mice were generated. After culturing for 7 days, 99% of FSP1-Cre cells were expressing GFP, showing a successful genetic recombination. Similar results were obtained for peritoneal macrophages.
In summary, the authors identify FSP1+ cells as a subset of bone marrow-derived inflammatory macrophages. The presented gene expression profiles and individual immunofluorescent stainings place these cells clearly in the myeloid-monocytic lineage. In the injured liver, FSP1+ cells do not express markers typical for myofibroblasts. Moreover, FSP1+ liver cells do not express collagen or take part in ECM production.
These observations lead to challenging conclusions concerning EMT in liver injury. FSP1 is a marker of dermal fibroblasts and a subset of fibroblasts in some organs but is also expressed by cells of myeloid-monocytic lineage. Therefore, studies on EMT in liver fibrosis, which rely mainly on FSP1 expression to identify fibroblasts in undergoing tissue remodeling, are prone to interpretational pitfalls. Likewise, FSP1-Cre-mediated gene deletion will not specifically occur in mesenchymal cells only and needs to be evaluated carefully.




