Dual specific STAT3/5 degraders effectively block acute myeloid leukemia and natural killer/T cell lymphoma
Graphical Abstract
Abstract
The transcription factors STAT3, STAT5A, and STAT5B steer hematopoiesis and immunity, but their enhanced expression and activation promote acute myeloid leukemia (AML) or natural killer/T cell lymphoma (NKCL). Current therapeutic strategies focus on blocking upstream tyrosine kinases to inhibit STAT3/5, but these kinase blockers are not selective against STAT3/5 activation and frequent resistance causes relapse, emphasizing the need for targeted drugs. We evaluated the efficacy of JPX-0700 and JPX-0750 as dual STAT3/5 binding inhibitors promoting protein degradation. JPX-0700/−0750 decreased the mRNA and protein levels of STAT3/5 targets involved in cancer survival, metabolism, and cell cycle progression, exhibiting nanomolar to low micromolar efficacy. They induced cell death and growth arrest in both AML/NKCL cell lines and primary AML patient blasts. We found that both AML/NKCL cells hijack STAT3/5 signaling through either upstream activating mutations in kinases, activating mutations in STAT3, mutational loss of negative STAT regulators, or genetic gains in anti-apoptotic, pro-proliferative, or epigenetic-modifying STAT3/5 targets. This emphasizes a vicious cycle for proliferation and survival through STAT3/5. Both JPX-0700/−0750 treatment reduced leukemic cell growth in human AML or NKCL xenograft mouse models significantly, being well tolerated by mice. Synergistic cell death was induced upon combinatorial use with approved chemotherapeutics in AML/NKCL cells.
INTRODUCTION
STAT3 and the closely related STAT5A and STAT5B transcription factors are encoded by three different genes on human chromosome 17q. Importantly, 17q gain is genetically a frequent chromosomal abnormality in cancer, observed in 2.5% of all cancer cases,1 which can promote the overexpression of STAT3/5. Enhanced STAT3/5 expression, gain-of-function (GOF) mutations, or prolonged activation frequently occurs in blood cancer and is associated with higher sensitivity to external stimuli, culminating in cancer proliferation and leukemic stem cell (LSC) maintenance, as shown in cutaneous T cell lymphoma (CTCL) and acute myeloid leukemia (AML).2-5 Interestingly, among the many different blood cancers, two classes present with particularly strong STAT3/5 gene expression signature, namely AML and natural killer/T cell lymphoma (NKCL).6-8
AML is a genetically heterogeneous blood cancer, which is classified into two main groups: (i) AML with defining genetic abnormalities (AML-DGA) and (ii) AML defined by differentiation. AML-DGA patients harbor driver mutations in 76 genes, out of which several can lead to increased STAT3/5 signaling.9 Upstream driver mutations in tyrosine kinases (TK), such as FLT3, JAK2, BCR::ABL1, KIT, mutations in the E3 ubiquitin-protein ligase CBL,9-11 and the hyperactivation of GTPases, such as HRAS, KRAS, NRAS, or RHOA, can promote either nuclear shuttling or STAT3/5-mediated alterations to cellular metabolism.12 FLT3-internal tandem duplication (ITD) rearrangements lead to enhanced STAT5 activation, due to additional docking sites for STAT5 on the receptor (from 8 to >800 bp) boosting STAT5-induced gene transcription and resulting in a poorer prognosis.13 Furthermore, somatic GOF mutations in STAT3/5 are common in lymphoid malignancies, such as NKCL, in which patients frequently harbor GOF variants of STAT3 (~30%), STAT5A/B (~10%), and JAK3 (~10%) in addition to mutations in tumor suppressors, such as PRDM1 (~40%), TP53 (~10%), chromatin modifiers KMT2D (~15%), KMT2C (~15%), and the family of DEAD−box helicases (~45%).8, 14 Due to the high prevalence of mutations in epigenetic modifiers, chromatin remodeling proteins, signaling TK, cell cycle regulators, and upregulation in drug efflux pumps, many AML/NKCL patients do not achieve long-term remission, and chemotherapy often has severe side effects, which compromises the patient's quality of life.11, 15, 16 Therefore, novel and targeted drugs are needed to prevent the relapse of patients and fight myeloid as well as lymphoid cancers.
Using AML and NKCL as model systems, we investigated the potential of directly targeting STAT3/5 as an effective therapeutic strategy to combat cancer growth and survival. We employed two covalent small molecule STAT3/5 degraders from the JPX-based series of compounds, JPX-0700 and JPX-0750, which were identified from the observation of the binding potential of an electrophilic warhead (pentafluorobenzene) to covalently modify Cys residues of the STAT proteins.17 Biophysical analysis of these compounds revealed that they could engage with STAT proteins in an irreversible manner via nucleophilic aromatic substitution. Furthermore, this covalent engagement directly led to perturbation of the local secondary and tertiary structure of the STAT proteins, ultimately leading to protein destabilization and degradation. This capacity for STAT degradation was recapitulated across models of leukemia in vitro, in cellulo, and in vivo and ultimately led to reduced tumor burden and suppression of leukemic dissemination.5 We screened a panel of blood cancer cell lines and demonstrated the sensitivity of particularly NKCL and AML cells to these compounds, both of which display high levels of STAT3/5 signaling. Profiling genetic hallmarks, we revealed chromosomal gains in proproliferative/anti-apoptotic STAT3/5 targets in NKCL, including MYC, MYCN, BCL2L1, MCL1, DNMT3A, and DNMT3B, or lost negative regulators, such as TP53, CDKN2A, and PTPN2, indicative of loss of negative regulatory network, as well as downstream amplification of STAT3/5 oncogene hubs. We showed that these inhibitors facilitate rapid STAT5 degradation, with a relatively slower STAT3 degradation profile, leading to the downregulation of oncogenic STAT3/5 targets in cellulo. AML and NKCL cancer cells and primary AML blasts showed significant loss of viability in the presence of STAT3/5 inhibitors. Tumor growth was reduced in xenograft models in the absence of toxic side effects. Moreover, we found synergistic cytotoxic effects when combining these STAT3/5 inhibitors with other clinically approved drugs used in AML or NKCL treatment.
RESULTS
Dual STAT3/5 degradation exhibits marked potency in AML and NKCL cell lines
Due to frequent mutations in STAT3/5 signaling in NKCL and AML (Table S1), as well as frequently occurring chemotherapy and tyrosine kinase inhibitor (TKI) resistance in NKCL and AML,11, 16 we hypothesized that the simultaneous inhibition of STAT3/5 might decrease the oncogenic signaling cascade to block proliferation and survival. For this, we tested our novel STAT3/5 inhibitors in diverse AML, chronic myeloid leukemia (CML), and peripheral T cell lymphoma (PTCL) cell lines. Cell viability assessment upon 72 h of JPX-0700/−0750 inhibitor treatment revealed low nM to µM IC50 values across the cell line panel for JPX-0700 (0.05–4.59 µM) and JPX-0750 (0.10–3.26 µM) (Figures 1A and S1A). Our analysis revealed that FLT3-ITD+ AML cell lines (MV4-11, MOLM-13, PL-21) were more sensitive toward the inhibitors compared to FLT3-ITD− AML cell lines (Figure S1B), and NKCL were the most sensitive PTCL cancer lines in our analysis, responding slightly better toward JPX-0700 treatment (Figures 1A and S1A). Next, we wanted to correlate cell line sensitivity to the corresponding STAT3/5 protein levels (Figure 1A,B). Interestingly, the IC50 values increased proportionally with higher total/phospho-STAT3 and total STAT5 protein levels in the cell lines, while lower IC50 values significantly correlated with lower protein levels (Figure S1C,D). The lung adenocarcinoma cell line (A549), normal human dermal fibroblasts (NHDF), and peripheral blood mononuclear cells (PBMC), used as controls, were due to their high IC50 values, regarded as predominantly resistant towards STAT3/5 inhibition (Figure 1A).
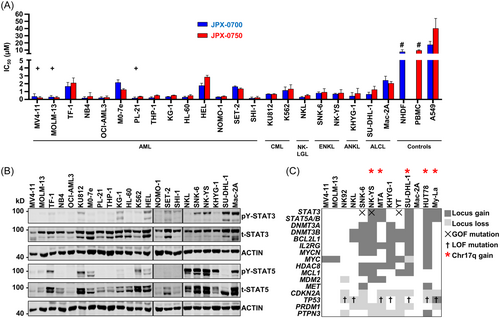
To validate this panel of patient-derived cell lines, which responded well to JPX-0700/−0750 inhibitors (Figures 1A and S1A), we investigated their common genomic aberrations promoting high STAT3/5 signaling in AML/NKCL (Figures 1B and S1E), additionally to reported GOF mutations in STAT3/5 (Table S1). Therefore, we conducted a genomic study using array comparative genome hybridization (aCGH). Usually, copy number gains or losses are often ignored or overlooked; however, gene dosage effects can culminate into vicious cycles of overactivation or enhanced survival, explaining partly the nature of cancer initiation or progression. We extended previous findings of the genetic profiling of NKCL6, 8 and compared them to AML profiles.18, 19 We observed recurrent copy number (CN) gains in STAT3, STAT5A/B, BCL2L1, MCL1, IL2RG, MYCN, MYC, HDAC8, MDM2, DNMT3A, and DNMT3B. Notably, frequent chromosomal losses in tumor suppressors were seen, namely PTPN3, TP53, CDKN2A, and PRDM1 (Figure 1C).
To evaluate the in cellulo target specificity of the novel dual STAT3/5 degraders, we used the IL-3-dependent murine cell line Ba/F3, modified to overexpress either STAT3, STAT5A, or STAT5B. Treatment of cells with JPX-0700/−0750 for 16 h led to loss of STAT3/5 activation and degradation of STAT3/5 total protein (Figure S1F–K). As expected, higher concentrations of inhibitors were required to degrade the STAT3/5 protein in the overexpressing cell lines compared with endogenous protein in the parental Ba/F3 cells.
Small molecule inhibitors induce cell death and block STAT3/5 downstream target gene expression in AML and NKCL
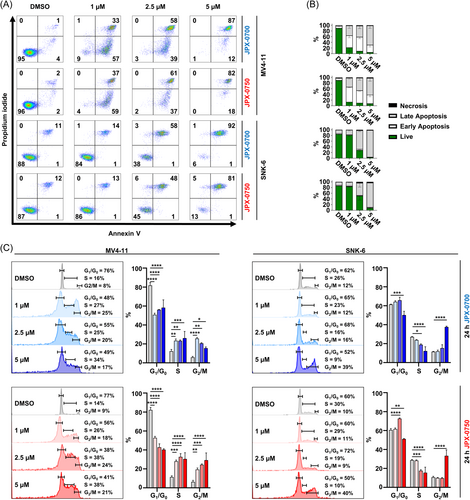
Analyzing the mode of action of cell killing revealed that the small molecule inhibitors reduced cell viability by inducing cell death, as demonstrated by Annexin V/Propidium iodide staining in the FLT3-ITD-driven MV4-11/MOLM-13 AML cells and SNK-6/NK-YS NKCL cell lines, upon 24 h treatment with JPX-0700/−0750 (Figures 2A,B and S2A,B). Measuring the kinetics of cell death induction revealed that both JPX-0700/−0750 are more efficacious in MV4-11 cells (9% and 4% live cells), compared to SNK-6 cells (84% and 86% live cells) at 1 µM concentration, which is most likely attributable to lower STAT3/5 levels and higher sensitivity toward STAT3/5 inhibition (Figure 1A,B). We observed an arrest of AML and NKCL cells primarily in the S and G2/M phase and a depletion of cells in the G1/G0 phase upon JPX-0700/−0750 treatment, indicating a growth arrest (Figure 2C). Furthermore, inhibitor-induced cell cycle arrest was accompanied by poly ADP-ribose polymerase (PARP) and Caspase-3 cleavage (CC3), key mediators of DNA damage/apoptosis pathways (Figure S2C,D).
In order to detect alterations in STAT3/5-target gene expression patterns blocked by JPX-0700/−0750, we conducted mRNA profiling from the 3′-end using QuantSeq analysis on the MV4-11 cell line. Treatment with JPX-0700 resulted in the downregulation of 814 and the upregulation of 2367 genes, whereas JPX-0750 led to decreased expression of 418 and the increased expression of 1653 genes (Figures 3A and S3A). Notably, our Gene Set Enrichment Analysis (GSEA) confirmed the downregulation of genes associated with oxidative phosphorylation, E2F targets, G2M checkpoint control, and MYC signaling, providing substantial evidence that STAT3/5 plays a pivotal role in cancer cell metabolism and cell-cycle progression of AML cells (Figures 3B and S3B). Genes associated with type I Interferon and TP53 signaling were also elevated, indicating an inhibition of cell proliferation due to STAT3/5 signaling blockade (Figures 3B and S3B). Prominent STAT5 target genes, such as PIM1, PIM2, PIM3, MYC, and CCND3, in addition to important STAT3 targets CDC25A, CCND1, and IL10RB were efficiently downregulated after inhibitor treatment, thereby confirming the targeted suppression of STAT3/5 (padj < 0.05). These findings of STAT3/5 target gene downregulation in MV4-11 by QuantSeq analysis were also confirmed in the SNK-6 and NK-YS NKCL cell lines, but using a separate methodology of quantitative reverse-transcription polymerase chain reaction (RT-qPCR). Treatment with JPX-0700/−0750 led to a significant decrease in IL2RA, PIM1, CCND2, MYC, and CDC25A mRNA levels (Figure 3C).
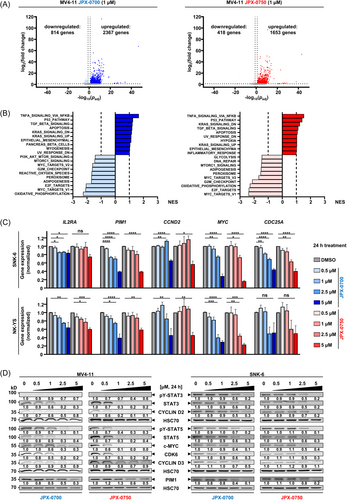
Next, we performed immunoblotting of STAT3/5, and their target gene products, to confirm the downstream impact on the signaling pathway in AML and NKCL cell lines. We focused on target genes that encode oncoproteins involved in cell proliferation and survival, such as c-MYC and PIM1, and proteins regulating the cell cycle, such as CYCLIN D2, D3, and CDK6.5, 7, 20 JPX-0700/−0750 reduced STAT3/5 proteins effectively in the AML and NKCL cells (Figures 3D and S3C). Degradation was stronger in AML cells at 1 µM after 24 h treatment, in line with the more efficient induction of cell death in these cells. Importantly, JPX-0750 completely abrogated c-MYC, CYCLIN D2, and CDK6 levels at 1 µM, but surprisingly CYCLIN D3 and PIM1 were only slightly downregulated, suggesting either a longer protein half-life or alternative routes of activation/mutation maintaining expression of these oncogenes. In summary, JPX-0700/−0750 are potent, cell death-inducing compounds that inhibit STAT3/5 and block downstream oncoprotein nodes in AML and NKCL cells.
JPX-0700 and JPX-0750 are well tolerated by mice and effectively suppress AML and NKCL xenograft growth
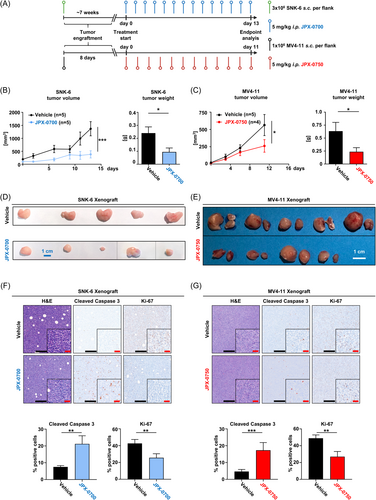
To assess whether JPX-0700/−0750 can inhibit AML and NKCL tumor growth in vivo, we established xenograft models using NKCL and AML cell lines. Leukemic cells were subcutaneously injected into both flanks of NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(CMV-IL3,CSF2,KITLG)1Eav/MloySzJ (NSGS) mice. Based on our previous in vitro data, we chose to treat the SNK-6/NK-YS xenografts with JPX-0700 and MV4-11 xenografts with JPX-0750. JPX-0700 or JPX-0750 (5 mg/kg) was daily administered intraperitoneal, beginning once tumors were palpable (Figures 4A and S4A). SNK-6 and MV4-11 formed tumors on the flanks of injected mice, whereas the NK-YS cells relocated to and established tumors in the vicinity of the inguinal lymph node (Figure S4B). Treatment with STAT3/5 inhibitors resulted in a significant reduction in SNK-6 and MV4-11 tumor growth, compared to vehicle treatment (Figure 4B–E), whereas the NK-YS tumors were only moderately reduced in size (Figure S4C,D). On average, SNK-6 tumor volume was reduced by 70% with JPX-0700 (Figure 4B,D), whereas MV4-11 tumor volume was reduced by 50% upon JPX-0750 treatment (Figure 4C,E). Endpoint analysis revealed a decreased Ki-67 index in SNK-6 and MV4-11 xenografts, whereas increased signals of cleaved Caspase-3 were evident in SNK-6, MV4-11, and NK-YS tumors, suggesting reduced cell growth and induction of cell death in JPX-0700/−0750 treated mice (Figures 4F,G and S5E). SNK-6 and MV4-11 tumor lysates were immunoblotted and probed for total STAT3 and STAT5 levels. Total STAT5 levels were reduced by 40% in MV4-11 tumors from inhibitor-treated mice and by 30% in SNK-6 tumors, whereas total STAT3 protein levels were reduced by 60% in SNK-6 xenografted tumors, indicating the on-target activity of the compounds (Figure S5F,G).
Importantly, we did a comprehensive toxicological analysis of inhibitor-treated mice and detected no statistically significant loss of body weight or obvious defects in overall hematopoiesis (Figure S4H–K). The compounds did not cause gross morphological changes to organs as indicated by liver/kidney damage parameter measurements (Figure S4L,M), and histological analysis (Figure S5A,B), indicating no toxic side effects. Blood urea nitrogen was slightly elevated in JPX-0700-treated mice, but was still within normal physiological range (Figure S4L). The safety of the administration of JPX-0700 was also evaluated in earlier in vivo experiments, using immunocompetent C57BL/6 mice, where we did not observe any significant changes in body weight, organ morphology, blood count, and liver/kidney damage parameters during the two weeks of daily treatment (Figure S5C–G). Thus, we conclude that the compounds can be effective in inhibiting AML and NKCL tumor growth in vivo, and are being well tolerated by mice with no adverse toxicity, but further optimization of the pharmacokinetic properties of the compounds is needed.
Combination of dual pharmacologic STAT3/5 inhibition with standard-of-care antineoplastic drugs achieves additive and synergistic effects in AML and NKCL cells
Due to the aggressiveness and heterogeneity of AML and NKCL, combinatorial therapies, if tolerable, are likely to overcome resistance, which leads to longer-term disease control and/or improved quality of life of patients. To identify synergies between the STAT3/5 inhibitors and clinically approved drugs for both AML and NKCL,11, 21 we tested JPX-0700/−0750 in combination with a DNA-damaging drug (doxorubicin), a DNA-methyltransferase inhibitor (azacitidine), a BCL-2 inhibitor/senolytic drug (venetoclax), and a multikinase inhibitor (cabozantinib), in vitro. Additionally, disease-specific drugs were tested in AML (gilteritinib, midostaurin, cytarabine, daunorubicin) and NKCL cell lines (vorinostat, belinostat, methotrexate, l-asparaginase) in combination with our STAT3/5 inhibitors. In general, we observed JPX-0750 having stronger combinatorial effects with anti-neoplastic drugs in MV4-11 and MOLM-13 cells, whereas JPX-0700 demonstrated superior synergistic and additive effects in SNK-6 and NK-YS cells (Figures 5A and S6A). When combined with JPX-0750, venetoclax, cytarabine, and doxorubicin displayed the highest synergy scores in MV4-11 cells. The highest synergistic effects were observed in SNK-6 cells when cabozantinib and venetoclax were combined with JPX-0700. Additionally, we confirmed the synergy results of our combination treatments with a second, independent software and observed similar synergistic and combinatorial effects in our cell lines (Figure S6B–D).
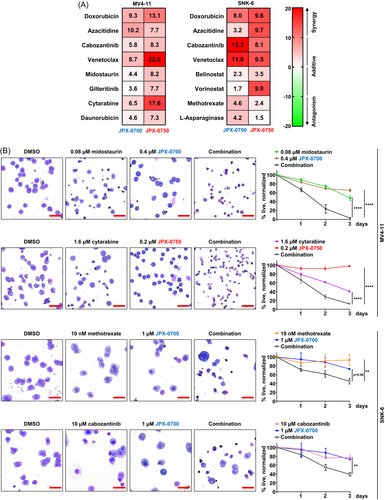
For better visualization of synergistic effects, cytospins were prepared from MV4-11 and SNK-6 cells treated with selected drugs in single treatments and in combination (Figure 5B). Two drug combinations per cell line were chosen: one drug being a first-line treatment commonly used for the disease and the second targeting a genetic abnormality present in the respective cell line. In this case, FLT3-ITD-driven MV4-11 cells were treated with cytarabine or midostaurin, while MET-mutated SNK-6 cells were treated with methotrexate or cabozantinib (Figure 1C), in combination with the STAT3/5 inhibitors. After 72 h of combinatorial treatment, we observed that cells were reduced in size, and cell membrane blebbing and disintegration was visible, indicating cell death, whereas single drug treatments did not lead to such significant changes in cell morphology (Figure 5B). In conclusion, the JPX class of compounds exhibited remarkable in vitro synergies when combined with clinically approved drugs, particularly JPX-0700 in NKCL and JPX-0750 in AML.
STAT3/5 inhibitors effectively kill primary AML blasts and can be combined with approved chemotherapeutics to achieve synergistic effects
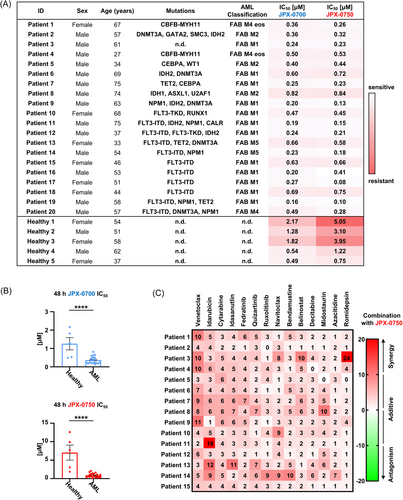
To test the translational potential of these JPX inhibitors, we focused on the treatment of primary AML cells. We were unable to obtain viable NKCL patient samples due to their rarity and the manner in which patient tumor samples are commonly processed after resection. Clinical specimens are solid tumor masses processed via fixation and paraffin embedding, leaving no sampling possibility to isolate primary NKCL cells from patient blood. Thus, we used AML blasts from 20 patients and healthy bone marrow cells from five donors. These were cultured and treated for 48 h with different concentrations of STAT3/5 inhibitors, and cell viability was determined. Remarkably, the administration of both STAT3/5 inhibitors as a single agent resulted in significant inhibition of AML blast growth, indicating their potent antileukemic effect (Figure 6A). Both JPX-0700/−0750 exhibited equivalent potency, and the mutational status of patient samples did not significantly impact the observed sensitivity, underlining the importance of STAT3/5 for AML survival and proliferation. Importantly, our findings demonstrated that AML blasts have a 4- and 10-fold higher sensitivity toward JPX-0700/−0750, respectively, in comparison to healthy bone marrow cells, revealing a therapeutic window (Figure 6B). Strikingly, when JPX-0750 was administered together with approved chemotherapeutics, including DNA damaging chemotherapies, DNA demethylating agents, BH3-mimetics, drugs that target FLT3, HDAC-, JAK-, or MDM2 inhibitors, synergistic effects were observed (Figure 6C). The combinations of JPX-0750 with venetoclax, idarubicine, idasanutlin, bendamustin, midostaurin, or romidepsin resulted in enhanced cell killing compared to single-agent therapy. Taken together, our results demonstrate the effective inhibition of primary AML blast survival and proliferation by dual STAT3/5 inhibition as mono- and combinatorial therapies.
DISCUSSION
We investigated the efficacy of novel, dual STAT3/5 inhibitors JPX-0700/−0750 in AML and NKCL. These electrophilic STAT3/5-targeting inhibitors facilitate the degradation of both phospho- and total STAT3/5, leading to the downregulation of pro-proliferative and metabolic cancer genes. Dual degradation of STAT3/5 was highly efficacious in inducing cell death and cell cycle arrest in both AML and NKCL cell lines. They displayed in vivo antitumor efficacy while being well tolerated by mice, demonstrating therapeutic activity against ex vivo AML patient blasts, but sparing healthy bone marrow cells. Moreover, the compounds displayed synergistic effects promoting more efficient killing of tumor cells in combination with standard-of-care drugs/chemotherapy in AML and NKCL, as well as primary AML patient blasts.
AML patients harboring inv(3) and TP53 mutations have statistically the worst prognosis in survival (hazard ratios: 2.9 and 1.7, respectively), but mutations in FLT3-ITD, BRAF, SRSF2, chromosomal alterations: +21, −5/5q, −17/17p, +13, −7, −9q, +22 and a complex karyotype are also negative prognostic for patients (hazard ratios: 1.2–1.5).9 Strikingly, genetic alterations in FLT3 are found in more than 30% of AML patients, significantly reducing the median overall survival time (hazard ratios: FLT3ITD [1.4], NPM1-DNMT3A-FLT3ITD [1.5], MLLPTD-FLTTKD [1.4]) to less than 1 year.9, 22 Past efforts to inhibit STAT3/5 signaling in AML focused on TKI acting upstream to block them.11, 23 However, AML patients frequently develop resistance to TKI and chemotherapy due to de novo mutations in TP53 and IDH1/2, the overexpression of drug efflux pumps (P-glycoprotein, ABCC1), the protective bone marrow niche and the metabolic reprograming or epigenetic alterations in LSCs.24 Moreover, enhanced expression of STAT3/5 or enhanced expression/mutations in downstream effector molecules, such as c-MYC, n-MYC, D-type Cyclins, AXL, or BCL-2 family members, often lead to therapy failure in AML.15, 25 In contrast, NKCL patients often relapse after initial chemotherapy and NKCL patients with systemic dissemination have a median survival time of less than 58 days.16, 26 Due to the increased expression of multidrug resistance proteins (MDR1, ABCC4) and mediators of cell proliferation (IL13, MYC), tumor cells circumvent the initiation of apoptosis.16 Relapsed patients are treated with L-asparaginase and respond initially well, but tumors upregulate IL2RA rapidly, which is mediated by STAT3/5, leading to resistance. Equally problematic are the side effects that late-stage NKCL patients experience during chemoradiotherapy. Therefore, novel and more targeted drugs are needed to prevent relapse and improve the quality of life of AML and NKCL patients.
Degradation-based therapeutic strategies could offer superior potential. We profiled two drugs from this class of compounds that displayed efficacy in degrading STAT3/5 proteins,17 thus reducing the expression of oncogenic STAT3/5 targets. Our JPX degraders could therefore potentially also block noncanonical protein functions, hence eliminating scaffolding activity or cofactor/corepressor recruitment,27 as well as mitochondrial STAT3/5 function, which was shown to be essential for oncogenic transformation mediated by RAS/RAF.12 Dual STAT3/5 degraders highlight the potential importance of blocking broader STAT functions beyond those mediated by tyrosine phosphorylation, affecting a larger spectrum of essential pathways in cancer. We could show in vivo efficacy of our inhibitors, effectively reducing the growth of MV4-11 and SNK-6 xenografted tumors. Furthermore, the reduced inhibitory effect of JPX-0700 on the in vivo growth of NK-YS tumors, which we hypothesize is due to the lesser penetration of the compound into the lymph nodes, advocates the further optimization of pharmacokinetic properties of our compounds.
Our drug synergy results using clinically approved therapeutics together with JPX-0700/−0750 highlight the possibility of directly targeting STAT3/5 to overcome drug resistance in AML/NKCL. The senolytic/BCL-2 inhibitor venetoclax highly synergized with JPX inhibitors in both AML/NKCL, suggesting a similar dependence on anti-apoptotic BCL-2 family members. In contrast, the high synergy of cabozantinib in SNK-6 cells could be explained by the underlying MET mutation, which is effectively targeted with the drug combination. Interestingly, while our synergy screen only predicted an additive effect of our compounds with the multikinase inhibitor midostaurin (targeting FLT3, PKCα/β/γ, SYK, FLK-1, AKT, PKA, c-KIT, c-FGR, c-SRC, PDGFRβ, VEGFR1/2 with IC50 values ranging from 22–500 nM), the viability of FLT3-ITD-driven MV4-11 cells was still effectively reduced by combining it with our compounds, implying that different experiments should be used to correctly judge the additive and synergistic effects of drugs.
In summary, the effectiveness of dual inhibition of phospho- and total STAT3/5 by JPX inhibitors in AML/NKCL emphasizes their essential roles in initiating and driving these cancers. Therefore, our findings here would provide a rationale for further clinical development of efficacious compounds toward their combinatorial use for blood cancer treatment.
MATERIALS AND METHODS
Cell culture and generating stable cell lines
Suspension cell lines were maintained in RPMI-1640 and adherent cell lines were grown in DMEM (both from Gibco) supplemented with 10% FBS (Biowest), 0.06 g/L penicillin, 0.1 g/L streptomycin (both from VWR), and 2 mM L-glutamine (Life Technologies). Culture media of NK92, NKL, SNK-6, NK-YS, and KHYG-1 cells were additionally supplemented with 5 ng/mL hIL-2 and M0-7e; TF-1 cell culture media was supplemented with 10 ng/mL hGM-CSF (both from ImmunoTools). Ba/F3 cells were grown in the presence of 1 ng/mL murine IL-3 (ImmunoTools). Mammalian expression constructs containing FLAG-tagged human STAT3, STAT5A, or STAT5B in a pMSCV-IRES-GFP plasmid were used to generate stable Ba/F3 cell lines. The procedure of retroviral transduction with these constructs was performed as previously described.28
Genetic analysis of cell lines
The array comparative genome hybridization analysis on the NK92, NKL, SNK-6, NK-YS, MTA, KHYG-1, YT, SU-DHL-1, Mac-2A, HUT78, and My-La cell lines was conducted as outlined previously.5 Additional mutational data of cell lines were data-mined from publicly available databases. aCGH analysis: MV4-11,18 MOLM-13.19 Sanger sequencing: NKL,29 NK92,29 SNK-6,29 NK-YS,29 MTA,30 KHYG-1,29 YT,29 SU-DHL-1,30, 31 Mac-2A,30, 31 My-La,5 HUT78,5 MV4-11,30, 31 MOLM-13,30, 31 THP-1,30, 31 OCI-AML3,30, 31 NB4,30, 31 M0-7e,30, 31 PL-21,30, 31 SHI-1,30, 31 KG-1,30-32 TF-1,30, 31 SET-2,30, 31 HL-60,30, 31 HEL,30, 31 NOMO-1,30, 31 KU812,30, 31 K562,30, 31 A549.30
Immunoblot analysis
Approximately 0.75 × 106 cells/mL were seeded into a six-well plate, and treated with the desired concentration of the drug of interest and the appropriate cytokine. Cells were harvested, lysed, and immunoblotted as described previously.5 Equal loading was confirmed by probing the same membranes with a specific antibody for human HSC70 or ACTIN. Information regarding the antibodies and their dilutions used for Western blotting is available in File S1.
QuantSeq and data analysis
Cells were incubated for 24 h with compounds and washed twice with phosphate-buffered saline. RNA extraction followed the standard protocol employing the RNeasy Mini Kit (QIAGEN). Subsequently, sequencing libraries were created utilizing the QUANT seq. 3′ mRNA-Seq Library Prep Kit (Lexogen). The combined libraries underwent processing at the Vienna Biocenter Next Generation Sequencing Facility in Vienna, Austria, and were then subjected to sequencing through single-read 70-bp chemistry on a NextSeq. 550 instrument (Illumina). Data analysis was performed as described previously.33
Cell death measurements
The induction of cell death by our inhibitors was assessed after 24, 48, or 72 h posttreatment using flow cytometric analysis of Annexin V and propidium iodide stained cells (FACSCanto II; BD Biosciences) according to the manufacturer´s instructions (eBioscience) as described previously.5
Cell viability assays
The experiments for the in vitro cytotoxicity and in vitro synergy screens were conducted as described previously.5 The analysis of drug synergies was performed using the SynergyFinder 3.0 web application or the Combenefit software.34, 35 For the cytospins: cells were treated for 72 h with the indicated inhibitor, washed once with PBS and after counting, approximately 5000 cells were spun on glass coverslips for 3 min at 800 rpm (Cytospin 3; Shandon). After drying, the cells were stained with Wright-Giemsa solution (Carl Roth) according to the vendor's instructions. Bortezomib (5 µM) was used as a positive control for cell death.
Cell cycle measurements
Approximately 1 × 106 cells were seeded in 1 mL of media and the cell cycles were synchronized with a double thymidine block.36 Following synchronization, cells were treated with inhibitors for 24 h and then analyzed using flow cytometry, as described previously.7
RT-qPCR
Per condition: Approximately 0.75 × 106 SNK-6 or NK-YS cells/mL were seeded in full media, supplemented with 5 ng/mL hIL-2. Cells were then treated either with DMSO (control) or with JPX-0700/−0750 for 24 h and were stimulated with 5 ng/mL hIL-2 for 4 h before harvesting. Cells were washed twice and RNA isolation, cDNA synthesis, and RT-qPCR were conducted as described previously.5, 37, 38
Patient samples
Bone marrow cells were isolated from the iliac crest of AML patients, located at the Helsinki University Hospital Comprehensive Cancer Center or the Medical University of Vienna, and were stored at −80°C until used. The percentage of blasts in the patient's bone marrow was on average 75%. Healthy bone marrow cells were isolated from individuals with suspected leukemia, revealing upon further diagnosis no presence of malignant cells in their bone marrow. The study was approved by the ethics committee of the Medical University of Vienna (1184/2014 and 1334/2021) and the Helsinki University Hospital (303/13/03/01/2011), and was conducted in accordance with the declaration of Helsinki. All patients gave written informed consent. AML samples were evaluated at accredited clinical laboratories, including a gene panel for myeloid mutations, chromosome analysis (Giemsa staining), FAB classification based on bone marrow morphology, and FLT3-ITD allelic ratio (ITD-AR) analysis. In cases where gene mutation panel results were unavailable, exome sequencing was employed as previously described.39
Ex vivo synergy screen
Primary cells were cultured in Mononuclear Cell Medium (#C-28 030; PromoCell) supplemented with 2.5 µg/mL amphotericin B (Sigma-Aldrich) and 10 µg/mL gentamicin (Thermo Fisher). Drug sensitivity testing was carried out at the FIMM High Throughput Biomedicine Unit, which is hosted by the University of Helsinki and supported by HiLIFE and Biocenter Finland as previously described.40-42
In vivo efficacy and safety evaluation
The animal research was conducted following the institutional regulations for animal welfare and with the approval of the Austrian Federal Ministry of Science, Research, and Economy (BMBWF-68.205/0130). A total number of 37 NSGS mice, aged 8–10 weeks, were used for the experiments. The MV4-11 xenograft cohort consisted of five females and four males, the NK-YS xenograft cohort of eight females and one male, and the SNK-6 xenograft cohort of seven male and 12 female mice. The SNK-6 xenograft experiment was repeated twice. After the tumors were palpable, mice were randomized according to average tumor volume and sex, and assigned to the vehicle (n[MV4-11] = 5, n[SNK-6 rep1] = 5, n[SNK-6 rep2] = 5, n[NK-YS] = 5) or treatment groups (n[MV4-11] = 5, n[SNK-6 rep1] = 4, n[SNK-6 rep2] = 5, n[NK-YS] = 4). Xenografted NSGS mice were treated with JPX-0700 (5 mg/kg), JPX-0750 (5 mg/kg) or vehicle (5% ethanol, 5% Kolliphor-EL [Carl Roth], 30% propylenglycol [Carl Roth], 20% HP-ß-cyclodextrin [Sigma Aldrich] in PBS) by intraperitoneal injection daily for 12 (MV4-11), 21 (SNK-6 rep1), 14 (SNK-6 rep2), or 22 (NK-YS) days. Tumor volumes were calculated using the formula: (tumor length × tumor width2): 2 for spherical tumors and the formula: 4/3 × π × (length: 2) × (width: 2) × (height: 2) was used for the calculation for flat tumors. Organs were fixed overnight in 4% phosphate-buffered formaldehyde solution (Roti® Histofix; Carl Roth), dehydrated, paraffin-embedded, and cut into 2 µm consecutive mouse organ or tumor sections. Sections were stained with Hematoxylin (Merck), Eosin G (Carl Roth), Ki-67 (MM1-L-CE; Leica Biosystems), and Cleaved Caspase 3 (#9664; Cell Signaling Technology). The stained slides were scanned with Evident Slideview VS200 (Olympus) and the percentage of Cleaved Caspase 3 and Ki-67 positive and negative cells were analyzed with QuPath (Version 0.4.3). No mice were excluded during the analysis. During the conduct and analysis of the in vivo experiments, the experimenters were not blinded.
Statistical analysis
GraphPad Prism 8 was used to perform the statistical analyses. Statistical tests are specified in Figure legends. The threshold for statistical significance was set to p < 0.05, unless otherwise specified. *p < 0.05, **p < 0.01,***p < 0.001, ****p < 0.0001.
ACKNOWLEDGMENTS
The authors wish to thank Safia Zahma, Emi Miyakoda, and Michael Machtinger from the Institute of Animal Breeding and Genetics at the University of Veterinary Medicine Vienna for technical support in histology and during in vivo experiments, and Stefan Stoiber from the Medical University of Vienna for technical support during in vivo experiments and for his support while designing figures. This research was supported using resources of the VetCore Facility (VetImaging | VetBiobank) of the University of Veterinary Medicine Vienna.
AUTHOR CONTRIBUTIONS
Daniel Pölöske, Helena Sorger, and Richard Moriggl: Conceptualization. Richard Moriggl: Resources. Daniel Pölöske, Helena Sorger: Data curation. Daniel Pölöske, Helena Sorger, Anna Orlova, Elisabeth Heyes, Anna Schönbichler, Marta Surbek, Sanna H. Timonen, Thomas Eder: Software. Daniel Pölöske, Helena Sorger, Anna Orlova, Anna Schönbichler, Marta Surbek, Elvin D. de Araujo: Formal analysis. Richard Moriggl: Supervision. Richard Moriggl: Funding acquisition. Daniel Pölöske and Helena Sorger: Validation. Daniel Pölöske and Helena Sorger: Investigation. Daniel Pölöske and Helena Sorger: Visualization. Daniel Pölöske, Helena Sorger, Anna Orlova, Elisabeth Heyes, Anna Schönbichler, Marta Surbek, Christina Wagner, Heidi A. Neubauer, Diaaeldin I. Abdallah, Aleksandr Ianevski, Heikki Kuusanmäki, Tobias Suske, Martin L. Metzelder, Michael Bergmann, Maik Dahlhoff, Florian Grebien, Roman Fleck, Sanna H. Timonen, Christine Pirker, Walter Berger, Wolfgang R. Sperr, Lukas Kenner, Peter Valent, Tero Aittokallio, Marco Herling, Satu Mustjoki, Patrick T. Gunning: Methodology. Daniel Pölöske, Helena Sorger, and Richard Moriggl: Writing—original draft. Richard Moriggl: Project administration. All authors: Writing—review and editing.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
FUNDING
Austrian Science Fund grant (FWF) SFB F04707-B20 (HS, RM). Austrian Science Fund grant (FWF) SFB-F06105 (RM, CW, HAN). Austrian Science Fund grant (FWF) P34781 (LK). The Christian-Doppler Lab for Applied Metabolomics (CDL-AM) in cooperation with Siemens Medical Solutions USA, Inc. (DP, LK). The European Union supported the Transcan-II initiative ERANET-PLL (MH, DP, RM). The European Union supported ERA PerMed JAKSTAT-TARGET (SM, MH, PTG, TA, HAN). The European Union Horizon 2020 Marie Sklodowska-Curie Innovative Training Network: “FANTOM” n. 101072735, “eRaDicate”, n. 101119427 (LK). The Vienna Science and Technology Fund (WWTF) LS19-018 (LK).
Open Research
DATA AVAILABILITY STATEMENT
All data associated with this study are present in the paper or the File S1. The QuantSeq data were deposited in the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) under the GEO-Accession GSE263984.





