Comparison of whole-genome and immunoglobulin-based circulating tumor DNA assays in diffuse large B-cell lymphoma
Minimal residual disease (MRD) has emerged as a promising biomarker in diffuse large B-cell lymphoma (DLBCL). Both pretreatment levels and dynamic changes in circulating tumor DNA (ctDNA) have been associated with clinical outcomes, and in many studies, ctDNA outperforms traditional response assessment tools.1-3 Newer panel-based approaches (CAPP-Seq, PhasED-Seq) appear to have improved sensitivity compared to immunoglobulin-based high-throughput sequencing (IgHTS).4, 5 There is growing interest in using ctDNA to guide response-adapted treatment approaches in DLBCL and ctDNA techniques that maximize sensitivity while retaining excellent specificity are needed to optimally guide such strategies.
To improve sensitivity, whole-genome sequencing (WGS) has been used to identify patient-specific tumor fingerprints which can be assayed within plasma cell-free DNA (cfDNA). This approach increases the likelihood of detecting tumor mutations but is challenging due to the massive excess of normal cfDNA in plasma. To address this, we developed minor-allele-enriched sequencing through recognition oligonucleotides (MAESTRO) which uses short allele-specific probes to enrich thousands of prespecified mutations and enable their detection using Duplex Sequencing with up to 100-fold fewer sequencing reads.6 We subsequently described a new implementation of this test called MAESTRO-Pool, wherein personalized MRD tests from all patients in cohort studies are pooled into a single test.7 This enables both the detection of MRD in each patient's plasma cfDNA and the assessment of the specificity of each bespoke MRD test using unmatched samples from other patients. We have further described an improved “dynamic MRD caller” that accounts for varied numbers of mutations and cfDNA molecules assayed and reports a probability score for MRD detection.7 This safeguards against false detection and enables the identification of borderline-negative samples for follow-up testing.
Here, we applied MAESTRO-Pool and our dynamic MRD caller in a pilot study, analyzing ctDNA in plasma samples from nine patients with relapsed/refractory DLBCL undergoing autologous stem cell transplantation (ASCT) and comparing our results with those obtained using conventional IgHTS MRD testing.
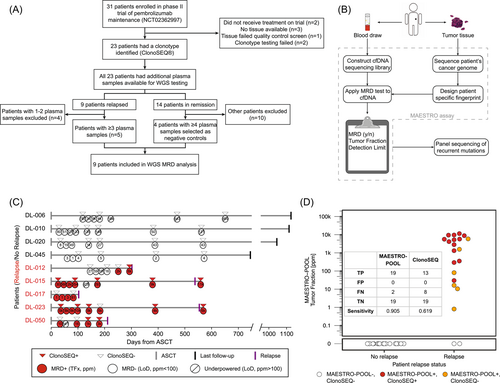
The study was performed using formalin-fixed paraffin-embedded (FFPE) tissue biopsies and serial post-ASCT peripheral blood samples from patients enrolled in a phase II trial testing pembrolizumab maintenance therapy following ASCT (NCT02362997).8 All patients signed informed consent. Plasma samples were previously analyzed using the IgHTS ClonoSEQ® assay (Adaptive Biotechnologies), and ctDNA was found to be a significant predictor of progression-free survival (PFS).9 Among 31 enrolled patients, nine had leftover genomic DNA (gDNA) from tissue samples and sufficient post-ASCT plasma samples. These patients were selected for additional analysis (Figure 1A).
FFPE tissue and CD3+/CD19− T cells (isolated from cryopreserved peripheral blood mononuclear cell samples via fluorescence-activated cell sorting) underwent gDNA extraction and WGS with a targeted raw depth of 30× and 15× for tumor and normal samples, respectively, as previously described.10, 11 A MAESTRO panel was designed for each patient and capped at a maximum of 5000 probes.6 Next, all patients' panels were combined into one large MAESTRO-Pool panel that was ordered from Integrated DNA Technologies and used for testing of all samples from all patients, as previously described.6, 7 For comparison, personalized MRD Tracker assays10 were also designed and applied to the same plasma samples, allowing us to compare MAESTRO-Pool to an orthogonally validated MRD test.
Additional baits targeting 63 unique loci within 12 genes were designed to capture single-nucleotide variants reported in relapsed DLBCL (Table S1). Additional details regarding sample collection, processing, germline DNA isolation, probe design, sequencing, and analysis are included in Supporting Information: Methods.
The baseline characteristics of the nine patients are summarized in Table S2. After a median follow-up of 36.6 months (range 30.0–39.6), five patients had relapsed. Patient-specific tumor mutations (median 422, range 81–1653) were identified from FFPE samples. Post-ASCT surveillance plasma samples were next analyzed using both MAESTRO-Pool and MRD Tracker (an orthogonal, whole-genome, tumor-informed MRD test that does not use mutation enrichment) for comparison.6 MAESTRO assays for each patient were pooled into a single test (6044 mutations total) and applied to plasma samples from all patients which were individually barcoded to enable demultiplexing. Meanwhile, MRD Tracker assays were capped at 1000 mutations per patient and included a similar median of 482 (range 55–949) mutations.
MRD detection was highly concordant for MAESTRO-Pool and MRD Tracker. Among 59 samples from the 9 selected patients, a discrepant MRD call was observed for a single sample (sample 4 from DL-12)—for which ctDNA was detected at 4 ppm using MRD Tracker, but not detected using MAESTRO-Pool (the sample-specific limit of detection [LoD] for that sample was 16 ppm). Estimated tumor fractions using MRD Tracker and MAESTRO-Pool were concordant (Figure S1). Even with a 13-fold reduction of required sequencing reads, we observed a similar median LoD for MRD Tracker (median LoD 40 ppm, range 1–4727) and MAESTRO-Pool (median LoD 30 ppm, range 1–18,243) (Figure S2).
MAESTRO-Pool also enables the empirical assessment of the specificity of each patient's bespoke MRD test using unmatched samples from the other patients in the cohort (Figure S3). For seven of nine patients', MRD tests, we observed 100% specificity among a median of 52 (range 51–54) unmatched samples from other patients. The remaining two patients' (DL-017 and DL-045) MRD tests showed 95% and 98% specificity, respectively, among 55 and 53 unmatched samples, with false detection occurring in four samples at 1, 23, 3, and 3 ppm. Notably, we observed slightly higher background base error rates in this cohort compared to a previous study,7 possibly explaining the four samples in which we observed low-level false detection (Figure S4).
Having shown that MAESTRO-Pool could identify and enrich low-frequency patient-specific mutant alleles in patients with DLBCL, we next sought to compare MAESTRO-Pool with IgHTS using ClonoSeq®, a Food and Drug Administration-approved, commercially available MRD assay (Figure 1C). MAESTRO-Pool identified ctDNA before recurrence for all five relapsing patients, including at the earliest available time point for four of five relapsing patients. The time from ctDNA detection to clinical relapse (lead time) was numerically longer for MAESTRO-Pool (median 178 days, range 69–518) compared to IgHTS (median 44 days, range not detected to 518 days) (p = 0.37). MAESTRO-Pool was associated with improved sensitivity compared to IgHTS (MAESTRO-Pool sensitivity of 80.6% for all samples and 90.5% for samples with matched ClonoSEQ results versus ClonoSEQ sensitivity of 61.9%) (p = 0.006), while maintaining high specificity (100% for both MAESTRO-Pool and ClonoSeq). Superior sensitivity as compared to ClonoSeq was primarily driven by MAESTRO-Pool's improved detection at ctDNA fractions below 1000 ppm. IgHTS detected ctDNA in 10/12 samples with a MAESTRO-Pool tumor fraction ≥1000 ppm, but in only 3/7 samples with a MAESTRO-Pool tumor fraction <1000 ppm (Figure 1D).
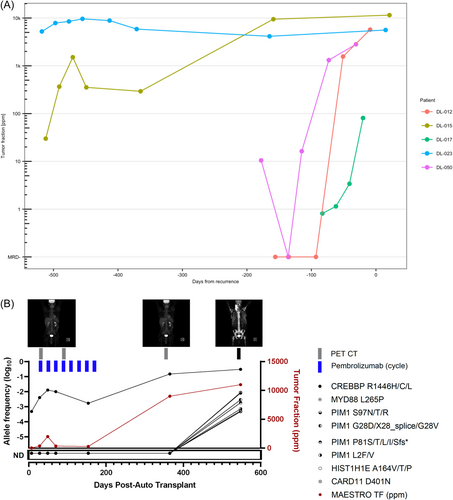
MRD generally increased with proximity to clinical relapse, but the rate of increase varied between patients (Figure 2A). Using an additional bait set of genes commonly mutated in relapsed DLBCL, we identified multiple novel mutations in relapsing patients. Notably, plasma from a patient who progressed 18.5 months post-ASCT (DL-015) manifested an emergent CREBBP R1446H mutation whose allele frequency increased from 0.048% after ASCT to 30.53% at relapse (Figure 2B).
In this pilot study, we show that ctDNA identified using MAESTRO-Pool is a sensitive marker of relapse among patients with relapsed/refractory DLBCL. Compared to IgHTS, MAESTRO-Pool appears to be associated with improved sensitivity while maintaining high specificity. By enriching low-frequency patient-specific mutant alleles, MAESTRO-Pool yielded similar results to an orthogonal assay (MRD Tracker) but required 13-fold less sequencing. Finally, our results demonstrate that this approach can also track genetic evolution by using complementary targeted sequencing approaches.
Maximizing sensitivity while maintaining near-perfect specificity is critical for the development of MRD-adapted treatment strategies in DLBCL. Newer panel-based MRD approaches have demonstrated improvements in sensitivity, but may still be insufficiently sensitive. For example, early ctDNA dynamics assessed using the AVENIO CAPP-Seq assay in the phase III POLARIX trial were associated with PFS, but the effect difference was relatively modest (−20% difference in 2-year PFS).12 Ultrasensitive MRD techniques, like MAESTRO-Pool, which rely upon more tumor reporters, may be better suited for response-adapted treatment approaches in DLBCL which could escalate or de-escalate treatment based on individual patient risk.
One potential downside of personalized WGS-based assays is that they do not capture new mutations at disease progression. Here, we show that the MAESTRO-Pool approach can be supplemented by adding a panel of relevant genes to look for treatment-emergent alterations. Even in the small panel tested, we identified novel mutations arising at the time of relapse, including a hotspot mutation within CREBBP's lysine acetyltransferase domain that has been associated with increased MYC expression13 and decreased MHC class II expression14 in prior studies.
Personalized MRD approaches do present unique challenges to clinical implementation. MAESTRO-Pool requires WGS and individualized probe design, but the cost of each continues to decline. While this approach entails upfront costs, MAESTRO-Pool enables the detection of low-abundance mutations with significantly fewer reads, and the upfront cost can be amortized over serial MRD samples.
The small number of patients studied and the retrospective approach are clear limitations of our work. Despite this, our results suggest that bespoke WGS liquid biopsy in DLBCL may offer unique advantages in optimizing the sensitivity of MRD detection, maintaining very high specificity, and allowing tracking of clonal evolution. The results of our pilot study support additional investigation using MAESTRO-Pool in DLBCL.
ACKNOWLEDGMENTS
The authors acknowledge the Gerstner Family Foundation for its generous support of this work. Reid W. Merryman, Mikaela McDonough, and Philippe Armand would like to acknowledge support from the Harold and Virginia Lash Grant Program. Reid W. Merryman would like to acknowledge support from an American Society for Transplantation and Cellular Therapy New Investigator Award and a Lymphoma Research Foundation Clinical Investigator Career Development Award.
AUTHOR CONTRIBUTIONS
Reid W. Merryman designed the research, performed research, analyzed the data, and wrote the paper. Justin Rhoades, Kan Xiong, Mark Murakami, and Viktor A. Adalsteinsson designed the research, performed research, analyzed the data, and edited the paper. Robert A. Redd analyzed the data and reviewed the paper. Katherine Antel, Hyun Hwan An, Mikaela McDonough, Liliana Guerrero, Andela Crnjac, Sainetra Sridhar, Timothy Blewett, Ju Cheng, Parastoo B. Dahi, Yago Nieto, Robin M. Joyce, Yi-Bin Chen, Alex F. Herrera, and Philippe Armand performed research and reviewed the paper.
CONFLICT OF INTEREST STATEMENT
Reid W. Merryman—Advisory board: Genmab, Adaptive Biotechnologies, Bristol Myers Squibb, Abbvie, Inteillia, Epizyme; consulting: Alphasights; institutional research funding: Merck, Bristol Myers Squibb, Genmab, Genentech/Roche. Yago Nieto—Consulting: Affimed, Novonordisk; research funding: Novartis, Biosecura, Astra-Zeneca, Affimed, Takeda. Yi-Bin Chen—Consulting: Incyte, Jasper, Gamida Cell, Daiichi, Celularity, Equilium, Actinium. Alex F. Herrera—Consulting: Bristol Myers Squibb, Genentech, Merck, Seattle Genetics, AstraZeneca, Karyopharm, ADC Therapeutics, Takeda, Tubulis, Genmab, Regeneron; research funding: Bristol Myers Squibb, Genentech, Merck, Seattle Genetics, KiTE Pharma, Gilead Sciences, AstraZeneca, ADC Therapeutics. Philippe Armand—Consulting: Merck, BMS, Pfizer, Affimed, Adaptive, Infinity, ADC Therapeutics, Celgene, Morphosys, Daiichi Sankyo, Miltenyi, Tessa, GenMab, C4, Enterome, Regeneron, Epizyme, AstraZeneca, Genentech; research funding (institutional): Merck, BMS, Affimed, Adaptive, Roche, Tensha, Otsuka, Sigma Tau, Genentech, IGM, Kite; Honoraria: Merck, BMS. Mark Murakami—Advisory board: Novartis, CancerModels.org; research funding: Generate Biomedicines, Genentech/Roche. Viktor A. Adalsteinsson is a co-inventor on a patent application covering MAESTRO (US 2023/0203568, pending) which has been licensed to Exact Sciences which was not involved in this study; receives research funding from Exact Sciences; and is a co-founder and advisor to Amplifyer Bio which was not involved in this study. The remaining authors declare no conflict of interest.
FUNDING
This research received no funding.
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.