MicroRNA-122, a tumor suppressor microRNA that regulates intrahepatic metastasis of hepatocellular carcinoma†
Potential conflict of interest: Nothing to report.
fax: (886)-2-28264092.
Abstract
MicroRNAs (miRNAs), which are inhibitors of gene expression, participate in diverse biological functions and in carcinogenesis. In this study, we show that liver-specific microRNA-122 (miR-122) is significantly down-regulated in liver cancers with intrahepatic metastastasis and negatively regulates tumorigenesis. Restoration of miR-122 in metastatic Mahlavu and SK-HEP-1 cells significantly reduced in vitro migration, invasion, and anchorage-independent growth as well as in vivo tumorigenesis, angiogenesis, and intrahepatic metastasis in an orthotopic liver cancer model. Because an inverse expression pattern is often present between an miRNA and its target genes, we used a computational approach and identified multiple miR-122 candidate target genes from two independent expression microarray datasets. Thirty-two target genes were empirically verified, and this group of genes was enriched with genes regulating cell movement, cell morphology, cell-cell signaling, and transcription. We further showed that one of the miR-122 targets, ADAM17 (a disintegrin and metalloprotease 17) is involved in metastasis. Silencing of ADAM17 resulted in a dramatic reduction of in vitro migration, invasion, in vivo tumorigenesis, angiogenesis, and local invasion in the livers of nude mice, which is similar to that which occurs with the restoration of miR-122. Conclusion: Our study suggests that miR-122, a tumor suppressor microRNA affecting hepatocellular carcinoma intrahepatic metastasis by angiogenesis suppression, exerts some of its action via regulation of ADAM17. Restoration of miR-122 has a far-reaching effect on the cell. Using the concomitant down-regulation of its targets, including ADAM17, a rational therapeutic strategy based on miR-122 may prove to be beneficial for patients with hepatocellular carcinoma. (HEPATOLOGY 2009.)
Since their discovery in Caenorhabditis elegans as important regulators of development timing,1 endogenous small hairpin microRNAs (miRNAs) have been found to have a broad impact on vertebrates and invertebrates by regulating gene activities through translational repression2 or posttranscription suppression.3-5 Rapid advances in the molecular characterization of miRNAs has not only revealed their physiological functions but also uncovered abnormal patterns of miRNA expression linked to many disease states including cancer.6, 7 Hepatocellular carcinoma (HCC) is one of the most common human malignancies; this disease shows exceptional heterogeneity in cause and outcome. It is especially prevalent among Asian populations. Despite successful local therapies such as surgery or transcatheter arterial chemoembolization, patients with HCC develop a high rate of recurrence due to local invasion and intrahepatic metastasis. Liver cancer is a complex disease involving epigenetic instability, chromosomal instability,8 and expression abnormalities of both coding9, 10 and noncoding genes; the latter includes miRNAs.11-14 Profiling of miRNA expression has identified signatures associated with HCC formation and progression that are potential cancer biomarkers. One such metastasis signature, consisting of 20 miRNAs, has been recognized as an independent predictor of survival.12 Deregulation of these 20 miRNAs in metastatic HCC implies that altered expression of their pertinent target genes may contribute to the development of metastasis or to recurrence. MicroRNA-122 (miR-122) is a member of this metastasis signature.
Recent work on tissue-specific miRNAs has demonstrated their participation in tissue specification and cell lineage decisions.15, 16 MiR-122 is a liver-specific miRNA known to modulate lipid metabolism,4, 17 hepatitis C virus replication,18 and antiapoptosis.19 Persistent expression of miR-122 has been detected during liver specialization into adult liver,20 yet miR-122 is down-regulated in HCC.21 Currently, only a handful of confirmed target genes of miR-122 have been reported;4, 17, 20, 22 hence, involvement of miR-122 as an effector of hepatocyte differentiation remains to be established.
In this study, we demonstrated that the loss of miR-122 expression in the intrahepatic metastasis of liver tumors is highly significant. We determined a functional role for miR-122 in liver tumor progression using both in vitro and in vivo models. Furthermore, we applied a bioinformatics-based computational prediction approach to investigate the miR-122 targets based on two microarray datasets where miR-122 is significantly down-regulated, one with local invasion of HCC and the other involving antagomir-122–treated mouse livers. Finally, we carried out target verification by 3′ untranslated region (3′UTR) reporter assay and expression analysis. Based on involvement in regulating cell adhesion, migration, and metastasis, a disintegrin and metalloprotease 17 (ADAM17), an empirically verified target gene of miR-122, was chosen for in-depth investigation.
Abbreviations
ADAM17, a disintegrin and metalloprotease 17; HCC, hepatocellular carcinoma; miRNA, microRNA; miR-122, microRNA-122; shRNA, small-hairpin interfering RNA; 3′UTR, 3′ untranslated region; T1, stage I HCC; T3, stage III HCC.
Patients and Methods
Cell Lines and Human Liver Tissues.
The human HCC cell lines Mahlavu, HuH-7, and SK-HEP-1 as well as the human embryonic kidney cell line HEK293T were cultured as described.23 Paired samples of tumor/nontumorous liver tissues were obtained from patients who had undergone primary HCC curative hepatic resection at Taipei Veterans General Hospital, Taiwan. The study was approved by the Committee for the Conduct of Human Research, and patient informed consent was obtained. This study consisted of 48 HCC samples from two tumor-node-metastasis (TNM) stages, stage I (T1, n = 25) and stage III (T3, n = 23), according to the American Joint Committee on Cancer 6th edition TNM classification. Immediately after surgical resection, all tissues were snap-frozen in liquid nitrogen and stored at −80°C.
Gene Expression Analysis.
Expression analysis of miR-122 was done by low stringency northern blotting and TaqMan MicroRNA Assay (Applied Biosystems, Foster City, CA). RNA (20 μg) was separated on a 15% denaturing polyacrylamide gel, transferred to Hybond-N+ membrane (Amersham Bioscience, Piscataway, NJ), and hybridized with a [γ-32P]-labeled 5′-ACAAACACCATTGTCACACTCCA-3′ probe. The most stringent wash was carried out using 2× standard sodium citrate, 0.5% sodium, dodecyl sulfate at 42°C before autoradiography. The same blot was reprobed for U6 snRNAl. TaqMan MicroRNA Assay, a quantitative stem-loop polymerase chain reaction was performed according to the manufacturer's specifications with U6 RNA as an internal control. Expression of pri-miR-122 was detected by RT-PCR using the primers 5′-GCTCTTCCCATTGCTCAAGATG-3′ and 5′-GTATGTAACAACAGCATGTG-3′. Expression levels of miR-122 target genes were also examined by reverse transcription polymerase chain reaction (RT-PCR) (primer sequences in Supporting Table 6).
Plasmid Constructs.
A partial human pri-miR-122 gene (a 562–base pair complementary DNA fragment encompassing 54,269,034 base pairs to 54,269,595 base pairs of 18q21.31, University of California Santa Cruz Genome Browser) was subcloned into the lentiviral expression vector, pPGK-GFP (National RNAi Core Facility, Academia Sinica, Taiwan) in place of GFP and called pPGK-122. pPGK-GFP, a positive control for the lentiviral infection, contains necessary cis-elements for packaging, reverse transcription and integration. PGK stands for the human phosphoglycerate kinase promoter sequences. pPGK-122 was used to make lentiviruses expressing miR-122 (lenti-122). A miR-122 seed-region (5′-GGAGTGTG-3′ to 5′-ATGACTGA-3′) mutant (lenti-122M) was constructed using a QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA).
Ectopic Expression of miR-122.
Stable HCC cell cultures expressing miR-122 were generated by coinfecting HCC cells with lenti-122 and short hairpin Luciferase (shLuc) at 100:1 ratio using 8 μg/mL protamine sulfate for 24 hours and puromycin (2 μg/mL) selection for 3-5 days. The shLuc targeting the luciferase gene provides the puromycin resistance. Expression of mature miR-122 was confirmed by northern blotting.
Prediction of miR-122 Targets Among Differentially Expressed Genes.
For miR-122 target prediction, we applied miRanda24 to energetically detect the most probable targets against the 3′UTR among all human and mouse coding genes (BioMart from the Ensembl database25). The miRNA/target duplexes with miRanda scores greater than 120 and minimum free energy values less than −10 kcal/mol were identified as miR-122 target sites. The analysis strategy is presented in Supporting Table 2.
3′UTR Reporter Assay.
The 3′UTR fragments of the candidate target genes were subcloned into the Xba1 site downstream of the luciferase gene in pGL3-Control Vector (Promega, Madison, WI). Under the control of SV40 promoter and enhancer, pGL3-Control Vector expresses strong luciferase activity in many types of mammalian cells. The negative controls were lenti-122M and lenti-GFP. HEK293T cells were infected with lenti-GFP and lenti-122 or lenti-122M for 24 hours. Cells were then seeded into 24-well plates and cotransfected with 0.5 μg of the respective pGL3-3′UTR construct and 0.05 μg of the pRL-TK vector (Promega) using jetPEI (Polyplus-Transfection, France). pRL-TK vector was used as an internal control reporter. After 48 hours, luciferase activity was measured using the Dual-Luciferase Reporter Assay System Kit (Promega). The effect of miR-122 was expressed relative to the average value from cells infected with lenti-GFP virus. A mutant of the single miR-122 binding site (5′-ACACUCCA-3′ to 5′-ACCCGCAA-3′) in the 3′UTR of ADAM17 was included. The mutagenesis primers are listed (Supporting Table 7) together with the nucleotide sequences of all the PCR cloning primers (Supporting Table 8).
RNA Interference with shADAM17.
HCC cells were plated and infected with lentiviruses expressing shADAM17 or shLuc in the presence of 8 μg/mL protamine sulfate for 24 hours followed by puromycin (2 μg/mL; 48 hours) selection. RT-PCR and/or western blotting were performed to validate the knockdown efficiency. The shRNA constructs are described in Supporting Fig. 4.
Cell Migration Assay and Invasion Assay.
Cells were tested for their migration and invasion abilities in vitro in a Minicell (Millipore) according to the methods described previously.23
Soft-Agar Colony Assay.
Cells were seeded at 5 × 103 cells/well in a six-well plate containing 0.3% low melting temperature agarose (Sigma, St. Louis, MO). Colonies were counted after 14 days. The Integrated Morphometry analysis function of MetaMorph software (Molecular Devices, Sunnyvale, CA) was used to measure the surface areas of the colonies and derive the size of the colonies.
In Vivo Tumorigenesis Assay.
Mahlavu and SK-Hep1 cells (5 × 106) infected with shLuc, lenti-122, or lenti-shADAM17 lentiviruses were implanted subcutaneously into the flanks of nude mice. Primary tumor growth was analyzed by measuring tumor length (L) and width (W), and tumor volume was calculated according to V = 0.4×ab2 as described.23 Orthotopical intrahepatic injection was conducted as described27 by using 20 μL of phosphate-buffered saline containing 50% Matrigel (BD Biosciences, MA); mixed with 5 × 106 tumor cells, and slowly injected into the livers of anesthetized mice using a 27-gauge needle. Mice were sacrificed 6-7 weeks later for histopathological evaluation. The methods used for the immunohistochemical staining of anti-CD31, the evaluation of microvessel density, and the quantification of tumor size are described in Supporting Methods. Only the results using Mahlavu cells are presented. The animal studies were approved by Institutional Animal Care and Use Committee (IACUC) of National Yang Ming University.
Statistical Analyses.
All data are expressed as mean ± standard deviation and compared between groups using the Student t test. Categorical variables were compared using the Wilcoxon Rank test. A P value <0.05 was considered statistically significant. *P < 0.05; **P < 0.01; ***P < 0.001.
Results
Specific Down-Regulation of miR-122 in HCC Correlates with Enhanced Transforming Activity.
Recently, miR-122 was found to be down-regulated in liver tumors of human and rodents21, 22 and is considered a member of a metastasis-related microRNA signature.12 However, the functional attributes of miR-122 associated with liver tumor progression have not been experimentally established. To substantiate the significance of miR-122 expression, a northern blot analysis of 20 HCC tumor pairs was conducted. Expression of miR-122 was remarkably lower in T3 tumors with intrahepatic metastasis (Fig. 1A). Validation by quantitative stem-loop PCR showed that the expression of miR-122 was significantly higher in the adjacent normal tissue (n = 48) and T1 tumors (n = 25). No statistically significant difference was found between T1 tumors and adjacent normal tissue. However, significant loss of miR-122 expression was found in T3 tumors (Fig. 1B), which implies that down-regulation of miR-122 significantly correlates with intrahepatic metastasis as reported.12
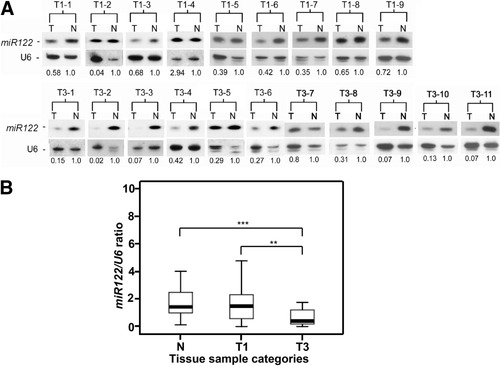
Expression of miR-122 was down-regulated in intrahepatic metastastic liver tumors. (A) Expression analysis by northern blotting. T: tumor part; N: normal adjacent tissue. The intensity of northern hybridization was retrieved using ImageQuant software. Each miR-122 intensity value was first divided by the corresponding intensity of U6 from the same tissue sample to adjust for sample variation. (B) Decreased miR-122 mRNA expression was present in an additional 48 pairs of HCC samples (T1, n = 25; T3, n = 23) and demonstrated by TaqMan MicroRNA Assay. Results were normalized against the expression level of U6 messenger RNA (mRNA) in each sample. The differences in miR-122 expression level between tissue sample categories were analyzed by Wilcoxon rank sum test. The box plot shows the data distribution across the group classification and presents the 75th and 25th percentile (upper and lower quartile) with the median value between. T1: primary HCC, T3: intrahepatic metastasis HCC; N: an average of expression level of all 48 normal adjacent tissues. **P < 0.01; ***P < 0.001.
To explore the functional role of miR-122 in HCC progression, ectopic overexpression of miR-122 was established using a lentiviral vector (lenti-122). When miR-122 was overexpressed in SK-HEP-1 or Mahlavu HCC cells (Fig. 2A), which are known to manifest invasion activity,23 a significant reduction of cell migration and invasive activity was observed (Fig. 2B). Overexpression of miR-122 also impaired anchorage-independent growth in soft agar, significantly reducing the number and size of the colonies (Fig. 2C). Consistent with the in vitro results, overexpression of miR-122 in Mahlavu cells significantly suppressed overall tumor growth and local invasion in vivo (Fig. 6). Restoration of miR-122 expression, however, did not influence HCC cell proliferation (Supporting Fig. 1). These in vitro and in vivo results imply that the gradual loss of miR-122 over a long period of time is likely to commit hepatocytes to tumorigenesis and intrahepatic metastasis.
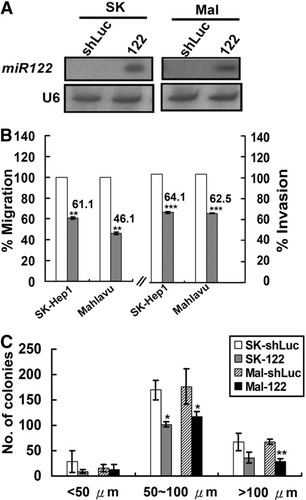
MiR-122 regulates the transforming activities of HCC cells. (A) Northern blots detected expression of mature miR-122 from cells infected with lenti-122 lentiviruses. (B) Restoration of miR-122 reduced cell migration and invasion. White bars, control cells infected with shLuc; gray bars, cells infected with lenti-122. (C) Anchorage-independent cell growth in soft agar of two HCC cell lines, SK-HEP-1 and Mahlavu, was reduced with miR-122 overexpression. Two lines of HCC cells were infected with lenti-122 or control shLuc. The Integrated Morphometry analysis function of MetaMorph was used to measure the surface areas of the colonies, which were used to derive the size and distribution of the colonies. *P < 0.05; **P < 0.01; ***P < 0.001.
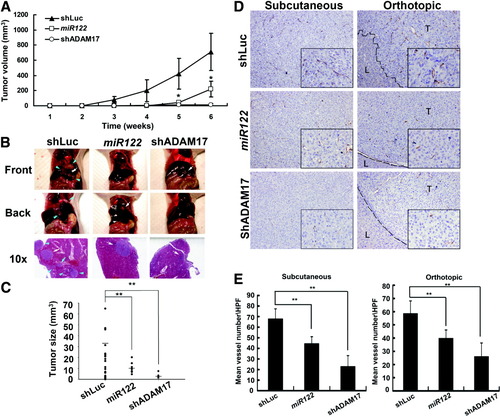
Effects of miR-122 overexpression or ADAM17-knockdown on subcutaneous tumors and orthotopic liver tumors. (A) In vivo subcutaneous tumor growth curves of shLuc-infected, miR-122-infected, or shADAM17-infected Mahlavu cells (n = 7). *P < 0.05. (B) Gross morphology of representative livers and abdominal cavity organs (upper and middle panels) as well as hematoxylin and eosin–stained images of representative tumors (lower panel) after intrahepatic injection of shLuc-infected, miR-122-infected, or shADAM17-infected Mahlavu cells. The liver tumors were examined 7 weeks after cell injection. The shLuc-infected Mahlavu cells showed extensive hepatic invasion, whereas only one tumor focus was found in the miR-122 and shADAM17 groups. The panels showed the morphology of the tumors from the injected site (white arrows) and the disseminating tumors (blue arrowheads) in the abdominal cavity in the shLuc group. It is important to note the presence of the tumor cells invading the hepatic vessels and forming small metastatic focus (blue arrow, shLuc). No metastatic tumors were found in the miR-122 group or in the shADAM17 group. Original magnification: 100×. (C) The graph shows the volumes and numbers of orthotopic and metastatic tumor foci in the livers after injecting shLuc-infected, miR-122-infected, or shADAM17-infected Mahlavu cells (n = 7, 31 tumor foci in shLuc group, seven foci in miR-122 group, and three foci in shADAM17 group). The lines show the average tumor sizes. Note that there are significant reductions in the tumor sizes when the experiment is carried out with miR-122-infected or shADAM17-infected Mahlavu cells compared to the shLuc control. The average volumes of the original orthotopic and metastatic tumors of the shLuc-treated group are significantly higher than those of the miR-122 and shADAM17-treated groups (35.5 ± 17.2 versus 10.7 ± 5.3 and 3.8 ± 2.6 mm3, respectively; P < 0.01). (**P < 0.01). (D) Blood vessel distributions of the subcutaneous and orthotopic tumors as revealed by immunohistochemistry staining using CD31 antibody. The left panel shows the vessel distributions in the subcutaneous tumors caused by shLuc-infected, miR-122-infected, and shADAM17-infected Mahlavu cells. Right panel shows the vessel distributions in the orthotopic tumors. The dotted lines show the edges of normal liver area (L) and tumor area (T). Note that the shLuc tumor has invasive edges while the miR-122 and shADAM17 tumors show smooth edges. The black frame indicates a representative 400× magnification field. The original magnification is 200×. (E) Bar chart shows comparisons of the mean CD31-positive vessel number per high-power field of the various tumor sections. Significant reductions in the number of CD31-positive blood vessels was found for the miR-122 and shADAM17 groups compared to shLuc control group when the subcutaneous tumors were examined (n = 20, 44.5 ± 6.5, 22.8 ± 10.5 versus 67.7 ± 9.7, P < 0.01, left panel). Similar results were found for the orthotopic tumors (n = 20, 39.7 ± 10.5, 26.0 ± 6.0 versus 58.5 ± 9.8, right panel). **P < 0.01.
Identification of miR-122 Target Genes.
MiR-122 regulates multiple target genes whose combined expression is associated with the metabolism of hepatic fatty acid and cholesterol as well as HCC pathogenesis.4, 17, 21 To identify putative metastasis genes that miR-122 suppresses, we integrated various computational prediction tools to track down genes where expression was increased by >1.5-fold in T3 HCC and in antago-miR-122–treated mouse livers. The salient feature common to these two types of liver tissues is that they both have extremely low levels of endogenous miR-122. In addition, the genes needed to have a putative miR-122 binding site mapped to their 3′UTR. A list of 45 most probable targets, including the known genes SLC7A120 and ALDOA,4 was compiled (Table 1). A reciprocal expression pattern between miR-122 and the candidate genes was identified by microarray (Supporting Table 1) and by RT-PCR (Supporting Fig. 2), which supports a role in their posttranscriptional suppression by miR-122.
| Gene Symbol | Human | Mouse | Molecular Function | No. Binding Sites | ||||
|---|---|---|---|---|---|---|---|---|
| T3/T1 | Sc-H | MFE-H | an/c | Sc-H | MFE-M | |||
| NUMBL | 3.1 | 181 | −20.5 | 4.5 | 131 | −14.5 | Protein binding | 8 |
| FOXJ3 | 1.6 | 176 | −21 | 1.7 | 133 | −10.6 | Transcription factor | 6 |
| XPO6 | 2.2 | 174 | −22.3 | 1.7 | 125 | −16.4 | Nuclear protein transport | 2 |
| SLC7A1 | 2.6 | 173 | −22.4 | 1.7 | 173 | −23.6 | Amino acid transport | 16 |
| STX6 | 1.6 | 169 | −20.9 | 2.5 | 135 | −17.2 | Protein transport | 5 |
| AP3M2 | 1.6 | 168 | −19.9 | 3.0 | 129 | −15.9 | Protein trafficking | 3 |
| G6PC3 | 2.1 | 167 | −18.4 | 6.1 | 158 | −13.1 | Glucose-6-phosphatase | 1 |
| GALNT10 | 1.6 | 167 | −20.4 | 1.9 | 152 | −20.5 | Calcium binding | 8 |
| RIPK5 | 1.8 | 166 | −18 | 1.6 | 150 | −16.6 | Kinase activity | 4 |
| TPD52L2 | 3.0 | 166 | −23.8 | 16.3 | 140 | −16.5 | Cell proliferation | 11 |
| AKT3 | 2.0 | 165 | −18.7 | 2.1 | 162 | −15.6 | Cell proliferation, apoptosis | 3 |
| FUNDC2 | 2.5 | 165 | −18 | 2.1 | 161 | −15.8 | HCV core binding protein | 4 |
| MAPK11 | 3.2 | 165 | −20.2 | 1.6 | 123 | −19.1 | MAPK activity | 2 |
| ALS2CR13 | 2.0 | 162 | −17.3 | 6.7 | 163 | −17 | Unknown | 7 |
| BACH2 | 1.8 | 162 | −15 | 2.7 | 140 | −13.5 | Transcription factor | 11 |
| ATP11A | 2.0 | 161 | −12.9 | 1.9 | 162 | −18.1 | Transport ions | 9 |
| SORT1 | 2.2 | 161 | −16.2 | 2.0 | 154 | −15.3 | Cell differentiation | 7 |
| ATP1A2 | 3.5 | 160 | −17 | 1.5 | 132 | −11.7 | Ion concentration balance | 7 |
| ADAM17 | 1.7 | 159 | −19 | 2.0 | 122 | −17 | Cell cell interaction | 1 |
| DUSP2 | 1.8 | 159 | −13.4 | 1.6 | 123 | −11.1 | MAPK phosphatase | 2 |
| OSMR | 2.5 | 159 | −15.8 | 2.9 | 142 | −15.5 | Cell proliferation | 4 |
| RABIF | 1.7 | 159 | −20.1 | 2.4 | 139 | −15.1 | Small GPT regulator activity | 2 |
| PALM | 2.3 | 156 | −15.9 | 16.1 | 161 | −18.7 | Cell mobility and cell shape | 3 |
| SPRED1 | 2.2 | 156 | −21.2 | 1.6 | 145 | −19.8 | Activate MAPK kinase | 1 |
| AACS | 1.8 | 155 | −13.8 | 1.8 | 163 | −17.8 | Isoprenoid biogenesis | 2 |
| TBX19 | 2.1 | 155 | −17.2 | 4.3 | 131 | −13.1 | Transcription factor | 5 |
| UBAP2 | 2.2 | 155 | −18.8 | 2.7 | 131 | −23.3 | Ubiquitin associated protein | 2 |
| EGLN3 | 3.9 | 154 | −17 | 1.6 | 137 | −15.7 | Apoptosis | 1 |
| NCAM1 | 1.9 | 154 | −15.2 | 3.8 | 154 | −15.2 | Cell differentiation | 4 |
| MECP2 | 2.2 | 153 | −12.1 | 3.1 | 160 | −26.2 | Transcription | 18 |
| CS | 1.6 | 152 | −12.6 | 1.8 | 148 | −11.9 | Catalyzes citrate synthesis | 3 |
| FOXP1 | 1.6 | 152 | −10.8 | 1.8 | 167 | −17.7 | Transcription factor | 7 |
| RAB11FIP1 | 1.5 | 152 | −20.1 | 1.9 | 133 | −13.3 | Protein transport | 5 |
| RAB6B | 1.5 | 152 | −13.3 | 1.6 | 164 | −19.6 | GTPase | 13 |
| TRIB1 | 2.1 | 152 | −17.4 | 1.8 | 134 | −12.8 | Kinase activity | 4 |
| TTYH3 | 1.8 | 152 | −17.1 | 1.9 | 160 | −16.6 | Chloride anion channel | 5 |
| ALDOA | 3.7 | 151 | −13.2 | 2.2 | 157 | −16.7 | Amino acid transport | 2 |
| ANXA11 | 2.0 | 151 | −18.6 | 4.3 | 139 | −17.2 | Calcium binding | 11 |
| CLDN18 | 1.7 | 151 | −18.5 | 1.9 | 130 | −15.1 | Cell and cell adhesion | 4 |
| ENTPD4 | 2.0 | 151 | −18.5 | 2.0 | 131 | −13.5 | Calcium binding | 3 |
| NFATC2IP | 2.7 | 151 | −19.9 | 2.2 | 135 | −15.2 | Protein modification | 4 |
| ANK2 | 2.1 | 150 | −12 | 3.1 | 137 | −17.9 | Cell proliferation | 5 |
| MEP1A | 4.3 | 150 | −17.8 | 1.6 | 132 | −12.8 | Peptidase | 3 |
| NFATC1 | 1.8 | 150 | −13.1 | 2.3 | 148 | −14.9 | Transcription factor | 6 |
| SLC7A11 | 9.5 | 150 | −19.5 | 1.6 | 164 | −14.4 | Amino acid transport | 4 |
- The relative gene expression level detected by microarray assay is shown as the mean ratios of T3/T1 for HCC samples and an/c for mice treated with antagomiR-122 (an) compared to the control mm-antagomiR122 (c). The criteria used for target selection are the prediction score of “seed match” (Sc) and minimum free energy (MFE). Scores for the human and mouse genes are labeled as Sc-H and Sc-M, respectively. MFE values for the human and mouse genes are labeled as MFE-H and MFE-M, respectively. Gene information is provided with Gene Symbol and Molecular Function. “No. Binding Sites” indicates number of putative miR-122 binding sites in the 3′UTR region of the putative target genes. Positions of the predicted binding sites are listed in Supporting Table 3.
Bioinformatic prediction recognized one to seven miR-122 binding sites in the 3′UTRs of these candidate genes (Table 1, Supporting Table 3). To test whether the predicted binding sites in the target genes could mediate repression of translation by miR-122, the 3′UTRs were subcloned downstream of the luciferase gene in pGL3-Control. These constructs were assayed in HEK293T cells infected with lenti-122 or a mutant miR-122 in which the seed region has been randomly mutated (miR122-M; Fig. 3A). As shown in Fig. 3A, the reporter with the 3′UTR of SLC7A1 showed a significantly lower luciferase activity in cells expressing miR-122 compared to miR122-M. Similar responses were detected for the reporter with the 3′UTR of AKT3. MiR-122 thus negatively regulates translation of these genes through the 3′UTR binding sites, and SLC7A1 and AKT3 are true miR-122 target genes. A significant reduction in luciferase activity (14%–54%) was detected with 32 of the 35 cloned reporter constructs (Fig. 3B). STX6 (syntaxin 6), SORT-1 (sortilin-1), and SPRED-1 (sprouty-related, EVH1 domain containing-1) failed to show a significant reduction in luciferase activity. No specific repression by miR122-M was detected (Supporting Tables 4 and 5). These 32 genes are therefore direct targets of endogenous miR-122.
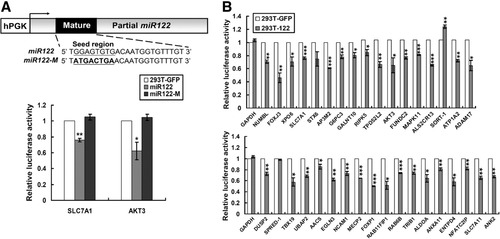
Confirmation of miR-122 target genes. (A) Diagram depicting the 3′UTR reporter assay. The miR-122 mutant (miR-122M) has the seed region mutated (upper panel). The specificity of the reporter assay was examined with parental miR-122 and miR122-M targeting the reporters for SLC7A1 and AKT3 (lower panel). The 3′UTR reporter assay was carried out in 293T cells overexpressing miR-122 or miR122-M. Lenti-GFP served as the negative control (293T-GFP). Cells were cotransfected with the pGL3-3′UTR construct and pRL-TK vector (Promega) using jetPEI. Luciferase assays were performed 48 hours after transfection using the Dual-Luciferase Reporter Assay System (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity. The effect of miR-122 was expressed relative to the average value from cells infected with the lenti-GFP viruses. (B) Overall, 32 of the 35 3′UTR constructs demonstrated a significant reduction in luciferase activity for 293T cells overexpressing miR-122 (293T-122).
To uncover the biological functions of the confirmed miR-122 targets, gene function analysis was carried out (Ingenuity Pathways Analysis 3.0; Ingenuity Systems, Redwood City, CA). Genes classified as having the molecular functions involved in “Cell Morphology” or “Cell Movement” (Supporting Fig. 3) were identified. The overlap between these two functions pinpointed three genes, ADAM17, MAPK11 (mitogen-activated protein kinase 11), and NCAM1 (neural cell adhesion molecule 1). ADAM17 plays a crucial role in the shedding of receptor ligands, such as certain epidermal growth factors (EGFs) and tumor necrosis factor alpha (TNFα), which are the major ligands for several signal transduction pathways.28 Previous reports have shown overexpression of ADAM17 in breast cancer,29 brain tumors,30 and non–small cell lung cancer,31 where the gene contributes to tumor invasiveness and metastasis. Although elevated ADAM17 expression has been identified as a pathological feature of HCC,32, 33 its function has not been determined. Therefore, we evaluated the functional role of ADAM17 in HCC tumorigenesis and progression.
ADAM17 Is a Critical Downstream Target of miR-122.
ADAM17 has one 3′UTR high-scoring binding site. The fact that expression from the 3′UTR reporter corresponding to the parental ADAM17 sequences (but not from one with a mutated binding sequences, ADAM17-M) had significantly suppressed luciferase activity in miR-122 transfected 293T cells confirmed the authenticity of this binding site (Fig. 4B). The knockdown of ADAM17 in SK-HEP-1 and Mahlavu cells (Fig. 4C) showed significant reduction in cell migration and invasion (Fig. 4D) as well as lower anchorage-independent growth in soft agar (Fig. 4E); this is similar to the phenotype detected with miR-122 restoration in these two lines of HCC cells (Fig. 2B,C). However, one difference from miR-122 restoration was that shADAM17 reduced the proliferation of Mahlavu cells (Supporting Fig. 4)
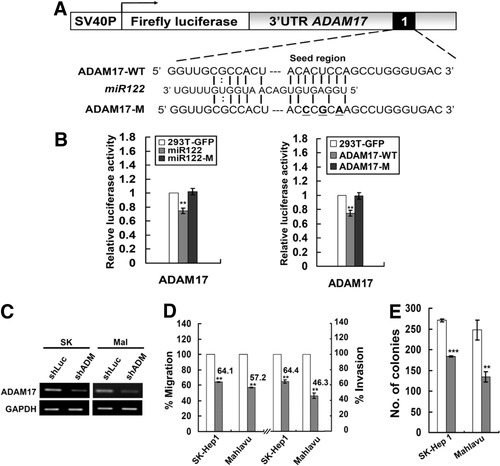
ADAM17 is a miR-122 target and regulates the transforming activities of HCC cells. (A) Diagram showing the miR-122 binding site mutant of ADAM17. (B) Verification of the binding site specificity. Reduction of luciferase activity driven by ADAM17-3′UTR construct was observed in cells expressing wild-type miR-122 (293T-122) but not in cells overexpressing mutant miR-122 (293T–122M) (left panel). Reporter constructs containing ADAM17-M failed to suppress the luciferase activity (right panel). (C) Suppression of ADAM17 mRNA expression by RNA interference. ADAM17 knockdown resulted in a reduction in (D) cell migration (left panel) and invasion (right panel) activity as well as (E) anchorage-independent growth in soft agar in two HCC cell lines. Cells were infected with either control shLuc (white bar) or shADAM17 (gray bar) for 24 hours, which was followed by 2-day puromycin (2 μg/mL) selection before the cells were plated for the migration, invasion assays, or soft-agar assay. Data are an average of triplicates for each condition. **P < 0.01; ***P < 0.001.
Moreover, overexpression of miR-122 or knockdown of ADAM17 brought about cell morphology changes and actin disorganization as shown by the confocal images of cells stained with the F-actin marker phalloidin (Fig. 5, Supporting Methods). Phalloidin-stained miR-122-Mahlavu cells showed a severe reduction of stress fibers (middle panel, Fig. 5A), which is similar to that of SK-N-SH cells treated with RhoA14 and forskolin.34 Surprisingly, there was an overwhelming reduction of membrane protrusions, and this was accompanied by a contracted cell shape in shADAM17-Mahlavu cells (right panel, Fig. 5A). Restoration of miR-122 impacted on the epithelial-mesenchymal transition as shown by a reduction in vimentin, a mesenchymal marker (Fig. 5B). Cell motility requires the formation of membrane protrusions such as lamellipodia or filapodia, a process that revolves around actin dynamics under the control of Rho–guanosine triphosphatases.35 Whether the restoration of miR-122 or the knockdown of ADAM17 disturbed the equilibrium of the Rho–guanosine triphosphatases remains to be elucidated.
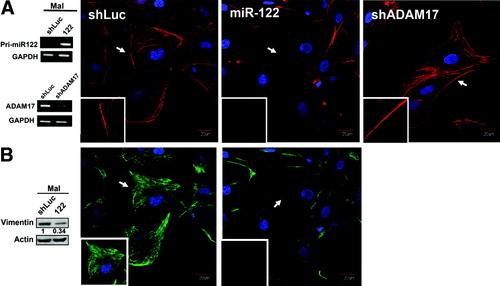
Restoration of miR-122 or suppression of ADAM17 expression affected the cytoskeleton of HCC cells. (A) Loss of actin filament was found in miR-122–overexpressing Malhlavu cells (miR-122), while a severe reduction in membrane protrusions was observed after ADAM17 knockdown (shADAM17). Inset: higher magnification view of the F-actin and the membrane protrusions. MiR-122 overexpression and ADAM17-knockdown were confirmed by RT-PCR (left panel). (B) Restoration of miR-122 resulted in differential expression of vimentin. As shown by western blot (left) and immunofluorescence (right), a significant loss of vimentin was detected upon miR-122 expression. shADAM17 did not affect expression of vimentin. Images were taken 5-7 days after infection. Cells were fixed and stained with rhodamine-phalloidin, DAPI (4′,6-diamidino-2-phenylindole), or anti-vimentin to detect F-actin, the nucleus, and vimentin, respectively. Bar, 20 μm. Original magnification: 60×.
Overexpression of miR-122 or Knockdown of ADAM17 Suppresses In Situ Tumor Formation and Metastasis.
To determine whether miR-122 and its target gene are associated with tumorgenesis or metastasis, Mahlavu cells were infected with shLuc, miR-122, or shADAM17 lentiviruses and implanted by either the subcutaneous route into the flanks of nude mice or orthotopically into the livers of nude mice. Restoration of miR-122 or silencing of ADAM17 significantly suppressed overall tumor growth, as assessed by tumor volume (Fig. 6A). Moreover, the orthotopic tumor model yielded a similar tumor growth trend in the original injection site. ShLuc-infected Mahlavu cells showed extensive hepatic invasion with a larger tumor volume; this contrasts with single smaller tumor focus that was found in the miR-122 and shADAM17 groups (Fig. 6B,C). The tumor growth reduction and metastasis shutdown may be speculated to be the results of angiogenesis inhibition. To confirm this speculation, subcutaneous and orthotopic tumors were subjected to neoangiogenesis analysis by staining with anti-CD31 antibody (Fig. 6D). Quantitative analysis showed that there was significant reductions in the blood vessels present in the miR-122 and shADAM17 groups compared to shLuc control for the subcutaneous tumors (Fig. 6E, left panel) and for the orthotopic tumors (Fig. 6E, right panel). Thus, miR-122 would seem to regulate tumorigenesis and intrahepatic metastasis via angiogenesis inhibition at least in part through its target ADAM17.
Discussion
Recent work has started to shed light on the importance of miRNA in hepatocarcinogensis. Deregulation of liver-specific miR-122 and a linkage to liver diseases has attracted intense interest; nonetheless, experimental affirmation of its role in liver pathology is lacking. In this study, we demonstrated that miR-122 acts as a tumor suppressor gene for hepatocarcinogenesis, particularly intrahepatic metastasis. Restoration of miR-122 has some impact on the epithelial-mesenchymal transition as shown by reduction in vimentin, a mesenchymal marker (Fig. 5B). Related to this, reduced in vitro cell migration and invasion, in vivo tumor growth, and local invasion can be detected upon restoration of miR-122. We are intrigued to find that restoration of miR-122 in highly migratory HCC cells neither triggered apoptosis nor inhibited in vitro cell proliferation (Supporting Fig. 1); instead, it led to a significant inhibition of angiogenesis (Fig. 6D,E).
Similar to a classical transcription factor, microRNAs exert their activity via regulation of their target genes. We report here 32 direct target genes of miR-122. This is the first time that 32 empirically verified target genes have been discovered in a single study. We attributed our success in target prediction to several tactics. First, in accordance with the consensus of posttranscriptional suppression by miRNAs, we correlated expression of miR-122 with its candidate targets from two microarray datasets constructed from mouse and human livers known to be low in endogenous miR-122 (Table 1). This complies with the species conservation principle and, as a result, greater confidence can be given to each target gene identified. Second, the use of both a high miRanda score and low miRanda minimum free energy was able to pinpoint most probable targets.36, 37 Finally, the false-positives from the prediction were eliminated by differential validation using both a wild-type and a “seed region” mutant of miR-122 via the 3′UTR reporter assay (Fig. 3B, Supporting Tables 4 and 5).
This set of 32 target genes is unique among those reported to-date. Lack of concordance may be attributed to several variables. The sources of liver tissues, either normal livers in which miR-122 was acutely depleted by an antisense approach or cancerous livers in which there is protracted down-regulation of miR-122 that has occurred naturally,12, 21 are likely to present different target gene pools. Many of the most affected genes from the studies using a loss-of-function approach involved hepatic lipid metabolism.4, 17, 38 In contrast, the molecular functions of the miR-122 targets presented here are affiliated with transcription regulation, amino acid transport, protein trafficking, and cell signaling (Table 1, Supporting Fig. 3). To unravel the biological role of miR-122 in HCC metastasis, ADAM17 was identified as a critical downstream target. Knockdown of ADAM17 in migratory HCC cells suppressed the in vitro proliferation, migration, invasion, and anchorage-independent growth of these HCC cells (Fig. 4D,E); this was in addition to suppressing in vivo tumor growth, angiogenesis, and local invasion in the liver milieu (Fig. 6). The collective results from RNA interference substantiate that ADAM17 is a bona fide oncogene involved in liver cancer. Up-regulation of ADAM17 has been detected in a wide variety of human cancers including HCC28, 32, 33 and colorectal liver metastasis.39 ADAM17 facilitates cleavage-activation of TNFα40 and selected ligands of EGF receptor (EGFR)41 as well as modulating integrin signaling during cell adhesion and migration.42, 43 In addition, coexpression of activated EGFR and ADAM17 has been observed in the neoplastic and endothelial cells from colon cancers,44 suggesting a role in angiogenesis. We postulate that ADAM17 is a likely component of the tumor stroma that facilitates angiogenesis and tissue remodeling during HCC progression. Further studies in these areas will be required.
We also noticed that the outcome of ADAM17 knockdown is reminiscent of the restoration of miR-122, both in vitro and in vivo, but that ADAM17 knockdown shows a stronger intensity; this may be explained because the net effect of miR-122 restoration represents an overall response from all target genes. MicroRNAs are pivotal modulators of gene expression and yet their involvement as effectors of cell signaling is largely unknown and is a matter of intense study at present. Our preliminary results showed that restoration of miR-122 greatly reduces activation of focal adhesion kinase (FAK), v-raf-1 murine leukemia viral oncogene homolog (Raf), mitogen-activated protein kinase(MEK), and extracellular-signal-regulated kinase (ERK) (Supporting Fig. 6). This finding provides clues as to how miR-122–mediated alteration of actin cytoskeleton occurs, but leaves open the question as to whether the expression of miR-122 controls hepatocyte differentiation. ADAM17 is probably not the only target of miR-122 that is relevant to tumorigenesis and metastasis. Identification of forkhead box P113 and SRY (sex determining region Y)-box 423 (unpublished data), two transcription factors known to regulate hepatocarcinogenesis and HCC metastasis, as the direct targets of miR-122, suggests a coordinated regulatory program involving miRNA and transcription factors during liver carcinogenesis. The input of the other targets needs to be evaluated in order to fully understand how miR-122 controls metastasis.
In summary, we have identified a miR-122–ADAM17 link that is a novel constituent in hepatocarcinogenesis. Recently, it has been shown by Zhou et al. that a selective inhibitor of ADAM17 effectively prevents activation of multiple EGFR ligands and suppresses resistance to the EGFR inhibitor gefitinib in non–small cell lung cancer.31 Moreover, amphiregulin, a preferred substrate of the ADAM17 sheddase, has been shown to contribute to the transformed phenotype of HCC cells33; thus, ADAM17 is a potential modulator of HCC cell proliferation. It will be necessary to gain a better understanding of whether miR-122 overexpression disrupts EGFR signaling by way of ADAM17 modulation. Although the efficacy of EGFR-targeted chemotherapies in HCC is less than satisfactory,45 restoration of miR-122 might have more far-reaching effects. With the concomitant down-regulation of its targets, including ADAM17, a rational therapeutic strategy based on miR-122 may be beneficial to patients with HCC.
Acknowledgements
We thank Drs. C. K. Chou and W. C. Lin for their invaluable comments, the Microarray and Gene Expression Analysis Core Facility of the VGH National Yang-Ming University Genome Research Center (VYMGC) for their technical support and the National RNAi Core Facility for providing the RNAi reagents.