Comparing functional and genomic-based precision medicine in blood cancer patients
Graphical Abstract
Abstract
Tumor-agnostic precision medicine (PM) strategies promise to support treatment decisions in relapsed/refractory blood cancer patients. Genomic-based PM (gPM) and drug screening-based functional PM (fPM) currently represent the most prominent PM methodologies. In this study, we report the feasibility analysis of the first 55 patients enrolled in the multicentric, randomized controlled EXALT-2 trial (NCT04470947) comparing treatment recommendations of gPM, fPM, and physicians' choice (PC) head to head. In 54 patients (98%), the diagnostic workflow was successfully implemented, resulting in treatment recommendations for 42 patients (76%), of whom 29 (69%) received the suggested individualized treatments. Actionable targets were identified in 65% by gPM and 80% by fPM (64% microscopy-based, 86% flow cytometry-based fPM). The median time to report was shorter for fPM than for gPM testing. The two strategies revealed overlapping drug targets in 60% of cases. Both, gPM and fPM can efficiently be integrated into the clinical routine to guide therapy decisions for the majority of patients.
INTRODUCTION
Precision medicine (PM) seeks customized treatment strategies on a patient-specific basis aiming to provide comprehensive personalized healthcare. In oncology, genomics has been the dominant tool for PM.1-3 Since many cancer patients lack actionable alterations to match patients to effective therapies accurately, there is a necessity to extend the advantages of PM to a larger proportion of cancer patients. Additional methods need to be explored, such as functional PM, a strategy by which living patient cancer cells are exposed to therapies and their sensitivity is measured to predict clinical response.4-9 It is incompletely understood how these strategies can be best exploited to maximize clinical benefit or if any of these methods are to be preferred in specific clinical settings.10, 11
The prognosis is dismal for aggressive hematologic cancer patients relapsing or refractory upon standard treatments.12, 13 If tumor-containing biopsies can be obtained in a timely and safe manner, these patients are candidates for PM programs or studies. We and others have previously demonstrated that functional PM (fPM) can provide clinical benefit to advanced hematologic cancer patients.8, 14-16 Moreover, molecular-guided therapies were reported to be effective in patients in uncontrolled studies.17-23 Prospective, controlled direct comparisons of PM methods are lacking.10, 11 In this study, we report the feasibility analysis of the multicentric, prospective, randomized-controlled EXALT-2 (NCT04470947) trial directly comparing genomic-based PM (gPM) and fPM with physicians' choice (PC).
MATERIALS AND METHODS
Study oversight and conduct
- -
Percentage of patients with progression-free survival (PFS) ratio ≥1.3 comparing the PFS on study treatment versus PFS of most prior treatment
- -
Overall response rate (ORR)
- -
Overall survival (OS)
- -
The number of treatable targets identified
- -
Successful bridging to hematopoietic stem cell transplant
- -
Time to: (i) informed consent signed; (ii) biopsy performed; (iii) PM results available; (iv) randomization; (v) board decision; and (vi) anti-tumor treatment started.
This study was approved by the independent ethics committee at the Medical University of Vienna (institutional review board vote: EK No: 2125/2018) and is conducted in accordance with the Declaration of Helsinki and the International Conference on Harmonization Guidelines for Good Clinical Practice. EXALT-2 is actively recruiting in five academic hospitals in Austria (Vienna, Graz, Innsbruck, Salzburg, and Linz) and is registered at clincaltrials.gov (NCT04470947).
Inclusion/exclusion criteria
EXALT-2 is enrolling patients with confirmed aggressive hematologic cancers according to the WHO classification, who had received at least two lines of treatment or have no standard therapy options in a relapse setting. Patients demonstrated a maximum response duration on the last previous treatment of 1 year and a relatively fit clinical performance as indicated by an Eastern Cooperative Oncology Group/World Health Organization Performance Status (ECOG/WHO PS) of less or equal to one. All patients were older than 18 years and provided written informed consent. Detailed inclusion/exclusion criteria are shown in Table 1.
| Inclusion criteria | Exclusion criteria |
|---|---|
| r/r aggressive hematologic malignancy | Other malignoma diagnosed <1 year before inclusion (except localized squamous cell carcinoma of the skin, basal cell carcinoma of the skin) |
| ≥2 prior lines of therapy or one therapy and no further standard of care available | Participation in another clinical trial |
| Best response to previous therapy is documented | Pregnancy |
| Response duration to previous therapy <1 year | Classical, nodular, or lymphocyte predominant Hodgkin's lymphoma |
| ECOG ≤ 1 | ECOG > 1 |
| Further therapy is medically feasible | Age < 18 years |
| Tumor cell-containing sample can be obtained |
- Abbreviations: ECOG, Eastern Cooperative Oncology Group; r/r, relapsed/refractory.
Sample size calculation
Sample size was calculated assuming true individual PFS ratio benefit proportions of 50% in fPM and gPM groups as well as 15% in the PC group. Thus, a sample size of 50 patients per group will result in 84% power for the comparisons fPM versus PC and gPM versus PC with chi-squared tests in the final efficacy analysis.
Randomization
Randomization was performed as a weighted, permuted block randomization in a 4:4:2 allocation ratio (fPM:gPM:PC) in blocks of 10 individuals. The random allocation sequence was generated using a software-supported algorithm (Randomizer®, version 2.1.0, supported by the Institute for Medical Informatics, Statistics and Documentation, Medical University of Graz, Austria).
EXALT-2 tumor board
The EXALT-2 tumor board convened after the results of the respective PM study arm were available, or as early as possible in the case of the PC arm. Only PM results of the respective PM study arm were available to the tumor board. The board consisted of at least two hemato-oncologists, a pathologist, a molecular biologist, and a pharmacist. For the PM study arms, treatment recommendation was given based on the respective PM test results. For gPM tests, this process involved a thorough board discussion if a detected genetic aberration could be deemed actionable based on the critical review of existing evidence. Likewise, top-scoring drug candidates from fPM testing were critically discussed and selected based on existing evidence. Treatment recommendations for specific substances (including dosage, duration of administration, etc.) were based on previously published protocols or following the protocols of ongoing clinical trials. Potential pharmacological interactions were checked patient-specifically by the participating clinical pharmacist.
If the EXALT-2 tumor board suggested a drug for use outside of the approved indication, treatment was performed as an “individualized healing attempt” according to Austrian law. A written board protocol was issued for every patient discussed in the EXALT-2 tumor board.
PM assays
Viable tumor cell-containing samples were collected as a “real-time biopsy” depending on the disease entity. In the case of leukemia or leukemic variants of lymphoma, blast cell-containing peripheral blood or bone marrow samples were collected according to routine clinical protocols. Tumor cells were then purified using density-gradient centrifugation (Ficoll-Paque PLUS®, GE Life Sciences) and used for fPM assays as described below. In parallel, blood or bone marrow was directly collected into EDTA-containing test tubes that were sent for gPM testing as described later. A pathologist review was required to ensure histological confirmation of relapse, tumor cell content, and surface marker expression.
In case of lymphoma or solid organ involvement, tumor tissue was collected from a biopsy, in most cases whole lymph node extirpation. However, depending on the tumor site and other patient specifics, needle biopsies were taken if deemed to yield sufficient viable material. At the institutional pathology department, samples were further processed to get (i) viable material for fPM testing and (ii) formaldehyde-fixed paraffin-embedded (FFPE) material for gPM testing, as well as histological confirmation of relapse and tumor cell surface marker expression. Downstream processing for fPM testing involved mechanical tissue disruption followed by filtering through a 70 µm cell strainer to obtain a single-cell suspension, as previously described.14
Based on previously14 reported promising clinical results obtained with the academic prototype of the image-based fPM assay, “pharmacoscopy,” the study was initiated using the further developed commercially available platform through allcyte®, then Exscientia®, Vienna, Austria. After the enrollment of the first 25 patients, the technical yield of delivered reports in 40% of cases (8/20 tests) prompted the study steering committee to deem the method too ineffective to proceed further in the clinical trial. Therefore, the ethics and steering committees approved an additional high-throughput (HT) flow cytometry-based platform established at the Medical University Vienna for fPM assays within the study. Subsequently, both platforms were used in parallel at the investigators' discretion for fPM testing. After improvements in the image-based fPM assay setup, the assay was kept in use, and efficacy increased to 64% by the censoring date of the current feasibility analysis. The testing of combination matrices was not planned as standard but secondary testing of drug combinations (e.g., to reflect common combination treatments) was possible based on initial hits with both fPM platforms.
Image-guided fPM
The image-guided fPM assay was performed as previously described.14, 15, 24 All tests were performed centralized on a commercial basis by Exscientia®, Vienna, Austria. Briefly, ~20 × 103 cells were plated in 384-well Cell Carrier Ultra plates (PerkinElmer) that allow analysis by a high-content microscope. Drugs were pre-printed on the microtiter plates with acoustic liquid handling (Echo 550, Beckman Coulter Life Sciences [formerly Labcyte Inc.]). FDA/EMA-approved anti-cancer agents were tested in triplicate at three different concentrations (100, 1000, and 10,000 nM) (Table S1). After a short-term culture (18–24 h, 37°C, 5% CO2) cells were fixed with formaldehyde and Triton X followed by staining with DAPI and a patient-specific antibody cocktail based on the surface expression of the tumor cells. After staining, plates were imaged with the Operetta CLS High-Content Analysis System (PerkinElmer). The analysis was performed using an automated image analysis algorithm by which the viability of each cell under drug exposure or vehicle (DMSO serving as a negative control) was quantified based on analysis of nuclear morphology, its staining pattern, and location in the well. Integrating this data on a cell population level, the relative viability of marker positive (diseased) and marker negative (healthy) cells was calculated in response to drug treatment and drugs prioritized for treatment by their ability to selectively kill diseased cells. The fraction of cancer cells after ex vivo drug exposure was compared with the fraction in DMSO controls, resulting in a relative cancer fraction (RCF). The RCFs were then averaged across different concentrations and replicates for each drug and subsequently transformed to 1-RCF with positive scores indicating selective cancer cell killing.
Flow cytometry-guided fPM
The HT flow cytometry-guided fPM assay was developed at the Medical University of Vienna, Austria. All tests were centrally performed at the Medical University of Vienna. Briefly, primary patient cells were seeded in 384-well microtiter plates containing FDA/EMA-approved anti-cancer drugs and incubated overnight at 37°C and 5% CO2. A complete list of drugs and tested concentrations is available in Table S1. The compounds were printed on tissue culture-treated 384-well plates (Corning) using an Echo 550 (Labcyte Inc.) acoustic dispenser in five different concentrations in 10-fold dilutions encompassing a 10,000-fold concentration range (either 0.1–1000 nM or 1–10,000 nM depending on prior published efficacy reports, expected compound's potency and selectivity for the intended target, and achievable plasma concentrations in vivo). DMSO served as a negative control with 34 wells per assay plate. After incubation, the plates were spun down (100 g for 5 min), and the supernatant from each well was aspirated using the Biotek MultiFlo FX Multi-Mode Dispenser. Cells were stained with fluorochrome-conjugated antibodies with a patient and/or indication-dependent panel at a 1:500 dilution in PBS for 1 h at room temperature. Cell viability was assessed by the addition of DAPI (BioLegend, Lot B205177) to the staining panel. The assay plates were read on an iQue3 HT flow cytometer screener (Sartorius).
The data were first gated to remove noise, doublets, and dead cells. The gating was then continued on viable single cells, and the selection of population/s of interest was based on the staining panel used. The obtained cell counts were compared between cells exposed to the drugs and cells exposed to DMSO for each cell population of interest per each test compound concentration, allowing for a percent survival calculation. A four-parameter log-logistic model was fitted for the dose–response curve generation of each tested drug in each cell population of interest utilizing the drc R package,25 and a summary metric was calculated with the “compute AUC” function of the PharmacoGx R package26 facilitating drug ranking. Activity is calculated relative to vehicle controls (DMSO wells) per cell population, thereby eliminating bias from the overall cell viability of the sample during the incubation period with the test compounds. The computed AUC values range between 0 and 1, wherein a value higher or equal to 0.12 indicated that a test compound markedly affected cell viability in at least one tested concentration. Moreover, cancer-selective responses were calculated by contrasting the drug response profile of the target cell population to that of the bulk cell population (DAPI-cell population) or the non-disease cell population (e.g., drug response in healthy cells present in the patient specimen), thereby identifying truly disease(cell)-specific drug hits.
Normalized fPM scores (z-scores)
gPM
In the case of a solid tissue biopsy, the gPM assay was performed on formaldehyde-fixed paraffin-embedded (FFPE) material that was obtained according to routine protocols of the institutional pathology department after native material for fPM was put aside as described earlier. If blood or bone marrow samples were used, fresh material was directly collected into EDTA-coated test tubes. gPM was performed using the CE-certified FoundationOne® Heme test (Hoffman LaRoche), which comprises sequencing of DNA (406 genes) and RNA (265 genes; https://www.rochefoundationmedicine.com/home/services/heme.html). Tests were centrally performed at a Roche laboratory in Penzberg, Germany.
Genetic aberrations of the gPM assay were considered targetable if there are therapies that are either (i) proven beneficial in the patient's tumor type, (ii) proven beneficial in another tumor type, or (iii) deemed beneficial and are investigated in an active clinical trial based on sufficient preclinical evidence. The level of evidence for gPM-suggested therapies was classified according to the definitions of the National Comprehensive Cancer Network (NCCN).
RESULTS
Patients with confirmed relapsed/refractory (r/r) aggressive hematologic cancers were eligible. Detailed inclusion/exclusion criteria and details on study conduct are shown in Table 1 and “Materials and Methods” section.
A “real-time biopsy” (solid tissue biopsy, bone marrow aspirate, or peripheral blood draws) containing viable tumor cells was collected from each patient. Samples were subjected to image-based and/or HT flow cytometry-based fPM, as well as gPM testing.
The study design is illustrated in Figure 1A. Patients were randomized into three study arms: fPM, gPM, or PC (control arm) with a 4:4:2 allocation ratio. Regardless of the randomization, fPM and gPM assays were conducted for all patients. However, only the results from the randomized study arm were presented to the treatment-suggesting multicentric multidisciplinary molecular tumor board (EXALT-2 board). In the PC control arm, no PM support was available. The board was composed of at least two hemato-oncologists, a pathologist, a molecular biologist, and a pharmacist. If a PM assay failed because of technical reasons or did not identify a treatment rationale, the study protocol permitted changing to the other experimental arm. If both experimental arms failed or did not identify a treatment rationale, the patient switched to the PC arm. Study arm changes because of clinical, histological, demographic, or other reasons were not allowed per protocol.

Between June 10, 2020, and August 31, 2023, 56 patients were screened, of which 55 were enrolled. Approximately half of the patients had previously received more than three lines of systemic anticancer therapies. Detailed patient characteristics are shown in Table 2. Fifty-four patients (96% of screened patients) underwent real-time biopsy (Figure 1B). Eleven patients had to discontinue the study after real-time biopsy because of either inconclusive histology, failure to confirm relapse, or death/rapid progression of the disease. Forty-three patients (77% of screened patients) comprised the intention to treat (ITT) population and were randomized to the three study arms. Eighteen patients (42% of randomized patients) were randomized to fPM, 15 patients (35%) to gPM, and 10 patients (23%) to the PC arm. One patient had to discontinue the study after randomization due to death from tumor progression. In 14 instances (33% of randomized patients), a study arm change was required according to the criteria described earlier (Table S2). The response evaluation schedule is shown in Table S3. The primary endpoint is the individual PFS benefit for each patient. However, efficacy analysis will be reported only for the final study population of 150 patients according to the study protocol.
| EXALT-2 feasibility cohort (n = 55) | fPM randomized (n = 18) | fPM treated (n = 7) | gPM randomized (n = 15) | gPM treated (n = 8) | PC randomized (n = 10) | PC treated (n = 14) | |
|---|---|---|---|---|---|---|---|
| Age at screening (years) | |||||||
| Median | 65.3 | 64.9 | 55.0 | 69.7 | 71.3 | 68.9 | 66.4 |
| Range | 21.9–81.9 | 36.7–80.7 | 21.9–72.7 | 21.9–81.9 | 28.1–81.9 | 32.4–80.2 | 32.4–81.2 |
| Sex | |||||||
| Male | 35 (64%) | 12 (67%) | 5 (71%) | 11 (73%) | 8 (100%) | 7 (70%) | 9 (64%) |
| Female | 20 (36%) | 6 (33%) | 2 (29%) | 4 (27%) | 0 (0%) | 3 (30%) | 5 (36%) |
| Prior lines of therapy | |||||||
| 1 | 6 (11%) | 5 (28%) | 1 (14%) | 0 (0%) | 1 (13%) | 1 (10%) | 2 (14%) |
| 2 | 22 (40%) | 6 (33%) | 1 (14%) | 5 (34%) | 3 (37%) | 5 (50%) | 6 (44%) |
| 3 | 14 (25%) | 4 (22%) | 3 (44%) | 6 (40%) | 1 (13%) | 0 | 2 (14%) |
| 4 | 8 (15%) | 2 (11%) | 1 (14%) | 2 (13%) | 2 (25%) | 2 (20%) | 2 (14%) |
| >4 | 5 (9%) | 1 (6%) | 1 (14%) | 2 (13%) | 1 (13%) | 2 (20%) | 2 (14%) |
| Best response to previous therapy | |||||||
| CR | 19 (35%) | 6 (33%) | 2 (29%) | 4 (26%) | 3 (37%) | 3 (30%) | 4 (29%) |
| PR | 11 (20%) | 6 (33%) | 1 (14%) | 1 (7%) | 2 (25%) | 0 (0%) | 2 (14%) |
| MR | 3 (5%) | 1 (6%) | 1 (14%) | 1 (7%) | 0 (0%) | 1 (10%) | 2 (14%) |
| SD | 1 (2%) | 0 (0%) | 0 | 1 (7%) | 1 (13%) | 0 (0%) | 0 (0%) |
| PD/refractory | 21 (38%) | 5 (28%) | 3 (43%) | 8 (53%) | 2 (25%) | 6 (60%) | 6 (43%) |
- Abbreviations: CR, complete response; fPM, functional precision medicine; gPM, genomic-based precision medicine; MR, molecular response; PC, physicians' choice; PD, progressive disease; PR, partial response; SD, stable disease.
For forty-two patients (98% of the ITT population, 75% of screened patients), a treatment recommendation was enunciated by the EXALT-2 tumor board. Thirty-two of these patients suffered from malignant lymphoma (B-NHL: n = 17 patients, T-NHL: n = 15 patients) and 10 patients from leukemia (AML: n = 7, ALL: n = 2, CML: n = 1; Figure S1). Three patients (7% of randomized patients) could not start treatment due to clinical deterioration. Ten patients did not receive the full first cycle of recommended antineoplastic therapy because of clinical deterioration and/or disease progression (fPM: n = 6, gPM: n = 1, PC: n = 3) and were recorded as early drop-outs. Twenty-nine patients (68% of ITT population; fPM: n = 7, gPM: n = 8, PC: n = 14) completed at least one cycle of the respective therapy and were evaluable per protocol, representing the final efficacy analysis patient cohort (Figure 1B). Treatment was initiated after a median of 25 days (range: 7–46 days) after biopsy and a median of 7 days (range: 0–28 days) following the weekly EXALT-2 board with no significant difference between study arms (Figure 1C,D). Median time from biopsy to report was shorter for fPM than for gPM tests (microscopy-based fPM: 7 days, flow cytometry-based fPM: 6.5 days, gPM: 19 days, p < 0.0001, Figure 1E).
The proportion of patients that received a therapy recommendation from the EXALT-2 board and subsequently started study therapy was similar between subgroups (B-NHL [n = 24]: 71%/67%, T-NHL [n = 18]: 83%/78%, and leukemia (n = 14): 71%/64%, respectively). However, a higher proportion of leukemia patients had to discontinue therapy before full administration of the first cycle (B-NHL: 2/16, 13%, T-NHL: 2/14, 14%, leukemia: 6/9, 67%, Figure S2).
Overall, PM assays were technically feasible in most instances: flow cytometry-based fPM: 86%; microscopy-based fPM: 64%; gPM: 86% of tests. Actionable hits were detected with fPM assays in 100% and with gPM in 76% of feasible tests. Thus, PM assays identified a treatment rationale in 86% (fPM, flow cytometry), 64% (fPM, microscopy), and 65% (gPM) of tested patient samples, respectively (Figure 2A).
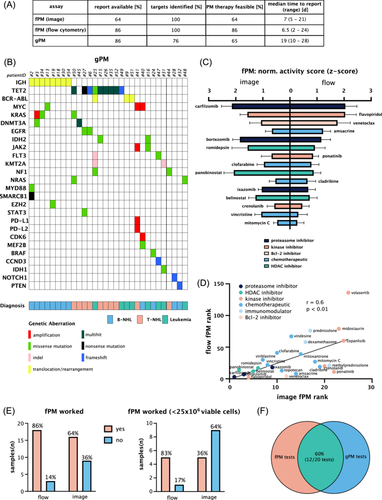
gPM identified a median of 5 (range: 1–13) genetic aberrations per patient, of which a median of 1 (range: 0–5) aberration was conceived as an actionable genetic target (Figure 2B). A summary of overall and targetable genetic aberrations in B-NHL, T-NHL, and leukemia subgroups is shown in Figure 3A–D.

For fPM tests, we observed a high level of concordance between image- and flow cytometry-based assays in terms of technical applicability and top-scoring drugs (Figure 2C,D and Table S4). However, the flow cytometry-based fPM assay showed a favorable performance on biopsy samples with low cell yield (<25 × 106 cells, Figure 2E). When both fPM and gPM results were available, overlapping results were identified in 60% of patients (12/20 patients, Figure 2F and Table S5). Concordance was defined as positive if at least one drug targeting the detected genetic aberration featured in the top 10 drug hits identified by the fPM assay. Accordingly, treatment suggestions by the EXALT-2 tumor board were comparable between PM study arms (Table S6).
Moreover, there was a clear relationship between the biopsy method and viable cell yield that impacted assay performance (Figure S3A,B). Needle biopsy-based procedures yielded lower cell numbers and frequently were insufficient for functional tests (insufficient material in 5/6 (fPM) and 1/6 (gPM) instances; Figure S4). The lowest total cell count that resulted in a technically valid fPM test result was 5 × 106 cells (Figure S3A,B). However, to allow for screening with our fPM test setup of 112 drugs in different concentrations (see “Materials and Methods” section and Table S1) on two assay plates, we established a threshold requirement of approximately 10 × 106 total cells with cell viability of more than 50% to guarantee a technically valid fPM assay (Figure S3C,D). There was no relationship between disease entity and fPM assay performance.
DISCUSSION
The feasibility analysis of the randomized EXALT-2 trial demonstrates that either gPM or fPM approaches can efficiently and safely be integrated into clinical routine. The study is not powered for efficacy interim analysis. Hence, it remains to be answered whether one PM approach is to be preferred over the other. However, the observation that the study arm changes due to difficulties with at least one of the assays occurred in approximately 30% patients, suggests that further improvements are warranted in terms of biopsy procedural guidelines, laboratory protocols, and technical assay performance. Based on these findings, we suggest performing a combination of tests to minimize the risk of assay failure in an individual patient and maximize the chances of providing PM-guided clinical benefit.
This is in line with the increasing evidence that a unidirectional, mutational-based approach to PM might be underpowered to bring long-lasting clinical benefit to individual patients. The recently published results from the NCI-MATCH trial indicate that a purely genomics-driven PM approach resulted in positive results in approximately 25% of substudies.20 This aligns with data from the large-scale WINTHER, i-PREDICT, and Beat AML trials.19, 21, 22 Results of trials using functional testing encourage expectations that the addition of fPM can provide further information in this situation and improve the clinical benefit that is conferred by genomic testing alone.14, 16
Lessons that can be learned from this feasibility analysis concern assay requirements and biopsy methods. We observed that a minimum of 5–10 × 106 viable cells were required for our setup to produce technically valid fPM results. As a consequence, biopsy methods that are unlikely to meet these criteria (such as endobronchial ultrasound-guided biopsy, endoscopic biopsy, skin biopsy) should be avoided, if possible, when seeking fPM guidance. In our experience, total lymph node extirpation in the case of lymphoma and bone marrow biopsy in the case of leukemia remain the best available options to ensure technically valid fPM results.
Taken together, PM can be integrated as a clinical decision-making tool in advanced aggressive hematologic malignancies. Future studies will reveal if the detection of actionable genetic targets and treatment suggestions from functional assays predict efficacy. The upcoming outcome and efficacy analysis of this study will further explore if PM support translates into a clinical benefit.
AUTHOR CONTRIBUTIONS
Lukas Kazianka: Writing—original draft; investigation; methodology; visualization; writing—review and editing; formal analysis; project administration; data curation; resources; validation. Alexander Pichler: Writing—review and editing; methodology. Christiane Agreiter: Methodology; project administration; formal analysis. Johannes Rohrbeck: Writing—review and editing; formal analysis; methodology; resources. Christoph Kornauth: Conceptualization; investigation; methodology; writing—review and editing. Edit Porpaczy: Investigation; writing—review and editing; resources. Christian Sillaber: Investigation; writing—review and editing; resources. Wolfgang R. Sperr: Investigation; writing—review and editing; resources. Karoline V. Gleixner: Investigation; writing—review and editing; resources. Alexander Hauswirth: Investigation; writing—review and editing; resources. Ulrich Jäger: Investigation; writing—review and editing; resources. Peter Valent: Investigation; writing—review and editing; resources. Constanze Jonak: Investigation; writing—review and editing; resources. Stefanie Porkert: Writing—review and editing; resources. Ruth Exner: Writing—review and editing; resources. Wolfgang Willenbacher: Investigation; writing—review and editing; resources. Dominik Wolf: Investigation; writing—review and editing; resources. Peter Neumeister: Investigation; writing—review and editing; resources. Katharina Prochazka: Investigation; writing—review and editing; resources. Alexander Deutsch: Writing—review and editing; resources; project administration; methodology. Richard Greil: Investigation; writing—review and editing; resources. Clemens Schmitt: Writing—review and editing; resources; investigation. Robin Ristl: Methodology; writing—review and editing; conceptualization. Marius Mayerhoefer: Investigation; writing—review and editing. Ingrid Simonitsch-Klupp: Methodology; writing—review and editing; resources; formal analysis. Tea Pemovska: Investigation; writing—original draft; methodology; validation; visualization; writing—review and editing; software; formal analysis; project administration; data curation; supervision; resources. Philipp B. Staber: Conceptualization; investigation; funding acquisition; writing—original draft; methodology; writing—review and editing; supervision; resources.
CONFLICT OF INTEREST STATEMENT
A. P. reports being an employee, founder, and shareholder of exalt®FlexCo. Experimental work was performed while being a PhD student at Medical University Vienna. Manuscript drafting and data interpretation were performed when being employed by and shareholder/founder of exalt®FlexCo, which uses the techniques described in this manuscript for translational research (not involved in clinical data collection). T. P. reports being a founder, and shareholder of exalt®FlexCo. She has patents for EP24158079.4 and EP 24169882.8 pending and licensed to the Medical University of Vienna. T. P. reports a grant unrelated to the submitted work by the Comprehensive Cancer Center (CCC) Vienna. P. B. S. reports being a founder, and shareholder of exalt®FlexCo. He has patents for EP24158079.4 and EP 24169882.8 pending and licensed to the Medical University Vienna.
P. B. S. reports grants from the Vienna Science and Technology Fund, the Austrian Science Fund, as well as grants and personal fees from F. Hoffmann-La Roche AG. P. B. S. reports personal fees unrelated to the submitted work from Amgen, AstraZeneca, Takeda, AbbVie, Janssen Cilag, Medical University of Vienna, Incyte, BeiGene, and BMS. L. K. reports grants unrelated to the submitted work by the Comprehensive Cancer Center (CCC) Vienna and the Austrian Academy of Sciences (OeAW).
All other authors have no conflicts of interest to declare.
FUNDING
This study is sponsored by the Medical University of Vienna and received funding for study conduct and operating costs from Roche Austria.
Open Research
DATA AVAILABILITY STATEMENT
The data of the current feasibility analysis are not publicly available due to reasons of sensitivity and ongoing conduct of the trial. Data are available from the corresponding author upon reasonable request and are stored under controlled access at Medical University of Vienna.