Quantifying growth and transformation frequency of oncogene-expressing mouse hepatocytes in vivo†
Potential conflict of interest: Nothing to report.
Abstract
Gene changes can affect cancer cells in many ways, but changes that increase disease severity—by allowing cells to proliferate when they should be quiescent, by enhancing their rate of growth under growth permissive conditions, or by increasing the risk that they will accumulate additional carcinogenic alterations—must be identified so that strategies to counter their effects can be developed. We describe a novel in vivo assay system based on hepatocyte transplantation that permits us to accomplish this objective for genetically modified hepatocytes. We find that the oncogenes c-myc and transforming growth factor α, but not simian virus 40 T-antigen, increase the rate of hepatocyte growth under growth permissive conditions. However, no single oncogene can induce hepatocyte growth in quiescent liver. In contrast, at least one oncogene combination, transforming growth factor α/T-antigen, was sufficient to direct cell autonomous growth even in this nonpermissive environment. Furthermore, we could quantify risk for progression to neoplasia associated with oncogene expression; increased transformation frequency was the principal carcinogenic effect of T-antigen. Conclusion: This system identifies biological mechanistic role(s) in carcinogenesis for candidate genetic changes implicated in development of human liver cancer. The quantitative and comparative evaluation of gene effects on liver cancer allows us to prioritize targets for therapeutic intervention. (HEPATOLOGY 2010;)
Several categories of genetic alterations have been identified in human liver tumors, including inactivation of tumor suppressor genes, mutation or increased expression of protooncogenes, and increased activity of growth factor/receptor signaling loops. Identifying the precise influence of each of these genetic changes on liver cancer development remains a crucial endeavor, both to increase understanding of how cancer initiates and progresses and to direct the development of appropriate therapies.
Transgenic mice and, more recently, gene-targeted or knockout mice, have been employed to begin to address this need.1, 2 Cancer initiation events no longer are random, as occurs in chemical carcinogenesis. Instead, these models permit specification of the genetic alteration used to direct the onset of carcinogenesis. Therefore, a specific disease latency, multiplicity, pattern of progression, and tumor histotope can be assigned to oncogenic changes commonly associated with human liver cancer. For example, overexpression of the transcription factor c-myc and of the epidermal growth factor receptor ligand transforming growth factor alpha (TGF-α) have been identified in a large fraction of human liver cancers. In early transgenic mouse models, hepatocyte-targeted c-myc expression induced benign liver neoplasms in mice older than 1 year of age, with an incidence of 50%-65%.3, 4 TGFα induced a high incidence of benign and malignant liver tumors between 10 and 15 months of age.5-8 Simian virus 40 transforming antigen (TAg), in addition to other activities, binds to and inactivates the p53 and Rb tumor suppressor proteins,9 thereby inhibiting cell cycle arrest. Loss of normal p53 function is the most common genetic change observed in human liver tumors. In transgenic mice, TAg can induce benign and malignant liver neoplasms by 3 to 4 months of age with an incidence of 100%.3, 10 Transgenic mice coexpressing two oncogenic transgenes in hepatocytes displayed increased tumor multiplicity and decreased latency compared with single transgenic littermates.3, 4, 6, 11-13 However, the types of analyses performed using these models, which include gross and microscopic observation of lesion development and molecular examination of tumors, remain similar to earlier experimental designs. Furthermore, transgene regulatory elements target expression to most or all cells of a particular type, yet focal lesions develop. This finding indicates that additional genetic or epigenetic changes must accumulate in the target cell population that are able to complement transgene expression. As a consequence, though we can use transgenic animals to determine whether any genetic change predisposes a tissue to neoplasia, it remains difficult to identify the specific biological mechanism(s) by which that change increases carcinogenic risk.
We describe a hepatocyte transplantation system that allows us to measure directly the influence of oncogene expression on hepatocyte growth in both growth-stimulatory and growth-quiescent hepatic environments, and to quantify the frequency with which oncogene-expressing cells progress to malignancy. This experimental approach provides quantitative and mechanistic data regarding the role of specific oncogenes during hepatocarcinogenesis.
Abbreviations:
BrdU, bromodeoxuridine; CHeGA, comparative hepatocyte growth assay; EO, extreme outlier; hPAP, human placental alkaline phosphatase; lacZ, β-galactosidase; TAg, simian virus 40 T antigen; TGFα, transforming growth factor alpha; uPA, urokinase-type plasminogen activator.
Materials and Methods
Experimental Animals
Mice were housed and maintained according to The Guide for the Care and Use of Laboratory Animals in Association for Assessment and Accreditation of Laboratory Animal Care–accredited facilities. All experimental procedures were approved by the Institutional Animal Care and Use Committee. The transgenic lines used in these studies have been assigned the following genetic designations: major urinary protein uPA line 350-2, TgN(MupPlau)1Eps; hsMT-nLacZ line 379-4, TgN (Mt1nLacZ)4Eps; R26-hPAP line 808-6, TgN(R26ALPP)5Eps; AL-TAg line 522-8, TgN(Alb1SV)46Bri; MT-TGFα line 1745-8, TgN(Mt1Tgfa)149Bri; AL-c-myc line 741-3, TgN(Alb1Myc)82Bri.3, 5, 14
For these studies, mice were of the FVB, C57BL/6, or (FVB6)F1 strain background. One group of recipient mice were athymic Swiss nu/nu. Transplant recipient mice carrying metallothionein (MT)-TGFα donor hepatocytes were administered 25 mM zinc sulfate in drinking water starting at the time of transplant to induce transgene expression.5
Hepatocyte Isolation and Transplantation
β-Galactosidase (LacZ)-marked or human placental alkaline phosphatase (hPAP)-marked donor hepatocytes were isolated from 2-week-old to 5-week-old donor mice using a modified two-step ethylenediaminetetra-acetic acid/collagenase A protocol.14 In all cases, mice were excluded as donors if they displayed focal lesions visible on gross examination. Transgenic mouse livers lacking these alterations contain few or no areas of parenchyma that would be microscopically diagnosed as neoplastic, although dysplastic cells may be present.3, 5, 6, 12 The concentration of viable large cells (hepatocytes) was determined by trypan blue exclusion using a hemacytometer. Cells were maintained at 4°C until transplanted. Hepatocytes were transplanted surgically in 10 μl of L15 medium (Life Technologies, Rockville, MD) via intrasplenic injection into histocompatible recipients within 6 hours of isolation.14
Detection and Quantification of Donor Cell Liver Repopulation
Recipient mice were administered 0.1 mg/kg cadmium sulfate intraperitoneally to induce expression of the MT-nLacZ transgene, then 16 to 24 hours later liver was collected and a portion was fixed in 4% paraformaldehyde at 4°C for 1 hour then transferred to 70% ethanol. β-Galactosidase- and hPAP-expressing hepatocyte foci were identified as described,14 by incubating separate pieces of liver with an appropriate enzyme-selective substrate. Transgene-expressing cells displayed a blue reaction product. The cross-sectional surface area occupied by single blue-staining donor clones was determined on the liver surface under low-power magnification by measuring the major axes (a, b) of each stained focus, then calculating the area of an oval using the formula A= π[1/4(a+b)]2.14 The surface areas of up to 50 foci were measured per liver for each donor cell type. Mean focus volume was calculated using the formula V = 4/3πr3, taking r as [mean A/π]1/2. Median cell number per focus was calculated by dividing median focus volume by mean hepatocyte volume (8.2 × 10−6 mm3 for a 25-μm-diameter hepatocyte).14 Median cumulative cell doublings was calculated as the number of cell doublings needed to produce the median cell number per focus starting from a single progenitor cell (assumes no cell death). Comparative hepatocyte size or mean cross-sectional area (μm2) was determined microscopically.
Immunohistochemistry
To label cells undergoing DNA synthesis, mice were injected with 200 mg/kg bromodeoxyuridine (BrdU; Sigma-Aldrich, St. Louis, MO), a nucleotide analog that is incorporated into DNA during the S-phase of the cell cycle, 1 to 2 hours before euthanasia. For immunohistochemistry, we followed standard procedures using an anti-BrdU rat monoclonal (Accurate Scientific, Westbury, NY) applied to tissue sections at a dilution of 1:40, or an anti-TAg rat monoclonal (Pab101; Santa Cruz Biotech, Santa Cruz, CA) applied at a dilution of 1:200.
In Situ Hybridization
In situ hybridization on frozen tissue sections was performed as described.15 Digoxigenin-labeled riboprobes were prepared using the Roche DIG Nucleic Acid Detection Kit (Roche, Indianapolis, IN). Hybridization was performed in a humidified chamber at 55°C overnight using 0.5 ng/μL DIG-labeled sense (control) or antisense probes. Hybridization was detected using anti-DIG AP-conjugated antibody (Roche) diluted 1:5000, and color detection was using NBT/BCIP stock solution (Roche). Nonspecific background was removed by incubation in 95% ethanol for 1 hour. Marker hPAP was not detected in this assay.
Cell Turnover Analysis
The BrdU labeling index was calculated as the number of BrdU-positive hepatocyte nuclei as a percentage of all hepatocyte nuclei counted within a donor cell focus. Up to 1000 cells were examined per focus. Apoptotic indices in foci were calculated similarly, using morphological criteria to identify apoptotic cells: (1) chromatin condensation and nuclear fragmentation into apoptotic bodies, (2) eosinophilic cytoplasm, and (3) cell shrinkage.
Results
Experimental Design
We have developed a transplantation-based assay system to assess the effect of oncogene or growth factor expression on hepatocyte growth in vivo (the Comparative Hepatocyte Growth Assay, CHeGA; Fig. 1). A mixture of 3 × 104 cells from each of two populations of donor hepatocytes is transplanted into liver of 3-week-old to 4-week-old recipient mice with liver disease, and subsequent donor hepatocyte growth is compared. The first donor cell population contains normal (control) hepatocytes that express a lacZ marker transgene, and serves as a standard against which to compare growth of the second population. The second population contains hepatocytes coexpressing one or two growth-regulatory molecules (growth factor or oncogene) plus a human placental alkaline phosphatase (hPAP) marker transgene.14 Recipient animals express the major urinary protein uPA (urokinase-type plasminogen activator) transgene, which is hepatotoxic.14 The transient liver disease that is present in these mice provides a growth-stimulatory environment for transplanted donor hepatocytes, and the donor cells proliferate and expand as clonal foci for approximately 4 weeks. By this time, diseased hepatic parenchyma has been replaced by a combination of healthy donor hepatocytes and healthy endogenous hepatocytes that have inactivated uPA transgene expression, and the repopulated liver becomes quiescent.14, 16 Donor cell number was chosen so that donor repopulation of liver would be minimal (<5%),14 thereby enabling accurate measurement of donor focus size uncomplicated by the presence of multiple adjoining foci. We compare the size of foci from the two donor cell populations at 1, 2, 4, 8, and 12 weeks posttransplantation, using histochemistry to distinguish between foci expressing β-galactosidase (the lacZ gene product) or hPAP. For each oncogene evaluated, we use at least two donor cell preparations, and multiple recipients are examined at each time posttransplantation. We also performed histochemical, immunohistochemical, or in situ RNA hybridization assays to detect donor cell transgene expression in representative recipient animals. Coexpression of all transgenes was identified in 90%-99% of foci with each transgene combination (Table 1).
Comparative hepatocyte growth assay. See text for details.
| Transgenes (Number of Mice) | Foci Coexpressing All Transgenes, Mean ± SD (No. of Foci)* |
|---|---|
| TGFα/hPAP (4) | 99 ± 1% (461) |
| c-myc/hPAP (4) | 94 ± 5% (295) |
| TAg/hPAP (4) | 97 ± 9% (134) |
| c-myc/TGFα/hPAP (3) | 93 ± 8% (220)† |
| TAg/TGFα/hPAP (3) | 96 ± 5% (102) |
| TAg/c-myc/hPAP (2) | 90 ± 0.4% (128) |
- * hPAP expression determined by enzyme histochemistry. Oncogene expression determined by immunohistochemistry and/or in situ hybridization.
- † c-myc and TGFα mRNAs could not be distinguished by the in situ probe employed (specific for a shared human growth hormone 3′ noncoding region); therefore, this value represents the percentage of hPAP foci positive for either or both. However, growth of these foci was uniformly increased relative to TGFα or c-myc foci (Table 3), providing biological evidence for uniform transgene expression.
Marker Gene Expression Does Not Differentially Affect Hepatocyte Growth
In control experiments, we measured mean areas of transplant foci in recipients receiving normal hPAP (with no oncogenic transgene) and normal lacZ donor cells (Fig. 2A). We observed no significant differences in focus size except at 1 week posttransplantation. The difference at 1 week is likely a measurement artifact caused by the nuclear localization of β-gal versus the cell surface and cytoplasmic localization of hPAP. In larger (older) foci, this does not bias measured focus size. Thus, markers do not differentially affect focus growth. We identified two growth stages of normal donor cells in recipient livers (Fig. 2A). First was a “growth phase,” from weeks 1 to 4 posttransplantation, during which donor hepatocyte foci increase in size. Second was a “quiescence phase,” from weeks 4 to 12 posttransplantation, which is related to completion of parenchymal repopulation by healthy cells and corresponding loss of the growth stimulus.14, 16 During this stage, control donor focus size remains constant.
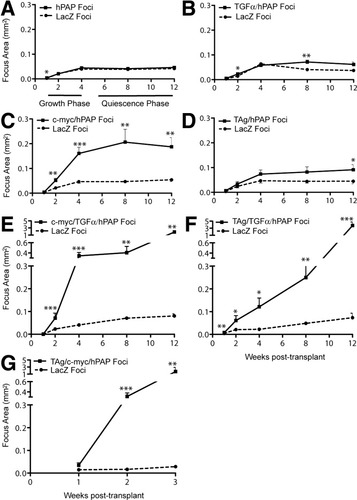
Transplant focus mean cross-sectional areas versus time posttransplantation. Mean focus cross-sectional area was determined for both donor cell populations in each recipient mouse. Data are plotted as mean ± standard error of the mean for all mice examined at each time posttransplantation. See Table 1 for number of mice. (A) Normal hPAP and lacZ donor hepatocyte foci; (B) TGF-α–expressing donor hepatocyte foci; (C) c-myc–expressing donor hepatocyte foci; (D) TAg-expressing donor hepatocyte foci; (E), c-myc- and TGF-α–coexpressing donor hepatocyte foci; (F), TAg- and TGF-α–coexpressing donor hepatocyte foci; (G), TAg- and c-myc–coexpressing donor hepatocyte foci (note different time scale). Means for lacZ-marked and hPAP-marked hepatocyte foci within each experiment were compared using the Mann-Whitney test. No asterisk, not significantly different (P ≥ 0.05); *, different at P < 0.05; **, different at P < 0.01; ***, different at P < 0.001.
We also calculated the number of hPAP-marked donor hepatocyte cell doublings required to generate the observed median focus sizes at 4 and 12 weeks posttransplantation (Table 2; for this and subsequent analyses we evaluate median data). As expected for a quiescent liver, cell doublings are not significantly different between 4 and 12 weeks for genetically normal control hepatocytes expressing only hPAP.
| Transgenes Expressed | Time Posttransplant (No. of Mice) | Median Cumulative Cell Doublings† |
|---|---|---|
| hPAP only | 4 weeks (15) | 9.0 ± 0.4 |
| 12 weeks (10) | 9.7 ± 0.4 | |
| TGFα/hPAP | 4 weeks (10) | 10.4 ± 0.2** |
| 12 weeks (8) | 9.9 ± 0.4 | |
| c-myc/hPAP | 4 weeks (11) | 11.8 ± 0.4*** |
| 12 weeks (9) | 11.9 ± 0.5** | |
| TAg/hPAP | 4 weeks (10) | 9.7 ± 0.4 |
| 12 weeks (9) | 10.0 ± 0.6 | |
| EO‡ (6) | 15.3 ± 0.4*** | |
| c-myc/TGFα/hPAP | 4 weeks (12) | 13.0 ± 0.5*** |
| 12 weeks (9) | 14.7 ± 0.4*** | |
| EO‡ (9) | 22.0 ± 0.5*** | |
| TAg/TGFα/hPAP | 4 weeks (5) | 10.6 ± 1.1 |
| 12 weeks (9) | 15.7 ± 0.7*** | |
| EO‡ (9) | 22.8 ± 0.6*** | |
| TAg/c-myc/hPAP | 2 weeks (8) | 12.3 ± 0.4*** |
| EO‡ (7) | 18.2 ± 0.8*** |
- † Calculated as described in Materials and Methods, starting from median focus cross-sectional area for the experimental groups shown in Fig. 2. No asterisk, not significantly different (P ≥ 0.05) compared to hPAP values using Mann-Whitney one-tailed comparison.
- *different at P < 0.05.
- ** different at P < 0.01.
- *** different at P < 0.001.
- ‡ EO = extreme outlier medians identified at 12 weeks (or 2 weeks for TAg/c-myc) posttransplant; Mann-Whitney statistics comparing EO with corresponding 12 week (or 2 week) median value.
Examination of scatter plots of focus area in individual recipient mice (Fig. 3A) indicated that focus growth varied considerably among recipients. Thus, we normalized the data by dividing each hPAP focus area by the mean area of lacZ foci for that mouse, obtaining a focus ratio distribution for each recipient (Fig. 3B). Normalization is possible because we compare data between two cell populations in the same recipient mouse liver, which therefore have been exposed throughout the study to the same hepatic and systemic environments. The average of median focus ratio distribution values for all mice at each time posttransplantation should equal “one” if there is no difference in the size of hPAP versus lacZ foci (Table 3). At 1 week posttransplantation, hPAP foci appear larger than lacZ foci (P = 0.049; likely because of a measurement artifact, as noted above), but at subsequent times the values are very close to 1. Note that Fig. 3 displays only representative data. All data are summarized in Table 3.
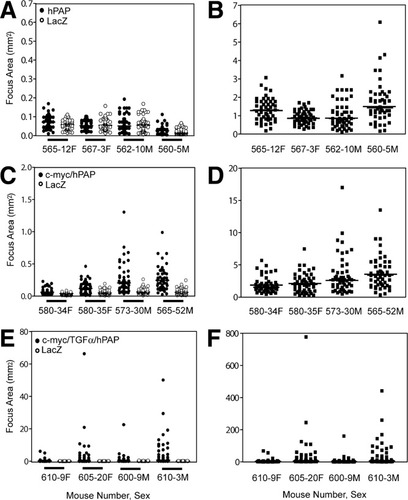
Size and hPAP/lacZ ratio distributions of transplant foci. Representative data at 12 weeks posttransplantation. (A), (C), and (E): Size (cross-sectional area) distribution of hPAP-marked foci (solid circles) and lacZ-marked foci (open circles) in representative recipients. Each distribution contains 50 data points (some not visible because of extensive overlap); (B), (D), and (F): hPAP/lacZ focus ratio distribution of (B) hPAP; (D) c-myc/hPAP, and (F) c-myc/TGFα/hPAP transplant foci in recipient mouse livers. The focus ratio distribution is generated by dividing each hPAP area value by the mean lacZ area value for that recipient mouse. In each ratio distribution, the horizontal line represents the median value.
| Transgene(s) | Focus Ratio Distribution Medians, Weeks Posttransplant†‡ | Extreme | ||||
|---|---|---|---|---|---|---|
| 1 | 2 | 4 | 8 | 12 | Outlier%‡, | |
| hPAP only | 1.70±0.28(5) | 0.97±0.08(13) | 1.0±0.05(15) | 1.02±0.10(11) | 1.06±0.07(10) | 0.4±0.3 |
| TGFα/hPAP | – | 1.49±0.13(7) | 1.36±0.09(10) | 1.92±0.45(6) | 1.80±0.24(7) | 0.3±0.8 |
| (0.003)** | (0.001)** | (0.007)** | (0.005)** | (0.4) | ||
| c-myc/hPAP | – | 1.92±0.19(6) | 2.99±0.40(11) | 2.33±0.45(6) | 2.91± 0.41(9) | 1.3±0.7 |
| (0.001)** | (0.0001)*** | (0.002)** | (0.0001)*** | (0.2) | ||
| TAg/hPAP | – | 1.27±0.25(5) | 1.28±0.24(10) | 1.40±0.19(9) | 1.47±0.25(7) | 2.1±1.2 |
| (0.13) | (0.23) | (0.07) | (0.10) | (0.007)** | ||
| c-myc/TGFα/hPAP | – | 2.3±0.15(6) | 8.7±2.8(12) | 8.2±1.8(7) | 6.0±0.84(9) | 7.0±4.8 |
| (0.0004)*** | (0.0001)*** | (0.0003)** | (0.0001)*** | (0.003)** | ||
| TAg/TGFα/hPAP | – | 2.1±0.4(6) | 4.6±1.8(5) | 4.4±1.6(9)‖ | 10.9±2.7(9)‖ | 8.4±3.3 |
| (0.005)** | (0.02)* | (0.0003)*** | (0.0001)*** | (0.0002)*** | ||
| TAg/c-myc/hPAP | 13.2±2.4(7) | 9.8±1.7(7) | 10.7±6.3(6)¶ | – | – | 4.6±4.9# |
| (0.002)** | (0.002)** | (0.0002)*** | (0.0003)*** | (0.005)** | ||
- † Data presented as average of individual ratio medians ± SEM (number of mice), followed by (P).
- ‡ P values obtained from one-tailed Mann-Whitney test comparisons between oncogene-expressing populations and control hPAP populations. No asterisk, not significantly different (P ≥ 0.05).
- * different at P < 0.05.
- ** different at P < 0.01.
- *** different at P < 0.001.
- Data presented as mean ± SD (P value) for percentage of Extreme Outliers within focus ratio distributions at 12 weeks post-transplant. The Mann-Whitney one-tailed test was used to compare oncogene-expressing populations with control hPAP population at 12 weeks post-transplant. See 12 week post-transplant column for number of mice.
- ‖ For the TAg/TGFα combination, 8 and 12 weeks were significantly different (P = 0.02); there were no other significant differences between 4 or 8 weeks and 12 weeks posttransplant within any other group.
- ¶ Mean ± SEM of ratio medians for TAg/c-myc foci at 3 weeks posttransplant. The P value was obtained from Mann-Whitney test comparison with 4 week posttransplant hPAP focus medians.
- # Extreme outlier percentage at 2 weeks posttransplant (compared statistically to hPAP extreme outliers at 2 weeks posttransplant).
Oncogenic Transgenes Have Distinct Effects on Hepatocyte Growth In Vivo
Single Transgenes.
We next examined growth of hepatocytes expressing transgenes that had been shown to increase the incidence of liver cancer in transgenic mice (Figs. 2B-D, 3C,D, and Table 3). Only TGFα and c-myc significantly increased the rate of focus growth during the growth phase in recipient livers compared with hPAP alone (Table 3). However, no single growth regulatory molecule induced continued focus growth during the quiescent phase, indicating that they were not sufficient to cause growth in an environment that was not growth-stimulatory.
To determine whether focus growth was affected by immune recognition of donor cells expressing the viral simian virus 40 T antigen (TAg), we also transplanted TAg/hPAP donor cells into athymic nu/nu recipient mice and measured focus size at 4 and 8 weeks posttransplantation. We found no significant focus ratio differences between nu/nu and immune-competent recipients (data not shown), indicating that immune rejection was not a major factor in these experiments. In addition, hPAP-marked donor parenchyma is stable for more than 18 months in recipient mice.14
Transgene Combinations.
Coexpression of growth regulatory molecules in donor hepatocytes produced dramatic differences in focus size at all times posttransplantation (Fig. 2E-G). Focus ratio distribution medians also were increased (Fig. 3F and Table 3), indicating that expression of each oncogene pair was sufficient to increase the rate of hepatocyte focus growth during the growth phase. Furthermore, TAg/TGFα donor focus growth continued during the quiescent phase (Table 3; compare weeks 8 and 12), so this combination of growth regulatory molecules induced cell-autonomous hepatocyte growth in the quiescent liver. The most dramatic growth was observed after coexpression of TAg and c-myc (Fig. 2G and Tables 2 and 3). Most recipients had to be euthanized before 4 weeks posttransplantation, so we could not evaluate the combined effect of these genetic alterations on growth within a quiescent liver environment.
To determine whether different genetic alterations influenced hepatocyte size within transplant foci, we measured microscopic hepatocyte cross-sectional area in foci (Table 4). Mean hepatocyte area did not significantly increase for any genetic alteration, although in foci expressing TAg plus either c-myc or TGFα, hepatocyte diameter sometimes was smaller than control hepatocytes. Thus, changes in focus growth were associated with hyperplasia, not cellular hypertrophy.
| Transgene(s) | Weeks Posttransplant (Number of Mice) | Mean Hepatocyte Cross-Sectional Area (μm2) Mean ± Standard Deviation† |
|---|---|---|
| hPAP only | 2 (7) | 740 ± 150 |
| 12 (5) | 760 ± 110 | |
| TGFα/hPAP | 2 (5) | 790 ± 130 |
| 12 (6) | 850 ± 93 | |
| c-myc/hPAP | 2 (6) | 830 ± 250 |
| 12 (3) | 830 ± 120 | |
| TAg/hPAP | 2 (5) | 710 ± 140 |
| 12 (3) | 640 ± 180 | |
| c-myc/TGFα/hPAP | 2 (3) | 940 ± 140 (P = 0.06) |
| 12 (4) | 660 ± 120 | |
| TAg/TGFα/hPAP | 2 (3) | 600 ± 60 |
| 12 (4) | 510 ± 80* | |
| TAg/c-myc/hPAP | 1 (4) | 590 ± 80‡ (P = 0.07) |
| 2 (3) | 560 ± 60‡ (P = 0.06) | |
| 3-4 (3) | 430 ± 18‡* |
- Mean hepatocyte cross-sectional area determined as described in Materials and Methods.
- † Statistical comparisons obtained from two-tailed Mann-Whitney comparisons between oncogene-expressing populations and control hPAP populations. No asterisk, not significantly different (P ≥ 0.05).
- * different at P < 0.05.
- ‡ The P value was obtained from comparison with 2 week posttransplant hPAP hepatocyte area.
Focus Growth Outliers.
Mean focus area plots (Fig. 2) can be influenced by two aspects of focus growth. First, oncogene expression may alter growth of all donor cells. Median focus size addresses this effect (Table 3) and reveals growth changes for which each oncogene or oncogene combination is sufficient. Second, oncogene expression may cause development of a few exceptionally large foci (outliers), which will increase mean focus size (Fig. 2) but not affect the distribution median. To quantitate outliers in focus ratio distributions at 12 weeks posttransplantation (the end-stage for these studies), we used the method of Tukey.17 In this method, extreme outliers (EOs) are defined as 3 × the interquartile range or more above the third quartile of a distribution, where the interquartile range is the distance between the first and third quartiles.
Among single oncogenes, only TAg expression was associated with a significant increase in outliers (Table 3). Coexpression of oncogenes increased outlier frequency further (even by 2 weeks for TAg/c-myc hepatocyte foci). Note that, for foci coexpressing TGFα and c-myc, all of the mean focus area increase from 4 to 12 weeks (Fig. 2E) was due to outliers, because median focus size did not increase (Table 3). There was no effect of recipient sex on outlier frequency (data not shown), consistent with the slight to minimal gender differences in disease latency in these lines of donor mice. When examined microscopically, most TGFα/c-myc median (non-outlier) foci at 12 weeks posttransplantation resembled normal hepatic parenchyma: only 2 of 10 non-outlier foci displayed a mildly atypical hepatocellular phenotype (two-cell-thick hepatic plates). In contrast, 11 of 11 EO foci were composed of dysplastic hepatocytes that were clearly distinguishable from adjacent parenchyma, including thickened hepatic plates, basophilic, and clear cell phenotypes. In this regard, most resembled classic altered cell foci that develop after carcinogen administration.
Transplanted hepatocytes enter the parenchyma by passing through or between endothelial cells to enter liver plates in the host liver, favoring transit and subsequent engraftment of single cells.18, 19 Nevertheless, to rule out differential transplantation of clumps of cells as a trivial explanation for increases in extreme outliers, we also evaluated EOs at 2 weeks posttransplantation. At 12 weeks posttransplantation, EOs had undergone five (TAg) to seven additional cell doublings compared with median foci of the same genotype (Table 2). If EOs resulted from transplantation of cell clumps, rather than enhanced growth, then EOs with this excess of cell doublings also would be present at 2 weeks. At 2 weeks posttransplantation, c-myc/TGFα distributions displayed no outliers, and TAg/TGFα distributions displayed only 0.7% outliers (versus 7% and 8.4%, respectively, at 12 weeks). TAg distributions showed 2.4% EOs, but these outliers displayed a median of only 1.1 additional cell doublings, compared with 2-week median TAg foci (9.8 versus 8.7). These data confirm that EOs are the result of increased focus growth after transplantation.
Cell Turnover Kinetics
To identify cell turnover characteristics in transplant foci, we determined hepatocyte DNA synthesis (BrdU labeling) and apoptotic indices in foci during the growth (2 weeks) and quiescent (8 or 12 weeks) phases posttransplantation (Table 5). Only TAg, alone or in combination, and c-myc/TGFα at 8 to 12 weeks, had an effect on apoptosis, significantly or nearly significantly increasing the index twofold to threefold. The most consistent effect on focus DNA synthesis was the expected decrease from 2 weeks to 8 to 12 weeks in most groups. At 2 weeks posttransplantation, only TAg/TGFα and TAg/c-myc caused increases in DNA synthesis compared with controls. At 8 to 12 weeks, TAg and c-myc/TGFα caused fourfold and threefold increases, but these were balanced by increases in apoptosis, consistent with lack of continued focus growth in the quiescent phase. In striking contrast, DNA synthesis in TAg/TGFα foci remained unchanged from 2 weeks to 8 to 12 weeks, explaining continued growth of these foci in the quiescent liver environment.
| Transgene(s) | Weeks Posttransplant (Number of Mice) | BrdU Labeling Index Mean ± SEM%† | Apoptosis Index Mean ± SEM%† |
|---|---|---|---|
| hPAP only | 2 (7) | 9.5 ± 1.6 | 0.70 ± 0.10 |
| 8-12 (7) | 1.14 ± 0.44 | 0.51 ± 0.14 | |
| TGFα/hPAP | 2 (5) | 8.3 ± 0.9 | 0.71 ± 0.34 |
| 8-12 (5) | 0.49 ± 0.11 | 0.48 ± 0.12 | |
| c-myc/hPAP | 2 (8) | 12.4 ± 2.3 | 0.99 ± 0.37 |
| 8-12 (6) | 1.09 ± 0.62 | 0.67 ± 0.24 | |
| TAg/hPAP | 2 (5) | 8.8 ± 1.9 | 1.60 ± 0.75 (P = 0.07) |
| 8-12 (7) | 4.4 ± 1.9* | 1.54 ± 0.62 (P = 0.08) | |
| c-myc/TGFα/hPAP | 2 (6) | 11.1 ± 1.8 | 1.18 ± 0.41 |
| 8-12 (4) | 3.17 ± 0.28* | 1.50 ± 0.64 (P = 0.05) | |
| TAg/TGFα/hPAP | 2 (3) | 14.0 ± 0.6 (P = 0.06) | 1.53 ± 0.48* |
| 8-12 (10) | 14.8 ± 1.5*** | 1.28 ± 0.19** | |
| TAg/c-myc/hPAP | 1-2 (7) | 28.4 ± 2.1*** | 1.96 ± 0.40** |
- BrdU and apoptotic indices were determined as described in Materials and Methods.
- † Statistical comparisons obtained from one-tailed Mann-Whitney comparisons between oncogene-expressing populations and control hPAP populations. No asterisk, not significantly different (P ≥ 0.05).
- * different at P < 0.05.
- ** different at P < 0.01.
- *** different at P < 0.001.
Discussion
Much of our current understanding of carcinogenesis is derived from animal models. Early experimental approaches, involving local or systemic administration of chemical carcinogens, defined the multistage model of carcinogenesis. This model implies that multiple cellular alterations are required for the development of neoplasia. Molecular analyses of both spontaneous and chemically induced tumors now have identified many genetic and epigenetic changes that accompany carcinogenesis. Subsequent approaches examined the carcinogenic influence of these identified genetic alterations in vivo using transgenic and gene-targeted mice.1, 2 These modeling approaches let us assign a specific increase in cancer susceptibility to the presence of a selected gene alteration and to identify carcinogenic interactions between gene alterations. Recently, experimental systems have been described in which a focal pattern of oncogene expression can be established in liver (reviewed by Marongiu et al.20). These systems require transplantation of marked hepatocytes into mice with diseased liver, induced by knockout of fumaryl-acetoacetate, partial hepatectomy after administration of the alkylating agent retrorsine, or hepatocyte-targeted expression of uPA. Reports describing transplantation of in vitro transduced fetal hepatoblasts21 or injection of oncogene-expressing transposon plasmids22 into mouse liver demonstrated feasibility of restricting oncogene expression to clones of hepatocytes, from which neoplasms arose, but were not quantitative.
The comparative hepatocyte growth assay (CHeGA) represents a complement to other experimental in vivo models of liver carcinogenesis. Unlike previous systems, which assess oncogene carcinogenicity, CHeGA allows us to separate and quantify the effects of gene alterations on cellular growth in growth stimulatory and quiescent environments. Furthermore, we can determine whether there is posttransplantation development of hepatocyte focus growth outliers. The presence of outliers implies that some transplanted cells possessed stable changes (genetic or epigenetic) in addition to oncogene expression at the time of transplantation, or developed these changes shortly after transplantation. Because outlier growth continues in quiescent liver, these underlying changes must create the potential in affected cells for cell-autonomous (environment-independent) growth, and in this way progenitors of outliers meet one criterion for preneoplastic cells. Using our growth assay, we can quantify, for any potential oncogene or oncogene combination, the associated risk of developing extreme outliers (EOs) with preneoplastic behavior. In fact, EO frequency is the best predictor of oncogene carcinogenicity in transgenic mice (see below), as expected if outliers are preneoplastic. This finding and our observation that EO microscopic anatomy is abnormal are consistent with suggestions by Laconi and colleagues20 that altered growth pattern is a principal marker of altered/nodular hepatocytes, although, in our system, these foci were identified by their ability to continue growth in a quiescent liver.
Our findings, together with published data regarding oncogene effects in transgenic mice, provide insight into the role of each oncogene in hepatocarcinogenesis. The principal effects of TGFα in transgenic mice are to stably increase hepatocyte number (liver mass increases up to twofold in transgenic mice),5, 8 and to increase the rate of hepatocyte replication after two-thirds partial hepatectomy and in 4-week-old but not 7-week-old mouse liver.7, 8 Consistent with these findings, TGFα quantifiably increases the rate at which hepatocytes can replicate under growth permissive conditions in CHeGA, but it does not uncouple replication from environmental controls in quiescent liver nor does it increase posttransplantation EOs. This liver phenotype is associated with a low risk for neoplastic progression on a per hepatocyte basis, because MT-TGFα transgenic mice develop a low tumor multiplicity with 10-month to 12-month latency.5, 8, 23 Similarly, c-myc acts to increase the rate at which hepatocytes replicate under growth-permissive conditions, both in transgenic mice24 and in CHeGA. However, c-myc–expressing hepatocytes remain tightly regulated by their environment and have a very low risk of escaping this regulation. This liver phenotype is consistent with the maintenance of normal liver mass, long tumor latency (>12 months), and low tumor incidence and multiplicity observed in AL-c-myc transgenic mice.3, 4 In contrast, although the viral TAg stably increases hepatocyte turnover (increased BrdU labeling and apoptosis) both in AL-TAg transgenic mice12 and in transplant foci, it does not directly increase net hepatocyte growth under permissive conditions. Rather, as demonstrated by an increase in EOs, it acts by measurably increasing the risk that a TAg-expressing hepatocyte will accumulate changes that allow it to escape normal growth controls. This finding is consistent with TAg's ability to cause hepatocyte genomic instability,3, 25 especially when coupled with the increased cell turnover that we detected. This liver phenotype results in both the shortest latency (3-4 months) and highest tumor multiplicity among single oncogenes in transgenic mice.3, 10
Oncogene coexpression provides important additional information about oncogene effects. In transgenic mice, coexpression of TGFα and c-myc induces hepatocyte aneuploidy, chromosomal breaks, and translocations, even by 3 weeks of age,26 reduces tumor latency (5-7 months), and increases tumor multiplicity.5, 6, 11, 13, 27 This combination also is associated with a pathway of hepatocarcinogenesis involving increased genomic instability.11, 13 Our data indicate that these oncogenes additively or synergistically increase posttransplantation hepatocyte growth in a permissive environment, but still cannot induce growth in quiescent liver. Nevertheless, as for TAg, they increase hepatocyte turnover and they dramatically increase EO frequency. In our transplantation system, we did not observe reduced apoptosis in foci expressing both oncogenes, in contrast to other data from mouse studies.27 The mechanisms underlying TGFα/c-myc oncogenesis appear to involve, first, increased risk for development of preneoplastic cells, likely the result of genomic instability. Second, once preneoplastic cells emerge that are unresponsive to normal growth inhibition, TGFα/c-myc can collaborate further to promote rapid cell autonomous outlier focus growth. In this sense, capacity for increased growth under permissive conditions remains a “silent trait” in quiescent liver that is revealed only if cells develop additional alterations.
The remaining oncogene pairs combine enhanced growth in a permissive environment (TGFα or c-myc) with inhibition of cell cycle arrest (TAg). These oncogene combinations decrease hepatocyte size in transplant foci, raising the possibility that partial cell dedifferentiation accompanies their expression. With respect to neoplasia, these pairings are remarkably potent, both in transgenic animals6, 12 and after hepatocyte transplantation. Both pairings are associated with synergistic growth increases in permissive liver, caused principally by changes in hepatocyte replication rather than apoptosis, and with elevated EO frequency. When paired with TAg, TGFα now can produce continued focus growth in the nonpermissive quiescent liver environment, the result of a continued high rate of DNA synthesis even when surrounding normal hepatocytes stop replicating. The TAg/c-myc interaction is the strongest: focus growth is so rapid that recipients do not survive to the quiescent liver phase. Thus, TGFα and c-myc increase the rate of hepatocyte growth not only in a permissive liver environment but also in cells rendered permissive for growth by other genetic changes.
Our data indicate that rate of focus hepatocyte turnover coupled with frequency of preneoplastic-like EOs in CHeGA provides a strong predictor of the risk for neoplastic progression associated with any oncogene or oncogene combination. Rate of hepatocyte growth under permissive conditions is not predictive. Our data further suggest that physiological maintenance of the normal quiescent liver environment has at least two components: tight control over activation of growth signaling pathways and stable capacity for cell cycle arrest. Interfering with either alone does not produce unregulated growth; however, interference with both may be sufficient to establish a “permissive” intracellular environment that allows cell autonomous hepatocyte replication, a defining characteristic of cancer cells. In fact, genes that regulate these two aspects of cellular growth control are strong candidate targets for “additional genetic changes” that may be present in growth outliers to permit their extreme growth. As shown above, by combining data on posttransplantation growth and transformation frequency, we can identify and quantify biological mechanisms by which candidate genetic changes contribute to liver cancer in the living organism.
Acknowledgements
The authors thank Meg Bowden, Adam Jochem, Tim Stein, Renee Szakaly, and Garrett Zielinski for technical assistance.




