Identification of metastasis-related microRNAs in hepatocellular carcinoma†
Potential conflict of interest: Nothing to report.
Abstract
MicroRNAs (miRNAs) have been used as cancer-related biomarkers. Hepatocellular carcinoma (HCC) is an aggressive cancer with a dismal outcome largely due to metastasis and postsurgical recurrence. We investigated whether the expression of certain miRNAs are associated with HCC metastasis. We examined the miRNA expression profiles of 482 cancerous and noncancerous specimens from radical resection of 241 patients with HCC. Using a supervised algorithm and a clinically well-defined cohort of 131 cases, we built a unique 20-miRNA metastasis signature that could significantly predict (P < 0.001) primary HCC tissues with venous metastases from metastasis-free solitary tumors with 10-fold cross-validation. However, significant miRNAs could not be identified from the corresponding noncancerous hepatic tissues. A survival risk prediction analysis revealed that a majority of the metastasis-related miRNAs were associated with survival. Furthermore, the 20-miRNA tumor signature was validated in 110 additional cases as a significant independent predictor of survival (P = 0.009) and was significantly associated with both survival and relapse in 89 cases of early stage HCC (P = 0.022 and 0.002, respectively). These 20 miRNAs may provide a simple profiling method to assist in identifying patients with HCC who are likely to develop metastases/recurrence. In addition, functional analysis of these miRNAs may enhance our biological understanding of HCC metastasis. (HEPATOLOGY 2008.)
Hepatocellular carcinoma (HCC) represents an extremely poor prognostic cancer that remains one of the most common and aggressive human malignancies worldwide.1, 2 The dismal outcome has been attributed to the major hallmarks of HCC, intrahepatic metastases or postsurgical recurrence. New tumor colonies frequently invade into the major branches of the portal vein and possibly other parts of the liver.3-6 Resection or liver transplantation are the best options for a potential cure; however, only about 10%–20% of patients with HCC, defined by parameters of relatively normal liver function and a manageable tumor lesion as determined by the available clinical staging systems, are currently eligible for surgical intervention. Moreover, patients who were resected often have a high frequency of metastasis/recurrence, and postoperative 5-year survival is only 30%–40%.
Metastasis is a complex process that involves multiple alterations.7, 8 Our understanding of such complexity has been improved by the advent of global microarray technology which allows for the molecular profiling of changes in gene expression that are associated with particular phenotypes, such as metastasis. In fact, several array-based metastasis markers have also been demonstrated to be useful as prognostic molecular biomarkers, potentially offering additional tools for advanced diagnosis of HCC. For example, using complementary DNA (cDNA) microarray technology, we developed a unique tumor messenger RNA (mRNA) gene expression signature to predict prognosis and metastasis in patients with HCC and identified osteopontin as a critical player in HCC metastasis.9, 10 The presence of a molecular prognostic mRNA signature in primary HCC clinical specimens was confirmed by several recent studies.11, 12 Because HCC is usually present in inflamed liver, we also developed a unique predictor based on the expression of mRNA genes in the liver microenvironment of patients with HCC, which was principally different from that of the tumor.13 Like many other prognostic signatures based on mRNA gene expression profiling, both the tumor and microenvironment signatures contain several hundred cellular coding genes. Therefore, it would be a challenging task to identify relevant biomarkers or potential pharmacological targets and interrogate scores of genes in clinical practice.
Recent studies indicate that expression profiling with small, noncoding RNA gene products (∼22 nucleotides) known as microRNAs (miRNAs) is a superior method for cancer subtype classification and prognostication.14-16 MicroRNAs exist in many organisms and play key regulatory roles in mRNA translation and degradation by base pairing to partially complementary sites of the mRNA, predominantly in the 3′-untranslated region.17-19 The miRNAs are expressed as long precursor RNAs that are processed by Drosha, a cellular nuclease, and subsequently transported to the cytoplasm by an Exportin-5–dependent mechanism.20, 21 The miRNAs are then cleaved by the DICER enzyme, resulting in mature miRNAs of ∼17–24 nucleotides in length that associate with a RNA-induced silencing–like complex.22, 23 The expression patterns, function and regulation of miRNAs in normal and neoplastic human cells are largely unknown but emerging data and their frequent location at fragile sites, common break-points or regions of amplification or loss of heterozygosity reveal that they may play significant roles in human carcinogenesis. The abnormal expression of several miRNAs have been observed in Burkitt's lymphomas, B cell chronic lymphocytic leukemia (CLL) and in many solid cancer types, including breast, liver, lung, ovarian, cervical, colorectal, and prostate cancer.14, 15, 24-31 Functional analysis has revealed the down-regulation of PTEN (phosphatase and tensin homolog) by miR-21, the tumor suppressor function of the let-7 family and the oncogenic function of the miR17-92 cluster.32-34 The biological and clinical relevance of miRNA expression patterns have been established in human B cell CLL and solid tumors, including breast cancers.15, 27, 35 Each miRNA has a distinct capability to potentially regulate the expression of hundreds of coding genes and thereby modulate several cellular pathways including proliferation, apoptosis and stress response.36 In addition, mature miRNAs are relatively stable. These phenomena make miRNAs superior molecular markers and targets for interrogation and as such, miRNA expression profiling can be used as a tool for cancer diagnosis.14, 37 In addition, the study of miRNAs is advantageous in improving our understanding of the mechanisms of cancer progression.
In this study we investigated the miRNA expression profile of HCC specimens from radical resection. We identified 20 miRNAs that are associated with HCC venous metastasis. In contrast to HCC staging systems, this 20-miRNA-based signature was capable of predicting survival and recurrence of HCC in patients with multinodular or solitary tumors, including those with early-stage disease. Moreover, this signature was an independent and significant predictor of patient prognosis when compared to other available clinical parameters. Our study suggests that these 20 miRNAs can assist in HCC prognosis and may have clinical utility for the advanced identification of patients with HCC with a propensity towards metastasis. Functional studies of these miRNAs may also help to elucidate the mechanism(s) leading to HCC metastasis.
Abbreviations
cDNA, complementary DNA; HCC, hepatocellular carcinoma; miRNA, microRNA; mRNA, messenger RNA.
Patients and Methods
Clinical Specimens.
Hepatic tissues were obtained with informed consent from patients who underwent radical resection between 2002 and 2003 at the Liver Cancer Institute and Zhongshan Hospital (Fudan University, Shanghai, China). The study was approved by the Institutional Review Board of the Liver Cancer Institute and the US National Institutes of Health. Gene expression profiles were conducted in primary HCC and corresponding noncancerous hepatic fresh frozen tissues from 241 Chinese patients with HCC. Among them, 93% had underlying cirrhosis and 68% had a serum alpha-fetoprotein level > 20 ng/mL (Table 1). The sample inclusion criteria are included in the supplemental text. The general strategy for partitioning cases and testing the miRNA signature is outlined in Fig. 1. A total of 131 metastasis or nonmetastasis cases were used to build a miRNA signature of metastasis (Fig. 1, Step 1). These cases were associated with clear outcomes (that is, those with accompanying metastasis at surgery and those without metastasis and recurrence at surgery and at 3-year follow-up). Among them, 29 had primary HCC lesions (19 solitary and 10 multinodular) accompanied by tumor emboli found in the major branches of the portal vein (n = 23), inferior vena cava (n = 2), or common bile duct (n = 4; one also with tumor thrombi in the inferior vena cava) and 102 had solitary HCC with no metastasis/recurrence found at follow-up (3 years). The median overall survival of cases with metastasis was 349 days whereas that of the nonmetastasis cases was 664 days. In the validation analysis, we used the remaining 110 independent cases (Fig. 1, Step 3). This group appeared to consist of cases whose prognosis could not be accurately determined at the time of resection by several HCC staging systems. The 110 cases included 43 multinodular and 67 solitary HCC. Of the 43 multinodular HCC cases, 18 developed intrahepatic recurrence and one developed extrahepatic metastasis in addition to an intrahepatic recurrence. Of the 67 solitary HCC cases, 4 patients had a solitary tumor with an appearance of aggregated nodules, 10 developed intrahepatic and/or extrahepatic metastases whereas 49 developed intrahepatic recurrence confirmed at follow-up (3 years). In patients with multinodular tumors, the largest tumor was used for miRNA profiling.
| Clinical Variable | Entire cohort (n = 241) Valuea | Training Set (n = 131) Value | Testing Set (n = 110) Value | P valueb (Train vs Test) |
|---|---|---|---|---|
| Gender | ||||
| Male | 211 (88) | 110 (84) | 101 (92) | |
| Female | 30 | 21 | 9 | 0.066 |
| Age–yr | ||||
| Median | 50 | 50 | 50 | |
| Range | 13–83 | 21–83 | 13–73 | 0.8735c |
| ALTd | ||||
| normal | 142 (59) | 82 (63) | 48 (44) | |
| abnormal | 99 (41) | 59 (45) | 51 (46) | 0.126 |
| Tumor size–cm | ||||
| ≤ 3 | 110 (46) | 62 (47) | 48 (44) | |
| >3 | 131 (54) | 69 (53) | 62 (56) | 0.823 |
| Multinodular | ||||
| No | 189 (78) | 121 (92) | 68 (62) | |
| Yes | 52 (22) | 10 (8) | 42 (38) | <0.0001 |
| Child-Pugh Class | ||||
| A | 231 (96) | 122 (93) | 109 (99) | |
| B | 10 (4) | 9 (7) | 1 (<1) | 0.021 |
| Portal Vein Tumor Thrombi | ||||
| No | 210 (87) | 102 (78) | 108 (98) | |
| Yes | 29 (12) | 29 (22) | 0 | <0.0001 |
| No data | 2 (<1) | 0 | 2 (<2) | |
| OKUDA stage | ||||
| 0 | 205 (85) | 106 (81) | 99 (90) | |
| 1 | 35 (14) | 24 (18) | 11 (10) | 0.064 |
| 2 | 9 (<1) | 1 (<1) | 0 | |
| CLIP stage | ||||
| 0 | 106 (44) | 61 (47) | 45 (41) | |
| 1 | 83 (34) | 43 (33) | 40 (36) | |
| >2 | 52 (22) | 27 (21) | 25 (23) | 0.002 |
| BCLC stage | ||||
| 0 | 26 (11) | 20 (15) | 6 (5) | |
| A | 160 (66) | 86 (66) | 74 (67) | |
| B | 27 (11) | 0 | 27 (25) | |
| C | 28 (12) | 25 (19) | 3(3) | <0.0001 |
| TNM Stage | ||||
| I | 99 (41) | 66 (50) | 33 (30) | |
| II | 87 (36) | 34 (26) | 53 (48) | |
| III | 55 (23) | 31 (24) | 24 (22) | 0.001 |
| Median Overall Survival Days | 590 | 581 | 0.0162 | |
| Median Disease-Free Survival Days | 566 | 359 | <0.0001 |
- NOTE. Bold indicates significant values.
- a Each value represents the number of patients (the % of patients).
- b χ2 test.
- c Unpaired student t test.
- d Normal: ≤50 (U/L); Abnormal: >50 (U/L).
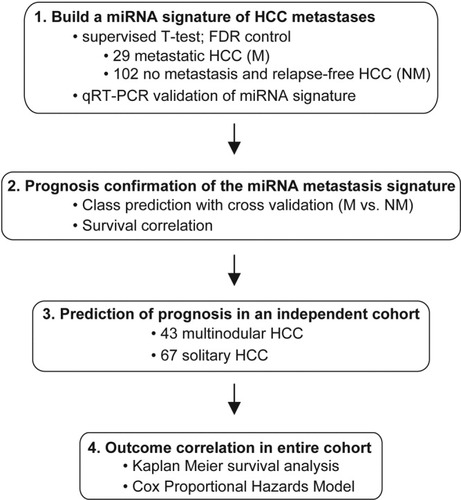
Schematic of the search for metastasis-related miRNAs and their association with HCC prognosis.
RNA Isolation,miRNA Arrays, and qRT-PCR.
The RNA isolation and miRNA array methodology were essentially as described.9, 14 In the analysis of the 241 HCC cases, RNA was isolated in a pairwise fashion from tumor or nontumor tissue and samples were selected in random order for miRNA analysis to avoid grouping bias. The tumor and nontumor tissues were run separately on a total of 482 single-channel microarrays (see Supplementary data). The microarray platform and data have been submitted to the Gene Expression Omnibus (GEO) public database at the National Center for Biotechnology Information, following MIAME guidelines. The accession numbers are GPL4700 (platform) and GSE6857 (samples; release date January 2008). In addition, REMARK guidelines have been followed to report the miRNA-based metastasis markers in this study.38 For quantitative reverse transcription polymerase chain reaction (qRT-PCR), 70 M or NM cases were randomly chosen from the 131-case training set and RNA was isolated and converted to cDNA using the High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA). Reactions were performed using ABI-purchased primer probe sets with the ABI-Prism 7700 Sequence Detector System (Applied Biosystems) (see Supplementary Methods). U6 RNA was used as a control. The statistical P value, generated by the student t-test, and the Spearman correlation constant were calculated using GraphPad Prism, version 5.
Statistical Analyses.
Unsupervised hierarchical clustering analysis was performed by the GENESIS software version 1.5 developed by Alexander Sturn (IBMT-TUG, Graz, Austria). The BRB ArrayTools software version 3.3 was used for supervised analysis to search for differentially expressed genes, as described9, 13 (see also Supplementary Methods). In the qRT-PCR-based profiling for class prediction of 70 M or NM cases utilizing the 4-miRNA signature, we used PAM (Prediction Analysis of Microarrays) developed by Tibshirani et al.39 PAM uses nearest shrunken centroids to predict unknown samples with cross-validated training and testing, which appears to be superior in performance to identify smaller sets of genes when compared to other class prediction algorithms. PAM permutation was performed using R version 2.5. The Kaplan-Meier survival analysis was used to compare patient survival based on prediction results, using Excel-based WinSTAT software. The statistical P value was generated by the Cox-Mantel log-rank test. Survival Risk Prediction analysis was performed using BRB ArrayTools software version 3.3. Cox proportional hazards regression was used to analyze the effect of fourteen clinical variables on patient survival using STATA 9.2 (College Station, TX) (see Supplementary Methods). The statistical significance was defined as p<0.05. TargetScan analysis was based on a website tool developed by Ben Lewis (http://genes.mit.edu/targetscan/index.html) (see Supplementary Methods).40 A search for experimentally proved targets of the 20 miRNAs was performed using the Tarbase database (www.diana.pcbi.upenn.edu/tarbase).
Results
The Search for Metastasis-Related miRNAs in HCC Tissues.
HCC-related mortality and recurrence could be attributed to at least 2 problems whose underlying biological activities can differ significantly, that is, metastasis and/or development of de novo HCC due to cirrhosis. Due to the limited number of HCC metastasis cases available for analysis, in a set of 241 archived HCC cases, we first searched for metastasis-related miRNAs based on the comparison between primary HCC or noncancerous tissues from all available cases with venous metastases (M; n = 29) at the time of surgery and 102 nonmetastasis cases (NM) with no evidence of metastasis at the time of surgery and after 3 year follow-up by a supervised class comparison approach (that is, the worst-prognosis and best-prognosis cases, respectively; see supplementary methods) (Fig. 1, Step 1; Table 1). We identified 20 miRNAs that could significantly discriminate the tumor tissues of M from NM cases (Fig. 2A; Table 2). It appeared that mir-219-1, mir-207, and mir-338 were most highly up-regulated, whereas mir-34a, mir-30c-1, and mir-148a were most highly down-regulated in metastasis cases. When the noncancerous tissue miRNA expression data were separately analyzed, we could not identify any miRNA capable of distinguishing M from NM at the same statistical significance. This may be a reflection of genetic make-up or epigenetic factors accounting for gene expression changes in the microenvironment versus gain or loss of a gene in the tumor or perhaps differences in the sensitivity between the mRNA and miRNA array platforms used to assay the liver microenvironment. Thus, unlike the mRNA profile associated with the HCC microenvironment,13 it appears that there are more measurable changes in miRNA expression in tumor cells compared to that of the surrounding tissue, and suggests that analysis of miRNA expression in tumor tissues may be better suited for differentiating groups of HCC patients. We note that significant miRNAs could not be identified when a comparison of these tissues was made with other clinical variables including multinodular status, microvascular invasion, and 4 clinical staging systems.
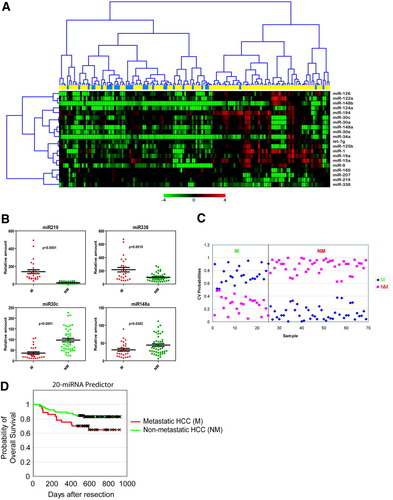
Significant differentially expressed miRNAs in metastatic versus nonmetastatic liver tissues from patients with HCC. (A) Hierarchical clustering of 20 miRNA genes whose expression was significantly (P < 0.001) altered in metastasis (M; blue bars; n = 29) and nonmetastasis samples (NM; yellow bars; n = 102) from class prediction analysis using the nearest neighbor algorithm employing 10-fold cross-validation to establish prediction accuracy. Each row represents an individual gene and each column represents an individual tissue sample. Genes were ordered by center correlation and complete linkage according to the ratios of abundance in each tissue sample compared to a normal liver tissue pool (n = 8), which were normalized to the mean abundance of genes. Pseudocolors indicate transcript levels below, equal, or above the mean (green, black, and red, respectively). The scale represents the gene expression ratios from −4 to 4 in log 2 scale. (B) qRT-PCR validation of significant differentially expressed genes. Relative expression fold of each miRNA (n = 3) normalized to U6 is shown for mir-219, mir-338, mir-30c, and mir-148a. Data are presented as the mean ± SEM, and the statistical significance calculated from the students t-test between M and NM samples is shown. (C) PAM analysis of M (n = 26; blue diamonds) and NM (n = 44; pink squares) samples used in the training set. (D) Kaplan-Meier survival analysis of metastasis and nonmetastasis samples based on prediction outcome of the 20 miRNAs.
| No | Metastasis-miRNA | Genomic Location | Parametric P-Value | Mean Intensities in M | Mean Intensities in NM | Ratio (M/NM) | Expressed in Metastatic HCC | Host targets* | Survival-miRNA |
|---|---|---|---|---|---|---|---|---|---|
| 1 | mir-338 | 17q25.3 | 0.0001 | 356 | 250 | 1.42 | up | IRF2 | mir-338 |
| 2 | mir-219-1 | 6p21.32 | 0.0002 | 578 | 391 | 1.48 | up | ADD2 | mir-219-1 |
| 3 | mir-207 | 9p21.1 | 0.0002 | 3676 | 2432 | 1.51 | up | n.a. | |
| 4 | mir-185 | 22q11.21 | 0.0009 | 461 | 346 | 1.33 | up | KCNN3 | |
| 5 | mir-30c-1 | 1p34.2 | 0.0001 | 813 | 1618 | 0.50 | down | KIAA0063 | mir-30c-1 |
| 6 | mir-1-2 | 18q11.2 | 0.0002 | 294 | 571 | 0.51 | down | G3BP2; GCLC; HAND2; TMSB4X; HDAC4; GJA1; KCNJ2 | mir-1-2 |
| 7 | mir-34a | 1p36.2 | 0.0004 | 261 | 539 | 0.48 | down | SPTBN2; E2F3, DLL1, NOTCH1 | |
| 8 | mir-19a | 13q31.3 | 0.0004 | 535 | 947 | 0.56 | down | PTEN | mir-19a |
| 9 | mir-148a | 7p15.2 | 0.0004 | 539 | 1084 | 0.50 | down | GTF2H1; PSCD3 | mir-148a |
| 10 | mir-124a-2 | 8q12.3 | 0.0004 | 236 | 448 | 0.53 | down | VAMP3; MTPN; MAPK14 | mir-124a-2 |
| 11 | mir-9-2 | 5q14.3 | 0.0005 | 197 | 347 | 0.57 | down | RAB8A; SLC20A2; VAMP3 | mir-9-2 |
| 12 | mir-148b | 12q13.13 | 0.0005 | 578 | 1063 | 0.54 | down | GTF2H1; PSCD3 | mir-148b |
| 13 | mir-122a | 18q21.31 | 0.0005 | 466 | 781 | 0.60 | down | GYS1; CAT-1 | mir-122a |
| 14 | mir-125b-2 | 21q21.1 | 0.0007 | 1346 | 2337 | 0.58 | down | ITGA9; YES1; LIN28 | mir-125b-2 |
| 15 | mir-194 | 1q41 | 0.0008 | 406 | 689 | 0.59 | down | HBEGF | mir-194 |
| 16 | mir-30a | 6q13 | 0.0008 | 2915 | 4572 | 0.64 | down | KIAA0063; VERATIN; TMEM2; THBS1, SLC7A6; PRO2730; TUBA3; CYR61; CDK6 | mir-30a |
| 17 | mir-126 | 9q34.3 | 0.0009 | 226 | 395 | 0.57 | down | n.a. | mir-126 |
| 18 | let-7g | 3p21.2 | 0.0009 | 582 | 838 | 0.69 | down | PSCD3; KRAS; NRAS | |
| 19 | mir-15a | 13q14.2 | 0.0010 | 294 | 461 | 0.64 | down | ASPH; SLC20A2; SPTBN2; DMTF1; BCL2 | mir-15a |
| 20 | mir-30e | 1p34.2 | 0.0010 | 960 | 1512 | 0.63 | down | KIAA0063 | mir-30e |
- * The experimentally proved host target genes are based on Tarbase. Potential host target genes in bold are based on TargetScan (release 3.1, November 2006) and are part of the 153-gene metastasis signature described in Ye et al., Nat Med 2003;9:416–423.
- Abbreviation: n.a., not available.
To validate the microarray data, we performed qRT-PCR of the top 2 up-regulated (mir-219 and mir-338) and the top 2 down-regulated miRNAs (mir-30c and mir-148a) in the M/NM comparison in a set of 70 randomly selected HCC cases. We found that each of these miRNAs could significantly discriminate M versus NM cases based on their RT-PCR based expression changes and significantly correlated with the miRNA array data (Fig. 2B; Supplementary Fig. 1). Next, we performed PAM analysis with 10-fold cross-validation and 1000 permutations of the class labels to assess the predictive capacity of these 4 miRNAs based on qRT-PCR readings. We found that the expression changes of these miRNAs could correctly classify M and NM samples with 100% accuracy (62% mean permutation accuracy, P < 0.0001) (Fig. 2C). Therefore, it appeared that the expression of these miRNAs were significantly associated with metastasis.
The Association of Metastasis-Related miRNAs with Survival.
To establish a classifier, we performed multivariate nearest neighbor class prediction with 10-fold cross-validation and 1000 permutations of the class label in the 131 M and NM cases (Fig. 1A, Step 2). In this analysis, 90% of the samples were randomly chosen to build a classifier which was then used to predict the remaining 10% of the cases. The accuracy of the prediction was calculated after 1000 repetitions of this random partitioning process (See Supplementary Methods). This analysis yielded a 20-miRNA signature that could significantly predict M and NM status with an overall accuracy of 76% (multivariate P < 0.001). Kaplan-Meier survival analysis based on the 20-miRNA prediction results revealed that the predicted metastasis group had a significantly shorter survival period when compared to the nonmetastasis group (P < 0.042) (Fig. 2D). Thus, this signature is associated with patient survival.
Next, we tested whether the predicted propensity for metastasis based on this signature was related to survival in 110 additional cases. Due to the limited number of patients with metastasis at surgery, we used these 110 cases who developed relapse within 3 years after surgery as patients with potential HCC metastasis to test our miRNA signature developed in “extreme” cases of the training set. We found that the miRNA predictor had a 72% overall prediction accuracy in these independent cases. The predicted M group had a significantly worse survival rate than the NM group (P = 0.009) (Fig. 3A). The 500-day cumulative survival rate based on the 20-miRNA predictor was 41% (95% confidence interval [CI] = 19% to 63%) for patients classified as M compared to 75% (95% CI = 64%–83%) for patients classified as NM (Fig. 3A). Interestingly, available HCC prognostic staging systems (that is, TNM, OKUDA, CLIP, or BCLC) were incapable of predicting patient survival in this independent case set (Supplementary Fig. 2).41-44
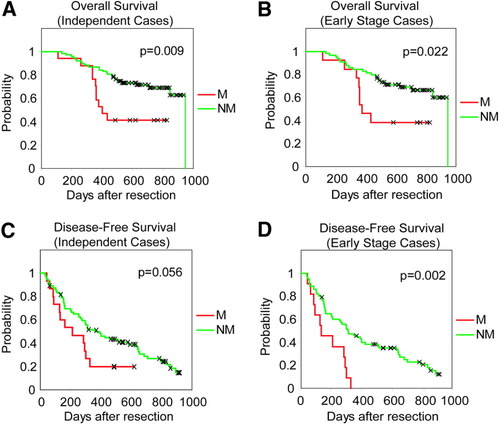
Analysis of the classification capacity of the 20-miRNA in an independent HCC case set or early-stage HCC based on TNM classification. Kaplan-Meier overall survival analysis of (A) 110 patients with HCC based on predicted classification by the 20-miRNA predictor or (B) 89 patients with early-stage HCC. (C) Kaplan-Meier disease-free survival analysis 110 patients with HCC based on predicted classification by the 20-miRNA predictor or (D) 89 patients with early-stage HCC.
Because the ability to predict risk of cancer spread at early stages of HCC could have a significant clinical impact, we also assayed the prognostic capacity of the 20-miRNA signature in patients with early stage HCC. Because long-term survival can be affected by tumor number and underlying liver disease, we performed univariate Cox proportional hazards regression using TNM, BCLC, CLIP or OKUDA staging of the test set to assess the affect of nodular status accounted for by stage on survival outcome in this cohort. We found that nodularity accounted for by staging systems did not affect outcome in this cohort and therefore, we assayed TNM stage I and II as early stage HCC (n = 89) (data not shown). A significantly worse survival was observed for the predicted M patients versus NM by the 20-miRNA signature in the early stage cases (P = 0.022) (Fig. 3B). The 500 day survival rate based on the 20-miRNA predictor was 38% (95% CI = 14%–63%) for patients classified as M compared to 73% (95% CI = 60%–82%) for patients classified as NM (Fig. 3B). In addition, we also tested the capacity of the miRNA signature to predict recurrence in the entire independent set or in early stage cases and found that the predicted M group based on the 20-miRNA signature had a higher recurrence rate (P = 0.056, or P = 0.002, respectively) than the NM group (Fig. 3C,D).
To further test whether metastasis-related miRNAs were those largely associated with survival, we searched for miRNAs whose expression was correlated with this outcome. A survival risk prediction based on two survival risk groups (that is, high versus low; cutoff defined by a 50% prognostic index; see Supplementary Methods) and a 10-fold cross validated permutation test (1000×) at a statistical risk prediction cutoff of P < 0.01 in the entire cohort yielded 46 miRNAs associated with survival (Table 3). We found that 16 (80%) of these survival-related miRNAs overlapped with the 20 metastasis-related miRNAs (Table 2). Kaplan-Meier survival analysis based on the survival risk prediction results revealed that the predicted high-risk group had a significantly shorter survival period when compared to the low-risk group (P = 0.014; 1000× permuted log-rank test) (Supplementary Fig. 3A). Similarly, in the entire cohort, the 20-miRNA-based predicted M group had a significantly worse survival rate than the NM group (P = 0.04) (Supplementary Fig. 3B). The 500 day cumulative survival rate based on the 20-miRNA predictor was 62% (95% CI = 48%–74%) for patients classified as M compared to 81% (95% CI = 74%–86%) for patients classified as NM in the entire cohort (Supplementary Fig. 3B). Therefore, it appeared that the miRNA metastasis signature identified in this study was significantly associated with patient prognosis, including patients with early-stage HCC.
| miRNA | Parametric P value | FDR | Permutation P value | Hazard Ratio | SD of log intensities |
|---|---|---|---|---|---|
| mir-338 | 0.00014 | 0.04634 | 1.00E-04 | 2.538 | 0.64 |
| mir-219-1 | 0.0015823 | 0.0529103 | 0.0024 | 2.036 | 0.729 |
| mir-206 | 0.0025752 | 0.0568261 | 0.003 | 1.963 | 0.622 |
| mir-183 | 0.0077901 | 0.0797369 | 0.0147 | 1.912 | 1.421 |
| mir-129-2 | 0.0062137 | 0.0719331 | 0.0096 | 1.789 | 0.639 |
| mir-192 | 0.0061754 | 0.0719331 | 0.0071 | 0.786 | 1.293 |
| mir-194 | 0.001596 | 0.0529103 | 0.0027 | 0.769 | 1.552 |
| mir-123 | 0.004512 | 0.0678394 | 0.0046 | 0.766 | 1.498 |
| mir-144 | 0.0079189 | 0.0797369 | 0.0078 | 0.756 | 1.205 |
| mir-215 | 0.0073086 | 0.0767983 | 0.0082 | 0.756 | 1.259 |
| mir-101-1/2 | 0.0010396 | 0.0529103 | 0.0026 | 0.755 | 1.163 |
| mir-148b | 0.0092462 | 0.0900145 | 0.0104 | 0.753 | 1.33 |
| mir-122a | 0.009978 | 0.0932232 | 0.0118 | 0.748 | 0.941 |
| mir-30c-1 | 0.0043015 | 0.0678394 | 0.0032 | 0.747 | 1.234 |
| mir-29b | 0.0068308 | 0.074131 | 0.0098 | 0.742 | 1.292 |
| mir-100 | 0.0045338 | 0.0678394 | 0.0048 | 0.738 | 1.315 |
| mir-148a | 0.0063023 | 0.0719331 | 0.0063 | 0.732 | 1.323 |
| mir-299 | 0.0097053 | 0.0931146 | 0.0116 | 0.731 | 1.145 |
| mir-26a-2 | 0.0035932 | 0.0642891 | 0.0034 | 0.725 | 1.114 |
| mir-29c | 0.0015432 | 0.0529103 | 0.0019 | 0.725 | 1.407 |
| mir-30b | 0.0047705 | 0.0678394 | 0.004 | 0.723 | 1.089 |
| mir-193 | 0.0067771 | 0.074131 | 0.0078 | 0.721 | 1.286 |
| mir-125b-2 | 0.0044504 | 0.0678394 | 0.0045 | 0.707 | 1.112 |
| mir-16-1 | 0.0061634 | 0.0719331 | 0.0062 | 0.705 | 1.014 |
| mir-124a-2 | 0.0022636 | 0.0534074 | 0.0023 | 0.696 | 1.185 |
| mir-15b | 0.0061564 | 0.0719331 | 0.0101 | 0.695 | 0.968 |
| mir-30e | 0.0034461 | 0.06337 | 0.0034 | 0.695 | 0.99 |
| mir-19a | 0.004595 | 0.0678394 | 0.0045 | 0.688 | 1.179 |
| mir-1-2 | 0.0052048 | 0.0678394 | 0.0055 | 0.687 | 1.176 |
| mir-126 | 0.0045176 | 0.0678394 | 0.0045 | 0.681 | 1.05 |
| mir-30a | 0.0057222 | 0.0719331 | 0.0069 | 0.679 | 0.875 |
| mir-103-2 | 0.0031911 | 0.0620029 | 0.0027 | 0.676 | 0.933 |
| mir-29a-2 | 0.0003649 | 0.0483128 | 4.00E-04 | 0.663 | 1.152 |
| mir-181a | 0.0049518 | 0.0678394 | 0.0043 | 0.614 | 0.744 |
| mir-200b | 0.0045435 | 0.0678394 | 0.0049 | 0.595 | 0.887 |
| mir-127 | 0.0019182 | 0.0529103 | 0.0018 | 0.588 | 0.865 |
| mir-345 | 0.0027926 | 0.0596355 | 0.0039 | 0.587 | 0.755 |
| mir-9-2 | 0.0011605 | 0.0529103 | 0.001 | 0.587 | 0.999 |
| mir-15a | 0.0020307 | 0.0534074 | 0.0023 | 0.579 | 0.853 |
| mir-340 | 0.0002841 | 0.0483128 | 4.00E-04 | 0.576 | 0.915 |
| mir-22 | 0.0066349 | 0.074131 | 0.0078 | 0.573 | 0.678 |
| mir-341 | 0.0016284 | 0.0529103 | 0.002 | 0.557 | 0.84 |
| mir-19b-1 | 0.0018824 | 0.0529103 | 0.0029 | 0.548 | 0.72 |
| mir-28 | 0.0052263 | 0.0678394 | 0.0069 | 0.546 | 0.594 |
| mir-152 | 0.0010018 | 0.0529103 | 0.0026 | 0.546 | 0.914 |
| mir-224 | 3.72E-05 | 0.0246264 | 1.00E-04 | 0.52 | 1.128 |
We further addressed the relationship of the 20-miRNA-based metastasis signature with survival outcome by performing a survival risk prediction analysis on the training and testing data sets. This analysis was performed using a geneset restricted to only the 20 metastasis miRNAs at a P value of 0.01 with 10-fold cross-validation and 1000 permutations for the significance of the log-rank test. The results demonstrated that these miRNAs could significantly differentiate patient groups based on survival outcome (log-rank P = 0.005 and permuted P = 0.03) (Fig. 4). This analysis provided further validation that these 20 miRNAs were associated with survival outcome.
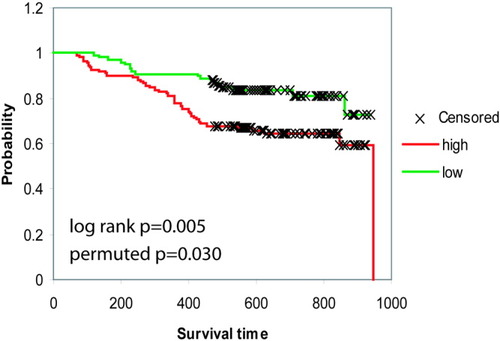
Metastasis miRNAs are associated with survival. (A) Survival Risk Prediction of 241 cases was performed using BRB ArrayTools at P = 0.01 with 2 risk groups (high versus low), 2 principal components, 1000 permutations of the significance of the log rank test and restricted to the 20-miRNA geneset. Kaplan-Meier overall survival analysis curves are shown for high-risk and low-risk survival groups with the log-rank P value and the permuted P value.
Comparison of the miRNA Predictor and Known Clinical Variables.
Next, we performed Cox proportional hazards regression analysis to determine whether the miRNA predictor was confounded by underlying clinical conditions. A univariate analysis revealed that the miRNA signature was a significant predictor of survival (P = 0.006) (Table 4). The multivariate survival model, which controlled for potential confounding covariates demonstrated that the miRNA predictor was associated with a significant 2.1-fold increased risk of death for patients with the M versus the NM expression profile (Table 4). We also assessed the miRNA signature in relation to other available staging information within this cohort. Although CLIP staging and OKUDA staging were significantly associated with survival in univariate and multivariate analysis, the 20-miRNA metastasis signature maintained its significant association with survival in final models including either CLIP (20miRNA HR =2.3 (CI: 1.3–4.1) and P = 0.007) or OKUDA staging systems (20miRNA HR = 2.3 (CI: 1.3–4.0) and P = 0.004) (data not shown). Analysis of BCLC staging was not significant in univariate analysis (Stage A versus O: P = 0.061; Stage B versus O: P = 0.063), perhaps due to the smaller number of cases available for analysis with known BCLC stage and survival data compared to that of TNM, CLIP, and OKUDA in the cohort studied (data not shown). Thus, the miRNA signature is an independent predictor for survival.
| Clinical Variable | Hazard Ratio (95% CIc) | P Value |
|---|---|---|
| UNIVARIATE ANALYSISb | ||
| miRNA predictor (M vs NM) | 2.1 (1.2–3.7) | 0.006 |
| Age | 1.0 (1.0–1.0) | 0.650 |
| Gender (M vs F) | 2.3 (0.8–6.4) | 0.108 |
| HBV (AVR-CC vs CC)e | 1.2 (0.7–2.1) | 0.527 |
| AFP (>20 ng/mL vs ≤20 ng/mL) | 1.7 (0.9–3.0) | 0.099 |
| Cirrhosis (Yes vs No) | 2.3 (0.6–9.6) | 0.238 |
| ALT (≥50U/L vs <50U/L) | 1.0 (0.6–1.8) | 0.895 |
| Ascites (Yes vs No) | 1.5 (0.7–2.9) | 0.268 |
| Total Bilirubin (≤17 μmol/L vs <17 μmol/L) | 0.9 (0.5–1.6) | 0.766 |
| Tumor size (>3 cm vs ≤3 cm) | 4.0 (1.9–8.1) | <0.0001 |
| Tumor encapsulation (None vs Complete) | 2.4 (1.3–4.4) | 0.006 |
| Multinodular (Yes vs No) | 0.8 (0.4–1.6) | 0.618 |
| Microvascular invasion (Yes vs No) | 3.0 (1.7–5.3) | <0.0001 |
| TNM stage (II vs I) | 1.9 (0.9–4.1) | 0.083 |
| TNM stage (III vs I) | 6.1 (3.0–12.3) | <0.0001 |
| MULTIVARIATE ANALYSISd | ||
| miRNA predictor (M vs NM) | 2.1 (1.2–3.6) | 0.011 |
| Tumor encapsulation (None vs Complete) | 1.9 (1.0–3.5) | 0.057 |
| TNM stage (II vs I) | 1.8 (0.8–3.8) | 0.128 |
| TNM stage (III vs I) | 4.8 (2.3–10.1) | <0.0001 |
- NOTE. Bold indicates significant values.
- a Analysis was performed on the entire cohort (n = 241).
- b Univariate analysis, Cox proportional hazards regression.
- c 95% CI, 95% confidence interval.
- d Multivariate analysis, Cox proportional hazards regression.
- e AVR-CC (active viral replication chronic carrier); CC (chronic carrier).
- Abbreviations: AFP, alpha-fetoprotein; ALT, alanine aminotransferase; HBV, hepatitis B virus.
A Search for Metastasis-Related miRNA Targets.
Of the 20 miRNAs, 4 were overexpressed in M whereas 16 were overexpressed in NM (Table 2). This suggested that miRNAs are mainly down-regulated in metastasis and hence, an upregulation of their targets would be found in samples bearing this phenotype. In a previous study using cDNA microarray we identified a 153-gene metastasis signature from hepatic tumors.9 Using a Tarbase bioinformatics approach, we searched for experimentally validated targets of the 20 miRNAs. We then generated a putative miRNA target gene list using TargetScan, whose output was restricted to those 153 tumor-associated metastasis genes with an inverse correlated expression level (Table 2). It appeared that the host targets SPTBN2, GTF2H1, PSCD3, VAMP3, and SLC20A2 are affected by multiple miRNAs related to metastasis and may be part of signaling pathways that significantly contribute to this phenotype.
Discussion
Metastasis and cirrhosis-related development of de novo tumors are two potential causes for the poor prognosis of HCC. In the study presented here, we have demonstrated that tumors with metastatic HCC have a significantly different miRNA expression profile when compared to tumors with nonmetastatic HCC. This 20 metastasis-related miRNA signature also significantly correlates with survival of patients with classically hard- to-distinguish HCC (solitary or multinodular tumors with recurrence/metastasis). Interestingly, several of the miRNAs in the signature have not been associated with the progression of any human malignancies reported and may therefore be uniquely associated with metastatic HCC.
Recent gene expression profiling studies have identified miRNAs associated with liver related disease. In a study of cholangiocarcinoma, miRNA expression was shown to be markedly different in malignant cholangiocytes, with higher expression of mir-21, mir-141, mir-200b which contributed to changes in tumor growth and response to chemotherapy.34 In another recent study comparing HCC samples and adjacent nontumor, 8 miRNAs were shown to be significantly altered, 5 of which were down-regulated in HCC. Classification of samples based on these miRNAs revealed an overall accuracy of >97%.45 In a comparison of the 8 miRNAs found by Murakami et al.45 with the 209 tumor versus nontumor miRNAs found in our analysis (data not shown), only 3 miRNAs overlap (mir-199a, mir-125, and mir-224). In addition, the mir-200 family is present in both signatures although mir-200b was found in our signature versus mir-200a in the signature of Murakami et al. The difference in miRNAs could be due to the differing platforms and particular samples used in either study, but it is affirming that several miRNAs are present in both signatures. We also performed a comparison of the metastasis-related miRNAs identified in this study to those in Murakami et al., which revealed distinct miRNA profiles with only a single overlapping miRNA target gene, the Ras-related G3BP2. This suggests that the alteration of miRNAs is significantly different in metastatic HCC than those distinguishing tumor from nontumor, although the regulation of certain target genes, particularly those related to the Ras family may be maintained.
To commence an understanding of how the expression changes of the 20 miRNAs affect outcome, we have attempted to identify candidate metastasis-associated miRNA targets that are differentially expressed in patients who develop metastases/recurrence. A combinatorial approach using miRNAs and mRNA gene expression profiling may enhance the accuracy of stratifying patients and remains to be assessed. Furthermore, the miRNA target genes may also serve as therapeutic targets to reverse the potential outcome of patients with a poor prognostic signature defined by miRNA classification. Reversion possibilities may occur through gene therapy options to alter the expression of miRNAs or their targets, perhaps through inactivation of oncogenic phenotypes by synthetic anti-sense oligonucleotides, generation of specific inhibitors to abrogate miRNA/target gene interaction or overexpression of tumor suppressive phenotypes using viral or liposomal delivery. In addition to clinical reversion, a comprehensive analysis of the miRNAs identified in this study as well as upstream regulators and downstream targets could shed light on the pathways and mechanisms associated with HCC pathogenesis. These approaches remain to be determined and will be the subject of future studies.
Factors other than metastasis, including poor liver condition, can affect survival of patients with HCC survival, and it is important to delineate the differences between metastatic pattern and survival outcome. It is difficult to ascertain whether tumors occurring after surgery are due to metastatic development or de novo tumorigenesis. Due to the limited number of patients with metastasis at surgery, we used patients who developed a relapse within 3 years after surgery as patients with potential HCC metastasis and poor prognosis. We have undertaken several approaches in this study to demonstrate that the 20-miRNAs and their related targets may be associated with HCC metastasis and significantly contribute to survival outcome. We note that although scarring of the liver enhances tumorigenesis and progression, our univariate results indicate that the poor prognosis within this cohort is not due to the presence of cirrhosis, but rather, metastatic proclivity. We also note that although a subset of the 46 survival-related miRNAs overlap with the 20-miRNA metastasis signature, the remaining miRNAs may affect HCC survival by an alternative mechanism which remains to be studied. In addition, due to the predominance of males, HBV positivity, and an Asian cohort, it remains to be determined whether the identified miRNA signature is also suitable for females, patients with HCC and other underlying liver diseases such as those related to hepatitis C and/or alcohol, or other ethnic groups with HCC.
Isolation, amplification and expression analysis techniques for miRNA are rapidly progressing, increasing the likelihood of feasible miRNA profiling in clinical tissue. Because miRNAs may provide a higher accuracy in subtype classification and our findings suggest a superior ability to distinguish poor-to-predict HCC patient cohorts, grouping patients according to their miRNA signature expression may have clinical utility. The advance identification of patients with poor prognosis (M) by the miRNA signature may allow for more personalized, directed or aggressive treatment regimens than patients classified in the good prognosis group (NM). For optimum clinical use and potentially more efficient diagnosis, it would be appropriate to have a minimum number of genes that could discriminate patients who are likely to develop more aggressive forms of the disease. A miRNA-based platform may provide such an advantage.
An accurate prognosis is essential, particularly in malignant diseases, to provide advice to patients and guidance for assessment and treatment. Clinical evaluation and therapeutic decisions in HCC is complex because they depend on both the grade of cancer spread (tumor staging) and residual liver function (chronic liver disease stage). A majority of patients with HCC are diagnosed at a late stage, and only a small percentage fit resection or transplantation criteria. Although well-defined and generally accepted staging systems are available for almost all cancers, HCC is an exception, with many different staging systems globally introduced to accommodate each stratum of the disease.42, 46-50 Although HCC staging systems may perform well in selecting appropriate surgical HCC candidates, they may still exclude many cases with less aggressive disease that could potentially be differentiated by their molecular portraits. Thus, an accurate predictor of prognosis and a sensible selection criterion that can be applied to patients with HCC, particularly with early stage HCC, for rational treatment decisions remains a challenging task. The miRNA signature identified in this study significantly correlates with survival of patients with HCC with relatively small tumors who were at an early stage of this disease. Because multinodular tumors were included in our analysis of early stage HCC, because of a lack of significant association with survival in this cohort, a more definitive claim of early stage utility of miRNAs in HCC will require larger cohorts with early stage parameters and will thus require further study. In contrast, the clinical HCC staging systems were unable to distinguish the outcome of these patients in this cohort. Our results suggest that a metastasis-associated miRNA signature may be a useful tool to classify patients with HCC at an early stage, assisting in their diagnosis and improving clinical outcome. Such advancements in early prognosis and associated interventional treatment may change the rather fatalistic approach to HCC.
Acknowledgements
We thank Drs. S. Ambs, C. Harris, and R. Simon for their invaluable comments, Zhipeng Yu and Elise Bowman for technical help, and Karen MacPherson for her bibliographic assistance.