Compound heterozygosity of a De novo 16q24.1 deletion and missense mutation in COX4I1 leads to developmental regression, intellectual disability, and seizures
Abstract
The COX4I1 is responsible for encoding a crucial component of cytochrome c oxidase, integral to electron transport in the mitochondrial respiratory chain. Mutations in COX4I1 can result in a rare autosomal recessive disorder characterized by growth retardation, slow weight gain, microcephaly, and potentially, hematologic symptoms such as Fanconi anemia or neurological impairments including developmental regression and severe epilepsy. In this study, we report the first case of COX4I1 deficiency in China, identified in a 6-year-old boy. The patient exhibited developmental regression, epilepsy, low body weight, microcephaly, generalized muscle hypotonia, and progressive cerebral atrophy, but without hematologic damage or short stature. Compound heterozygosity for a de novo 16q24.1 deletion and a P152T missense mutation in the COX4I1 was detected. The P152T missense mutation is previously reported in patients with similar clinical manifestations. Additionally, we provide the first instance of progressive brain atrophy observed through MRI in a COX4I1 deficiency patient, broadening our understanding of the mutation spectrum and clinical phenotype of this genetic disorder.
Plain Language Summary
We discovered the first case of COX4I1 deficiency in China, identified in a 6-year-old boy. The patient exhibited developmental regression, epilepsy, low body weight, microcephaly, generalized muscle hypotonia, and progressive cerebral atrophy, but without hematologic damage or short stature. Compound heterozygosity for a de novo 16q24.1 deletion and a P152T missense mutation in the COX4I1 was detected. Additionally, we provide the first instance of progressive brain atrophy observed through MRI in a COX4I1 deficiency patient, broadening our understanding of the mutation spectrum and clinical phenotype of this genetic disorder.
Key points
- Reported the first case of COX4I1 deficiency in China, expanding our knowledge of the mutation spectrum and clinical phenotype associated with this genetic disorder.
1 INTRODUCTION
Cytochrome c oxidase (COX), a key enzyme complex essential for cellular respiration, exists as a dimer and comprises 14 subunits in mammalian cells.1 A complex array of assembly proteins orchestrates the synthesis and assembly of COX, ensuring the cohesive integration of its subunits. Of particular note is COX4I1, which is prevalently expressed under standard physiological conditions and serves as a critical structural and functional regulator within the COX complex. The indispensable role of COX4I1 is underscored by its contributions to both the structural integrity and the regulatory capacity of the enzyme complex, which is pivotal for the efficient production of cellular energy, especially in the form of adenosine triphosphate (ATP).2
To date, there have only been two reports on disease caused by mutations in the COX4I1 worldwide, with marked clinical heterogeneity. Common clinical features include short stature, slow weight gain, and microcephaly. One of the patients showed clinical symptoms of Fanconi anemia and did not present symptoms of nervous system involvement.3 The other two siblings, from a family with consanguineous marriage, showed neurological symptoms such as developmental regression, epilepsy, and hypotonia, and magnetic resonance imaging (MRI) reveals changes resembling those of Leigh syndrome, but there were no signs of hematological system involvement.4
Here, we described a case of normal height, developmental regression, epilepsy, microcephaly, and without hematologic symptoms caused by compound heterozygosity of a de novo 16q24.1 deletion and missense mutation in the COX4I1.
2 METHODS
The patient's DNA sample was subjected to exome enrichment using the Nextera Rapid Capture Exome v1.2 kit (Illumina). Sequencing was performed on the HiSeq4000 platform (Illumina), generating 150-bp paired-end reads. Data analysis was conducted using Berry Genomics' proprietary Verita Trekker Variant Detection System and the Enliven Variant Annotation and Interpretation System, with reference to the human genome assembly hg19 (GRCh37). Informed consent for the DNA studies was provided by the patient's parents.
Variants were excluded if they failed to meet the following quality control criteria: a depth of coverage <10, an allele balance below 0.25, or a Phred quality score below 20. For variants subject to detailed analysis, we applied stringent parameters: (1) heterozygous variants with a minor allele frequency (MAF) below 0.1% in any of the following databases: 1000 Genomes (http: //browser.1000genomes.org), NHLBI-ESP project (esp6500s, http://evs.gs.washington.edu/EVS), The Genome Aggregation Database (GnomAD, http://gnomad.broadinstitute.org), Exome Aggregation Consortium (ExAC, http://exac.broadinstitute.org), and our in-house WES data; (2) homozygous or compound heterozygous variants with an MAF less than 1% in the above public databases; and (3) stop gain/loss, frameshift, splice-site variants and missense variants.5 Additionally, Copy Number Variation (CNV) analysis was performed using whole-exome sequencing data and validated by quantitative PCR (qPCR).
3 RESULTS
3.1 Case report
We present the case of a 6-year-old male child, initially diagnosed with developmental intellectual disability at the age of 5 years and 9 months. Born at full-term without incidents during gestation or delivery, his birth measurements included a head circumference of 39 cm (above the 97th percentile) and body weight of 3.95 kg. The first 4 months post-birth were marked by normal developmental milestones. However, at the age of 5 months, his parents observed a reduced interaction with his surroundings. A head MRI at this stage showed a slight widening of the subarachnoid space in the bilateral frontal and temporal regions (Figure 1B). By the eleventh month, the child began experiencing epileptic seizures characterized by upward eye deviation, spasm-like nodding, and embracing motions. The local hospital diagnosed him with infantile spasms and prescribed a treatment regimen including topiramate, valproic acid, and adrenocorticotropic hormone (ACTH). Despite these interventions, seizure control was not achieved. By the age of four, an increase in seizure frequency was observed, sometimes triggered by auditory stimuli. The addition of levetiracetam to his treatment regimen did not result in seizure cessation.
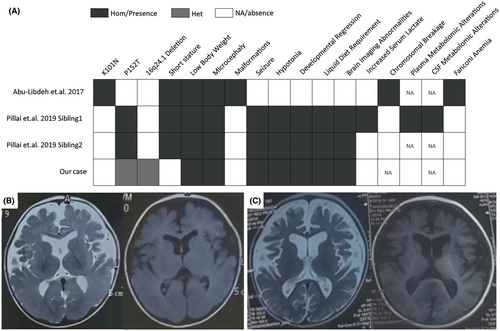
In the current assessment, the patient presents with normal stature (height of 117 cm, 50th percentile), microcephaly (head circumference of 45 cm, −3 SD), and body weight below the 3rd percentile (15 kg), resulting in a Body Mass Index (BMI) of 10.96 kg/m2. He exhibits global developmental delays and generalized hypotonia, with an inability to sit unassisted, speak, recognize family members, maintain visual tracking, or chew solid food. His nutritional needs are met through pureed food. Video electroencephalogram (EEG) readings show the presence of sleep spindles and occasional episodes of widespread multifocal and generalized intermediate to low amplitude spike waves, polyspike waves, spike-slow waves, polyspike-slow waves, spiking rhythms, irregular sharp waves, and sharp-slow waves during various stages of wakefulness and sleep. A consistent rhythm of 5–6 Hz is observed bilaterally from the frontal lobes during consciousness. Seizure episodes are characterized by frequent, generalized EEG patterns of intermediate to low amplitude multiphasic slow waves coupled with low amplitude fast rhythms, lasting approximately 1 s, with concurrent rapid tremors of the limbs, upper arms, or jaw visible on the video. MRI reveals significant enlargement of the ventricles and marked deepening and widening of the cerebral sulci, suggesting progressive cerebral atrophy (Figure 1B,C). Routine diagnostic results, including blood amino acids, carnitine, plasma amino acids, urine organic acids, and creatine kinase (CK) levels, were all within normal ranges.
3.2 Genetic results
Trio exome sequencing revealed a homozygous c.454C>A (p.P152T) (NM_001861.6) variant in the COX4I1 of the proband (II:2). Familial mutation testing established the presence of the c.454C>A variant in the father (I:1) in a heterozygous state, while it was absent in the mother (I:2). Sanger sequencing further confirmed these findings. In addition, the variant was not present in the proband's healthy brother (II:1, Figure 2A, B). The homozygous P152T variant, previously reported by Pillai et al. to be associated with clinical manifestations similar to those observed in the proband,4 is not listed in the public genetic databases mentioned in the methods section. Analysis using the VarCards database,6 which integrates predictions from 23 functional prediction software tools including SIFT, PolyPhen-2, VEST3, CADD, and REVEL, indicates that 21 of these tools predict the variant as pathogenic (Table S1). In alignment with the criteria set forth by the American College of Medical Genetics and Genomics (ACMG), the variant in question was classified as likely pathogenic (PP3_Moderate+PP1 + PS3_Supporting+PM2_Supporting+PM3_Supporting).7
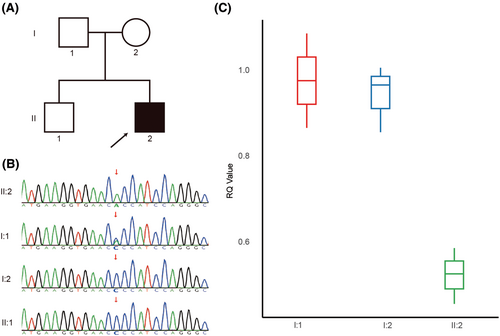
In an effort to elucidate the absence of the variant in the proband's mother despite the patient's homozygous state, we conducted a CNV analysis based on whole-exome sequencing data, focusing on the COX4I1 for potential CNV variants. The analysis revealed that the patient harbors a heterozygous deletion of approximately 0.13 Mb (chr16:85706011–85 840 495) in the 16q24.1 region, encompassing eight genes, including COX4I1 (Figure S1). We used three pairs of primers located on COX4I1 for qPCR analysis. The results indicated that the proband had only one copy, while the parents had two copies, confirming the presence of a heterozygous deletion (Figure 2C).
4 DISCUSSION
COX, known also as complex IV, COX comprises 14 subunits that catalyze the electron transfer from ferro-cytochrome c to molecular oxygen.1 This exergonic reaction is crucial not only significantly contributes to the electrochemical gradient necessary for ATP synthesis but also facilitates the transfer of protons across the inner membrane. COX4I1, a key protein within the mitochondrial electron transport chain,8 is vital due to its ubiquitous presence across species, highlighting its essential role in aerobic respiration and evolutionary conservation.
Our report is the second to describe pronounced neurological impairments associated with COX4I1 mutations, outlining a spectrum of distinct clinical features. Notably, our patient exhibits a homozygous variant due to a compound heterozygous missense mutation paired with a small deletion in the 16q24.1 region, expanding the known mutation spectrum of COX4I1. The identification of a deletion in the 16q24.1 region associated with the patient's condition is unprecedented, marking a novel genetic discovery in the context of COX4I1-related disorders. The duplications in the 16q24.1 region have been conclusively linked to intellectual disability, spastic paraplegia, and severe epilepsy, reinforcing the region's critical role in neural development.9 Remarkably, both deletions and duplications in this region lead to similar clinical outcomes, highlighting the delicate balance of genomic content in this area. Additionally, we observed that besides COX4I1, the region also contains genes such as GSE1, GINS2, C6orf74, MIR1910, EMC8, RNU-103P, and LOC01928557. Among these, RNU-103P is a pseudogene, and the MIR1910 encodes a microRNA. Currently, there is no evidence linking these two genes or LOC01928557 to neurodevelopmental phenotypes. According to the gnomAD database, the pLI values for GINS2, C6orf74, and EMC8 are relatively low (<0.03, Table S2), suggesting that heterozygous deletions are unlikely to cause significant symptoms. The GSE1, however, has a high pLI value of 1.0, and the DECIPHER database includes cases involving deletions of the GSE1. Nonetheless, there are no reported cases where a functional loss mutation of the GSE1 alone leads to symptoms like those observed in our patient.
Distinct from previous cases, our patient does not have short stature, aligning with the 50th percentile for height, which may be attributed to the genetic predisposition for taller stature based on parental heights (with a paternal height of 184 cm and a maternal height of 165 cm). The patient's low body weight and BMI may be attributed to the lack of chewing ability, necessitating a liquid diet, which can lead to inadequate nutritional intake. Additionally, the impact of the genetic mutation itself could be contributing to these observations. This emphasizes the importance of careful energy intake assessment in similar patients.
We provide evidence of MRI brain damage that progressively worsens with age, correlating with the patient's clinical symptoms of developmental regression. This demonstrates the ongoing progression of neurological damage, thus underlining the necessity for early diagnosis and the implementation of protective mitochondrial treatments, such as Coenzyme Q10 supplementation,4 as imperative.
The mechanisms underlying the broad clinical spectrum of COX4I1 deficiency are not fully understood. Accumulative DNA damage in Ki-67-positive cells associated with COX4I1 deficiency10 may partially explain Fanconi anemia-like symptoms, as observed with the homozygous K101N mutation. Both our case and Pillai's involve the P152T mutation,4 predominantly presenting neurological symptoms without Fanconi anemia. The reasons for these differences remain elusive, but there are still some clues available for reference. Pillai et al. discovered that the mitochondrial respiratory chain enzyme analysis conducted on a muscle biopsy specimen from a patient homozygous for the P152T mutation indicated that COX activity was reduced to 16% of residual activity in comparison to control values.4 Similarly, Abu-Libdeh et al.3 in fibroblasts from a patient with the homozygous K101N mutation exhibited COX activity reduced to approximately 24.8% (9.1/36.9) of control levels. COX plays a role in safeguarding neurons from hypoxic and neurotoxic damages.8, 11 Brain tissue has a lower tolerance to hypoxic events compared to other tissues, and functional abnormalities in COX4I1 impair this tolerance. The K101N mutation, which retains higher residual COX activity compared to the P152T mutation, may contribute to the absence of neurological symptoms in patients with the K101N mutation. Indeed, since the COX activity data was not derived from the same type of cells, the evaluation may not be entirely accurate. Future research into the phenotypic and metabolic changes caused by COX4I1 mutations in neural tissues will help to explain such clinical differences.
In summary, our findings demonstrate a case of COX deficiency caused by a missense mutation and a de novo 16q24.1 deletion in the COX4I1. The patient presents with developmental regression, intellectual disability, epileptic seizures, microcephaly, and progressive cerebral atrophy, yet maintains a normal stature and without the Fanconi anemia phenotype. This further expands the mutational and phenotypic spectrum of autosomal recessive COX4I1 deficiency.
AUTHOR CONTRIBUTIONS
Conceptualization: Zhen Liu and Xiao Mao. Methodology: Zhen Liu and Mei He. Data curation: Xuan Luo. Writing—original draft preparation: Zhen Liu. Writing—review and editing: Mei He, Xuan Luo, Hu Pan and Jinping Su. Supervision: Xiao Mao, Jinping Su. Project administration: Xiao Mao, Jinping Su. Funding acquisition: Xiao Mao, Jinping Su. All authors have read and agreed to the published version of the manuscript.
ACKNOWLEDGMENTS
We thank the child and his family for their support of this study. This research was funded by Natural Science Foundation of Hunan Province, China (grant no.2022JJ40206).
CONFLICT OF INTEREST STATEMENT
The authors declae no conflicts of interest. All the authors have read the Journal's position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
ETHICS STATEMENT
This study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board (IRB) of Hunan Provincial Maternal and Child Health Care Hospital. Written informed consent was obtained from the legal guardians of all participants involved in the study. To ensure the protection of our patient's and control subjects' privacy, all personal identifiers were removed from the study data.