Pax-5 is a key regulator of the B cell-restricted expression of the CD23a isoform
Abstract
Human CD23 (the low affinity IgE receptor) is a B cell differentiation marker involved in inflammatory responses. Two isoforms (CD23a and CD23b) are known, which differ only in their cytoplasmic domain. Whereas CD23b expression is specifically induced by IL-4 on B cells and cells of the myeloid lineage, CD23a expression is restricted to B cells. Each isoform is regulated by its own promoter. Pax-5 is a B-cell-restricted transcription factor with an essential role in early and late B cell development. Analyses of the CD23a promoter revealed a Pax-5-binding site, which can compete a high affinity Pax-5-binding site or directly bind Pax-5 protein in electrophoretic mobility shift assays. Introducing mutations into this site abrogates the binding. Expression of Pax-5 in 293 cellsresulted in a seven- to tenfold activation of a CD23a core promoter construct. Most importantly, ectopic expression of Pax-5 in the monocytic cell line U-937, which regularly expresses only the CD23b isoform, led to CD23a expression after stimulation with IL-4 and PMA. Our results suggest that Pax-5 is a key regulator of the B-cell-restricted expression of the CD23a isoform.
Abbreviation:
-
- EMSA:
-
Electrophoretic mobility shift assay
1 Introduction
The CD23 molecule is a type II membrane glycoprotein exhibiting substantial homology with several Ca2+-dependent animal lectins 1, 2. Two isoforms of human CD23 have been described: CD23a is restricted to B cells, whereas CD23b is induced by IL-4 on B cells and a variety of hematopoietic cells including monocytes/macrophages, T lymphocytes, eosinophils and platelets 3. The two isoforms differ only by a few residues in their N-terminal intracytoplasmic tail 4.
Initially characterized as the low affinity IgE receptor, CD23 is involved in a variety of biological processes such as IgE-dependent inflammatory processes on macrophages, eosinophils and platelets 5, 6, antigen presentation on B cells 7–9 and homotypic/heterotypic interactions between B and T cells 10, 11. CD23 is highly expressed on certain B cell neoplasms like B-CLL where the soluble form of CD23 can be used as a prognostic factor 12. In addition, it is involved in models of arthritic inflammation where inhibition of CD23 ameliorates disease 13.
It seems probable that the two isoforms have distinct functions, with CD23b controlling IgE-dependent cytotoxicity and CD23a, which contains an endocytosis signal in its intracytoplasmic tail,mediating endocytosis of bound ligands 14. This would correlate with the fact that CD23a and CD23b are connected to different signaling cascades. CD23b is involved in up-regulating cAMP and iNOS, whereas CD23a mediates an increase in intracellular calcium 15, 16. Additionally, recent observations show that there is distinct regulation of the two promoters 17.
The gene for CD23 is located on chromosome 19. The two isoforms are generated by using individual promoters and alternative RNA splicing. Given the genomic sequence of CD23a as a reference, CD23b mRNA lacks the first two exons and starts with an optional exon that is located within intron II 4. The CD23a promoter sequence was first described by Suter et al. 18 and the CD23b promoter was identified by Yokota et al. 4. The two promoters have been analyzed by several groups, and several transcription factors and binding sites have been described, among others the IL-4-responsive element 19–21.
The CD23a isoform is of particular interest since its selective expression in B-CLL correlates with a state of cell survival 22. CD23a is also closely associated with EBV mediated immortalization of B cells — EBNA2 binds and activates the CD23a proximal promoter 21. So far, nothing is known on how the B-cell-specific expression of CD23a is regulated. Among B-cell-specific transcription factors, Pax-5 was a good candidate; putative binding sites for Pax-5/BSAP have already been predicted in the CD23a proximal promoter 17, 23.
Pax-5 is a member of the Pax family of transcription factors with an essential role in early and late B cell development. In addition to all B lymphoid organs, Pax-5 can also be found in embryonic brain and adult testis of the mouse 24. During B cell development, the Pax-5 gene is expressed from the earliest B-lineage-committed precursor cell up to the mature B cell stage, but not in terminally differentiated plasma cells 25. Nutt et al. identified Pax-5 as an essential B lineage commitment factor, which exerts its function by activating the expression of B-lymphoid-specific genes and by repressing the transcription of lineage-inappropriate genes 26–28.
Detailed mutational analyses of Pax-5 revealed the bipartite structure of a paired domain and led to the identification of a consensus recognition sequence. Pax-5 is known to recognize DNA via its N-terminal paired domain 29 and to control gene transcription through a C-terminal regulatory module consisting of activating and inhibitory sequences 30. CD19, XBP-1, mb-1 and RAG-2 have all been shown to be Pax-5-responsive target genes 31–34. The generation of a mouse strain in which the Pax-5 gene can be conditionally inactivated allowed the analysis of Pax-5 function in mature B cells 35. Loss of Pax-5 in these cells resulted in a change of B cell subpopulations in the periphery, with down-regulation of several mature cell surface B cell markers, including CD23. This suggests a possible role for Pax-5 in the control of the CD23 gene.
Furthermore, the similar expression patterns between Pax-5 and CD23a, which are both restrictively expressed on B cells and lost after differentiation into plasma cells, encouraged us to investigate the potential role of Pax-5 as a regulator of CD23a expression. The sequence analysis of the CD23a core promoter revealed three putative Pax-5-binding sites. Electrophoretic mobility shift assays (EMSA) and competition experiments confirmed one site as a functional Pax-5-binding site, which could further be verified by mutational analysis. Luciferase assays in 293 cells demonstrate activation of the CD23a core promoter by Pax-5. Furthermore, we were able to show that stable transfection of Pax-5 via a retroviral transduction system in the monocytic cell line U-937, which regularly expresses only the CD23b isoform, allows inducible CD23a expression in these cells. These results point to Pax-5 as a key regulator of the B-cell-specific expression of CD23a.
2 Results
2.1 The CD23a core promoter contains three putative binding sites for Pax-5
The consensus motif for Pax-5 has been characterized by Czerny et al. in an extended comparison between natural ligands of Pax-5, including the recognition sequences that originate from the sea urchin H2A-2.2 gene and the human CD19 gene 29.
To identify putative Pax-5-binding sites in the CD23a promoter we performed multiple-sequence alignment between the CD23a core promoter (Fig. 1A) and the Pax-5 consensus sequence. Three putative Pax-5-binding sites were identified in positions –61 to –79, –78 to –96 and –215 to –233 of the CD23a promoter (Fig. 1) and named CD23-1, CD23-2 and CD23-3, respectively. Fig. 1B shows the sequence alignment of these three putative binding sites with the consensus motif and other previously identified Pax-5-binding sites from the sea urchin H2A-2.2 and the human CD19 genes. The CD23-3 putative binding site displayed the highest homology with the consensus motif (89%). A gap had to be introduced into the putative binding site CD23-1 in order to reconstitute the symmetry of the recognition sequence.
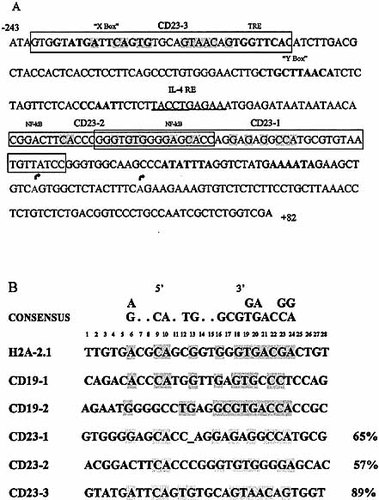
CD23a core promoter and consensus recognition sequences for Pax-5. (A) The CD23 core promoter (–243 to +82): the three putative Pax-5-binding sites are shown with the homologous nucleotides shaded. The boxes represent the synthetic oligonucleotides containing the three putative binding sites: CD23-1, CD23-2 and CD23-3. The STAT6-binding site (IL-4-responsive element) is underlined; the localization of NF-κB-binding sites is indicated by analogy with the murine CD23 promoter. The arrows (positions 1 and 16) show the beginning of exon Ia (5′ UTR). (B) Pax-5-recognition sequences: the three putative Pax-5-binding sites — CD23-1, CD23-2, CD23-3 — are shown in comparison with Pax-5-binding sites from the sea urchin H2A-2.2 and the human CD19 promoters. The orientation of the sequences corresponds to the natural orientation of the Pax-5-binding sites. Consensus sequences are shaded and the deduced consensus sequence 29 is shown above.
2.2 Pax-5 protein interacts with the CD23-1 binding site from the CD23a core promoter in vitro
We tested the three putative binding sites for their ability to bind Pax-5 protein. In the first-stage, synthetic oligonucleotides representing CD23-1, CD23-2 and CD23-3 — the three putative binding sites from the CD23a core promoter (see Sect. 4) — were used in EMSA competition experiments as cold competitors (Fig. 2A). EMSA were done using a high affinity Pax-5-binding site from the sea urchin H2A-2.2 gene and in-vitro-transcribed and translated Pax-5 protein. The plasmid construct used as template for Pax-5 translation generated an additional unspecific band (Fig. 2A, lane 1). The correct DNA–protein complex containing Pax-5 was verified by supershift experiments with a monoclonal antibody against Pax-5 (Fig. 2A, lanes 2, 3). Complex-formation was inhibited by a 10-fold excess of the oligonucleotide CD23-1, but not by an excess of oligonucleotides CD23-2 and CD23-3. For comparison, we also competed the high affinity Pax-5-binding site H2A-2.2 with itself (Fig. 2A, lanes 4–6). CD23-1 showed an approximately 10-times-lower affinity than the H2A-2.2 binding site. Both CD23-2 and CD23-3 oligonucleotides failed to compete the high affinity binding site.
To confirm the results obtained in competition assays the same oligonucleotides representing the putative CD23-1, CD23-2 and CD23-3 sites were appropriately labeled and checked for direct binding (Fig. 2B). The specificity of the complexes formed was confirmed by supershifts using a monoclonal antibody against Pax-5. CD23-1 was the only site at which we observed a specific complex formation with Pax-5 (Fig. 2B, lanes 4, 5).
To further analyze the CD23-1 site we constructed oligonucleotides in which the Pax-5-binding site was mutated and tested again their ability to interact with Pax-5, using EMSA. Since Pax-5 is able to interact with a panel of seemingly degenerate recognition sequences it is quite difficult to predict which nucleotides are essential in the protein–DNA interaction 29. For this reason we created two different mutated variants of the CD23-1 putative site (Fig. 3A). In CD23-1mu1, all nine nucleotides considered to be responsible for creating the Pax-5 binding site were mutated. CD23-1mu2 contains only two substitutions (C to A in position 4 and G to T in position 11 of the consensus motif), since these nucleotides seem to act like key points for Pax-5–DNA interaction 25, 29.
The ability of the two mutant oligonucleotides to bind to Pax-5 protein was subsequently tested by EMSA. As shown in Fig. 3B, both oligonucleotides CD23-1mu1 and CD23-1mu2 containing the mutated CD23-1 binding site were unable to compete, at any concentration, the sea urchin high affinity H2A-2.2 Pax-5-binding site. Direct binding assays showed that the oligonucleotides containing the mutated sites failed to bind Pax-5 protein (Fig. 3C). These results give additional evidence that CD23-1 is a Pax-5-binding site since mutations introduced in this site completely abrogate the binding of the protein.
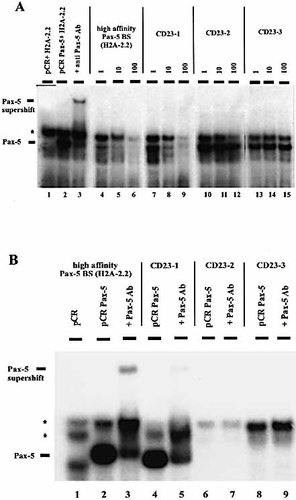
The CD23-1 binding site interacts with Pax-5. (A) An oligonucleotide containing the optimal Pax-5-binding site from the sea urchin H2A-2.2 gene was radioactively labeled and incubated with Pax-5 protein obtained by in vitro transcription and translation. Unlabeled oligonucleotides representing the optimal Pax-5-binding site H2A-2.2 (lanes 4–6) or containing the three putative Pax-5-binding sites from the CD23a promoter (lanes 7–15) were used as competitors (ratios of 1:1, 1:10 and 1:100 labeled H2A-2.2 : unlabeled competitor). Lane 1 shows the empty vector (pCR), used for in vitro transcription and translation of the Pax-5 protein, incubated with the labeled oligonucleotide, which forms an unspecific complex (marked by the asterisk). Lane 2: specific Pax-5 complex. Lane 3: Pax-5 supershift with anti-Pax-5 antibody. (B) Oligonucleotides containing the optimal Pax-5-binding site from the sea urchin H2A-2.2 gene (lanes 2, 3) and the three putative Pax-5-binding sites from the CD23a promoter (lanes 4–9) were radioactively labeled and incubated with Pax-5 protein obtained by in vitro transcription and translation. Pax-5 antibody was added to check the specificity of the bands (lanes 3, 5, 7 9). Lane 1 shows the empty vector (pCR), used for in vitro transcription and translation of the Pax-5 protein, incubated with the labeled high affinity site; the asterisks show unspecific complexes.
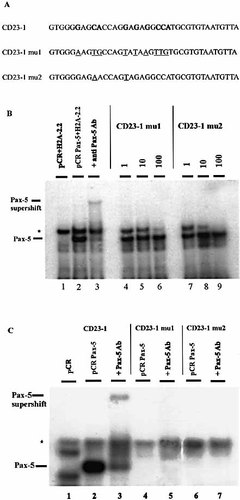
Mutations in the CD23-1 binding site prevent Pax-5 binding. (A) CD23-1mu1 and CD23-1mu2 oligonucleotides contain mutated variants of the CD23-1 putative Pax-5-binding site. Mutated nucleotides are underlined. (B) An oligonucleotide containing the high affinity Pax-5-binding site from the sea urchin H2A-2.2 gene was radioactively labeled and incubated with Pax-5 protein obtained by in vitro transcription and translation. Unlabeled oligonucleotides containing mutated variants of the CD23-1 putative binding sites (lanes 4–9) were used as competitors (ratios of 1:1, 1:10 and 1:100 labeled H2A-2.2:unlabeled competitor). Lane 1 shows the empty vector (pCR), used for in vitro transcription and translation of the Pax-5 protein, incubated with the labeled oligonucleotide, which forms an unspecific complex (asterisk). Lane 2: specific Pax-5 complex. Lane 3: Pax-5 supershift. (C) Oligonucleotides containing the putative CD23-1 binding site (lanes 2, 3) and mutated sites CD23-1mu1 and CD23-1mu2 (lanes 4–7) were radioactively labeled and incubated with Pax-5 protein obtained by in vitro transcription and translation. Pax-5 antibody was added to check the specificity of the bands (lanes 3, 5, 7). Lane 1 shows the empty vector, (pCR) used for in vitro transcription and translation of the Pax-5 protein, incubated with the labeled CD23-1 site; the asterisk shows an unspecific complex.
2.3 Pax-5 mediates the activation of the CD23a promoter in vitro
To examine the effect of Pax-5 on the activation of the CD23a promoter, a luciferase-reporter construct containing the CD23a core promoter (from position –203 to position +83) cloned in the pLuc+ vector was made and named pLuc+ACP (CD23a core promoter). Another construct, pcDNA3-Pax-5, was used for ectopic expression of Pax-5. The cell line 293 was transfected with the luciferase construct pLuc+ACP alone or together with pcDNA3-Pax-5, and luciferase activity was determined after 40 h under nonstimulatory conditions or after stimulation with IL-4 and PMA. As seen in Fig. 4B, Pax-5 activates the CD23a core promoter (pLuc+ACP) about sevenfold when compared with the activity of the promoter without Pax-5.
By site-directed mutagenesis, we mutated the CD23-1 putative binding site within the pLuc+ACP vector. The mutant construct, named pLuc+ACPmu, contains the same nucleotide substitutions as the oligonucleotide CD23-1mu1 (Fig. 3A). The pLuc+ACPmu construct containing the CD23a core promoter with the mutated Pax-5-binding site failed to activate as strongly as the pLuc+ACP construct containing the wild-type promoter. This is in line with our conclusion that the CD23-1 binding site behaves as a functional Pax-5-binding site.
A recent study 17 identified IL-4 as a main activator of the CD23a promoter, whereas the CD23b promoter showed a much wider range of responsiveness to extracellular stimuli. The authors described two STAT6-binding sites located close to each other in the –500 to –350 region of the CD23a promoter. Therefore we investigated whether Pax-5, besides controlling the specificity of CD23a expression in B cells, also cooperates with STAT6 in inducing a strong expression of the isoform.
For this purpose, another luciferase-reporter construct (pLuc+AP) that includes these STAT6-binding sites was made by cloning the whole CD23a promoter (from position –1216 to position +211) into the pLuc+ vector. For ectopic expression of Pax-5 and STAT6, pcDNA3-Pax-5 and pXM-STAT6, respectively, were used in 293 cells. As shown in Fig. 4C, Pax-5 and STAT6 induced a 14-fold and 12-fold activation of the CD23a promoter, respectively. When the cells are co-transfected with both transcription factors, however, a 40-fold activation of the CD23a promoter is induced. These results suggest that Pax-5 not only stimulates CD23a expression, but also cooperates with STAT6 in enhancing the level of CD23a transcription.
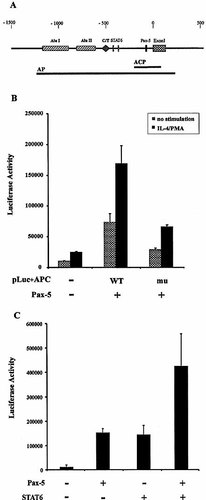
Pax-5 mediates the activation of the CD23a promoter. (A) Schematic representation of reporter constructs used. (B) 293 cells were transfected with the vector containing the CD23a core promoter (pLuc+ACP WT, positions –203 to +83) or the vector containing the CD23a core promoter in which the nucleotides of the putative Pax-5-binding site CD23-1 were mutated (pLuc+ACPmu). Where indicated cells were co-transfected with pcDNA3-Pax-5. Luciferase activity was assayed after 40 h under nonstimulatory conditions or after stimulation with IL-4 (50 ng/ml) and PMA (3 ng/ ml). The average luciferase value of three independent experiments is shown. Error bars indicate standard deviation of the mean. (C) 293 cells were transfected with the vector containing the CD23a promoter (pLuc+AP, positions –1216 to +211) together with pcDNA3-Pax-5 and pXM-STAT6 as indicated. Luciferase activity was assayed after 40 h. The average luciferase value of three independent experiments is shown. Error bars indicate standard deviation of the mean.
2.4 Pax-5 mediates CD23a expression in vivo
To further investigate whether the B-cell-restricted expression pattern of CD23a is determined by lineage-specific Pax-5 expression in vivo we used the monocytic cell line U-937 to determine the effects of ectopic Pax-5 expression on CD23a induction. U-937 cells regularly express only CD23b after appropriate stimulation with IL-4.
Human Pax-5 cDNA was stably inserted into the genome of U-937 cells by means of the recombinant retrovirus pEGZ 36. The Pax-5 protein was coordinately expressed with the chimeric selection marker composed of the enhanced green fluorescent protein (EGFP) and the Zeocin (Zeo) resistance protein. We isolated a Pax-5-expressing population (U-937/EGZ Pax-5) using Zeo selection. As a control, we generated populations expressing only the selection marker at equivalent levels (U-937/EGZ). Western-blot analysis of protein extracts from Pax-5 transfected U-937 cells showed positive Pax-5 protein expression with a band located around 40 kDa (Fig. 5B). As checked by sequencing, the full-length Pax-5 gene was successfully integrated into the U-937 genome without deletions or point mutations (data not shown). The smaller size of the Pax-5 expressed in U-937 cells might be due to splicing differences (see Sect. 3).
Since available anti-CD23 antibodies are directed against the extracellular region of the receptor, which is shared between the two isoforms, the expression of CD23a cannot be analyzed by FACS staining or Western-blot analyses. Therefore we performed RNase protection assays using a single RNA probe that is a full homologue to the CD23a mRNA and a 2/3 homologue to CD23b mRNA. The analysis within one sample allows a reliable quantification of CD23a and CD23b isoforms at the transcriptional level.
Fig. 5A shows the results of the RNase protection assay performed with RNA extracts from U-937 cells, U-937 expressing only the selection marker EGZ, and U-937 transfected with Pax-5. RNA extracts from WIL2 cells (a B cell line) were used as control for CD23a expression. CD23a/b isoform expression was assessed under nonstimulating conditions, as well as after 48 h of stimulation with IL-4, PMA or IL-4+PMA.
Both IL-4 and PMA stimulation induced readily detectable amounts of CD23b mRNA in U-937, U-937/EGZ and U-937/EGZ Pax-5 cells (Fig. 5A, lanes 2, 3, 6, 7, 10 and 11). CD23b expression was further increased by a combination of PMA and IL-4. However, in the Pax-5-transduced U-937 cells, stimulation by PMA plus IL-4 resulted in a strong and reproducible expression of CD23a mRNA (Fig. 5A, lane 12). IL-4 alone, as well as PMA stimulation alone, was unable to induce CD23a expression in this monocytic cell line. This experiment provides strong evidence that Pax-5 may be a limiting factor for enabling CD23a expression in vivo.
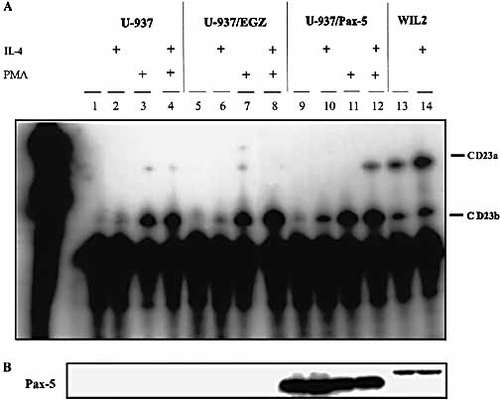
CD23a is expressed in U-937 cells transduced with Pax-5. (A) RNase protection assays were performed with RNA extracts from wild-type U-937 cells (lanes 1–4), U-937 transfected with the selection marker EGZ (lanes 5–8) and U-937 transfected with Pax-5 (lanes 9–12). Cells were stimulated with 50 ng/ml IL-4 and/or 3 ng/ml of PMA for 48 h as indicated. WIL2 cells (a B cell line) express CD23a and CD23b constitutively and were used as controls (lanes 13, 14). The extracted RNA was hybridized with a single CD23 mRNA probe for both CD23a and CD23b isoforms. (B) Western-blot analysis to assess Pax-5 expression.
3 Discussion
High CD23 expression is associated with various chronic diseases such as B-CLL, rheumatoid arthritis and lupus erythematosus. It is assumed that signaling through CD23 contributes to the pathogenesis of these diseases. In humans there are two CD23 isoforms that are differentially expressed. Whereas CD23b is widely detected on lymphocytes and myeloid cells, CD23a is restricted to B lymphocytes. The isoforms differ only in the short N-terminal intracytoplasmic part 4 and seem to be connected to different signal transduction pathways. The CD23a isoform specifically mediates endocytosis of bound ligands and can therefore influence B-cell-mediated antigen presentation 37. The two isoforms are each regulated by their own promoters. So far, IL-4 has been described as being the most important activator of CD23a and CD23b expression 4, 17. Binding sites for STAT6 and NF-AT transcription factors have been characterized in the CD23b promoter 19, 38. Studies of the mouse CD23 promoter, which is closely related in sequence to the human CD23a promoter, revealed functional STAT6- and NF-κB-binding sites 20. IL-4-responsive elements have been also documented inside the human CD23a promoter 17, 18. EBNA2 and Notch-2 can regulate the CD23a promoter by binding to CBF1 sites 23, 39. Both activatory elements may play a role in the immortalization of the cell by enhancing CD23a expression.
The question of how the B-cell-specific expression of the CD23a isoform is regulated has not been experimentally addressed so far. Among B-cell-specific transcription factors, Pax-5/BSAP-binding sites have been very recently predicted in the CD23a core promoter 17, 23. Pax-5 is a critical modulator of early B cell differentiation. Its expression is restricted to B cells, embryonic brain and testis. Binding sites for Pax-5 have been identified in promoters of several genes. Although it is a positive regulator of CD19, mb-1 and RAG-2 31–33, Pax-5 acts as a repressor for the immunoglobulin heavy-chain 3′Cα enhancer and the J-chain 40, 41. The importance of Pax-5 for development was demonstrated by knockout experiments. Pax-5 is important in early and late B cell development. Most Pax-5–/– mice die within 3 weeks and B cell development is blocked atthe pro-B cell stage 42. The loss of Pax-5 in mature B cells severely impairs B cell identity 35.
Here we demonstrate the presence of a functional Pax-5-binding site in the CD23a core promoter, which is able to induce and enhance CD23a expression. We also advance the hypothesis that Pax-5,being a B-cell-specific protein, plays a role in regulating the specific expression of CD23a on B cells. Since CD23 is expressed later than Pax-5 in B cell development, additional transcription factors must be involved in modulating CD23a expression. However, both CD23 and Pax-5 expression are lost when differentiation to plasma cells occurs.
The identification of putative binding sites for Pax-5 in the CD23a core promoter was initially done by sequence alignment with the Pax-5 consensus sequence. This consensus motif has been characterized by Czerny et al. in an extended comparison between known natural targets of Pax-5, including the recognition sequences that originate from the sea urchin H2A-2.2 gene and the human CD19 gene 29. As seen in Fig. 1B, the Pax-5-recognition sequence is divided in two halves — a more extensive 3′-consensus motif recognized by the N-terminal subdomain of the paired domain and a 5′-consensus motif recognized by the C-terminal part of the paired domain. One important observation was that all naturally occurring binding sites identified so far deviate from the consensus sequence and that Pax-5 is able to interact with a panel of seemingly degenerate recognition sequences. In testing the Pax-5-binding sites, we considered all putative binding sites with at least 50% identity to the consensus sequence. This resulted in three putative Pax-5-binding sites that we designated as CD23-1, CD23-2 and CD23-3.
The putative binding site CD23-1 was the only one able to compete the high affinity H2A-2.2 binding site and to directly bind the Pax-5 protein (Fig. 2). This was furtherconfirmed by the fact that oligonucleotides containing a mutated CD23-1 binding site completely lost the ability to bind the Pax-5 protein (Fig. 3). Interestingly, a gap had to be introduced in the CD23-1 binding site in order to reconstitute the symmetry of the site (Fig. 1B). Another putative binding site (CD23-3), which had 89% homology to the consensus sequence, failed to bind Pax-5 in the same assays (Fig. 2). These observations underline the known problems of predicting Pax-5-binding sites by sequence analysis.
Another approach to identify Pax-5-binding sites in the CD23a core promoter was to construct overlapping oligonucleotides covering the whole length of the promoter and to test their ability tocompete the sea urchin H2A-2.2 binding site (results not shown). Both strategies revealed CD23-1 as the only site in the CD23a core promoter able to bind Pax-5 protein in EMSA.
It is interesting to note that in our hands none of the two Pax-5 sites predicted in a recent study by Hubmann et al. 23 interacts directly with the Pax-5 protein. As this study used nuclear extracts from B-CLL cells, it is possible that Pax-5 was recruited by another factor into a protein complex, which interacts with these putative sites.
To test the function of the identified Pax-5-binding site in vitro we performed luciferase assays in 293 cells linking the CD23a core promoter or an extended CD23a promoter to a luciferase reporter gene. We chose this approach since previous work has shown that promoters of Pax-5-regulated genes, like CD19, are weakly active in transiently transfected B cells and can only be stimulated by ectopic Pax-5 expression in heterologous cell types 43. Overexpression of Pax-5 in 293 cells led to a 7–14-fold activation, depending on the CD23a promoter used (Fig. 4). However, reporter constructs with the mutated CD23-1 site were still slightly active. The remaining activation may be due to the recruitment of Pax-5 into protein complexes that can bind with lower efficiency but can still lead to a lower activation of the promoter. This may be indeed another mechanism by which Pax-5 regulates the CD23a promoter and which needs to be approached in future studies. Furthermore, in experiments using the extended promoter, which includes two reported STAT6-binding sites, Pax-5 acted in a cooperative manner together with STAT6 in further enhancing CD23a expression above the level of the induction provided by the two factors alone. These results suggest that Pax-5 not only determines the B cell specificity of CD23a, but also potentiates the STAT6-mediated stimulatory effect.
To further investigate the ability of Pax-5 to induce CD23a expression in vivo, we selected the monocytic cell line U-937, which normally expresses only CD23b after appropriate stimulation 4. Using a retroviral infection system we ectopically expressed Pax-5 in U-937 cells. Western-blot analyses demonstrated a strong expression of Pax-5, with a band located around 40 kDa (Fig. 5B). The B cell line WIL2 — used as a CD23a positive control — showed a Pax-5 band somewhat lower than the expected 50 kDa. As sequence analyses of the integrated Pax-5 construct revealed no partial deletions or point mutations creating a STOP codon we suppose that the size difference is due to splicing differences. Several Pax-5 isoforms generated by usage of a second distal start codon or by differential splicing were described in different B cell lines 44.
The expression of Pax-5 in the U-937 cell line enabled a clear CD23a expression after appropriate stimulation with IL-4 and PMA. In comparison with B cells, where the ratio of CD23a:CD23b expression is around 3–4:1, independently of the applied stimulus 37, CD23b exceeds CD23a expression in U-937/Pax-5 cells. This is likely due to the difference in the cellular environment between B cells and macrophages (e.g. specific transcription factors, availability of the promoter). In any case, the CD23a induction in Pax-5-transduced U-937 cells was consistent andhighly reproducible. Neither IL-4 nor PMA stimulation alone was able to induce CD23a expression in Pax-5-transfected U-937 cells. This would indicate IL-4 as being necessary but not sufficient for Pax-5-mediated expression of the CD23a isoform. Pax-5 seems to be a prerequisite transcriptional activator, which cooperates with other transcription factors during B-cell-specific expression of theCD23a isoform.
In conclusion, we identified a functional Pax-5-binding site in the CD23a core promoter. In combination with other transcription factors, Pax-5 is able to mediate CD23a expression even in cells that normally do not express CD23a. Therefore our results indicate Pax-5 being a key regulator of B-cell-specific expression of CD23a.
4 Materials and methods
4.1 Plasmid constructs
The following plasmids were used in mammalian cell transfections / reporter gene assays. pcDNA3-Pax-5 contains the human Pax-5 gene cloned in the EcoRI site of the vector. In pLuc+ACPand pLuc+AP, PCR-generated fragments representing the CD23a core promoter (–203 to +83) and CD23a promoter (–1216 to +211) were cloned in the SalI site of the pLuc+ vector. pXM-STAT6 was kindly provided by Dr E. Pfitzner.
The bicistronic retroviral vector pEGZ/MCS contains the human Pax-5 gene inserted between the EcoRI and SmiI sites. The vector is based on the pczCFG2 hCD8 IEYZ retroviral vector 36. In this vector the CMV enhancer replaces the U3 region of the 5′ long terminal repeat (LTR) of the murine leukemia virus (MuLV). Because of the reverse transcription process, expression now becomes MuLV-LTR-dependent in target cells. The coordinate expression of the chimeric marker gene (EGFP/Zeo) and the gene of interest is achieved via an internal ribosomal entry site derivedfrom encephalo-myocarditis virus 36.
The following plasmids were used for in vitro transcription. The construction of pBS-23A and pBS-β-actin was previously described 37. Briefly, pBS-23A contains a 299 bp BstXI / DraIII fragment of the CD23a cDNA cloned into the SmaI site of the pBlueScriptSK+ vector (the CD23a cDNA fragment was obtained by RT-PCR from tonsilar B cell total RNA, using 5′-GCCATGGAGGAAGGTCAATATTCA-3′ as 5′ primer and 5′-GACTTGAAGCTGCTCAGATCTGCT-3′ as 3′ primer). pBS-β-actin contains a 540 bp fragment (primers: 5′-GTGGGGCGCCCCAGGCACCA-3′ and 5′-CTCCTTAATGTCACGCACGATTTC-3′) cloned in the same vector.
The pCR-Pax-5 vector was used for in vitro transcription and translation. It contains the full-length human Pax-5 in the MCS. Human Pax-5 was generated by RT-PCR using 5′-TTCCCTGTCCATTCCATCAA-3′ and 5′-TCATGGGCTCTCTGGCTA-3′ as 5′ and 3′ primers and B cells total RNA as template.
4.2 Site-directed mutagenesis
To mutate the Pax-5-binding site CD23-1 within the CD23a core promoter cloned in the pLuc+ vector, we used site-specific mutagenesis by overlap extension 45, 46. Two sets of mutagenic primers were used: R2/FM and RM/F2. The sequences of the primers were:
R2–5′-TGTATCTTATCATGTCTGGATCTCGAAGCTTGC–3′; FM–5′-CACGCACAACTTATACTGGCACTTCCCACACCC-3′; RM–5′-GTGTGGGAAGTGCCAGTATAAGTTGTGCGTGTAAT-3′; and F2–5′-TTTACCAACAGTACCGGAATGCCAAGCTCAG-3′.
The conditions for all PCR reactions were: 25 cycles of 1 min at 94°C denaturation, 1 min at 67°C annealing and 1 min at 72°C elongation and a last cycle of 1 min at 94°C denaturation and 10 min at 72°C annealing, using pLuc+ACP plasmid as a template. The mutagenesis product was cloned between the HindIII and BamHI sites of the pLuc+ vector.
4.3 Cell preparation and Western-blot analyses
Whole cell lysates were prepared from 1×108 cells in 200μl 4×SDS Buffer (Roth). Proteins were separated by SDS-PAGE, transferred to a nitrocellulose membrane and analyzed by Western blots.
4.4 Cell culture, transfection and reporter gene assay
The cell line 293 was transfected using Gene Porter (GTS Inc.) in DMEM (Gibco) supplemented with L-glutamine (2 mM), antibiotics and 10% FCS. For each transfection 100 ng of the pLuc+vectors and 500 ng of the pcDNA3-Pax-5 and pXM-STAT6 vectors were used. The DNA concentration was brought to a total of 2 μg DNA per transfection using salmon sperm DNA. Four hours after transfection the cells were induced with IL-4 (50 ng/ml) and PMA (3 ng/ml). Extracts were prepared from transfected cells after 40 h in 1× reporter lysis buffer (Promega). The luciferase activity wasassessed with a luminometer (Berthold) and the results were normalized for equal concentration of total protein.
4.5 EMSA
Human Pax-5 protein was obtained by in vitro transcription and translation using TNTR Quick Coupled Transcription / Translation Systems (Promega) and pCR-Pax-5 plasmid as a template. In-vitro-translated Pax-5 protein (2 μl) was incubated with 40,000 cpm (equivalent to approximately 0.5–1.0 ng) of [32P]-labeled oligonucleotides as probes and 1 μg poly(dIdC) as a nonspecific competitor in Pax-5 binding buffer 29. The probes were labeled with [α-32P]dCTP. In competition assays double-stranded oligonucleotides described below were used as unlabeled competitors in 1-, 10- or 100-fold excess. The samples were run on nondenaturing 5% polyacrylamide gels at 15 V/cm followed by autoradiography. In supershift assays 1 μg of human Pax-5 monoclonal antibody (BD Biosciences) was used.
The following oligonucleotides were used as probes or as unlabeled competitors: (1) Pax-5 high affinity binding site from the sea urchin H2A-2.2 gene 29 was obtained by annealing the oligonucleotide pair M118 and M119 (5′-CAGGGTTGTGACGCAGCGGTGGGTGACGACTGT-3′ and 5′-GCCACAGTCGTCACCCACCGCTGCGTCACAACC-3′); (2) the putative Pax-5-binding site CD23-1was obtained by annealing the oligonucleotide pair M304 and M305 (5′-GGGTGTGGGGAGCACCAGGAGAGGCCATGCGTGTAATGTTA-3′ and 5′-GGATAACATTACACGCATGGCCTCTCCTGGTGCTCC-3′); (3) the putative Pax-5-binding site CD23-2 was obtained by annealing the oligonucleotide pair M306 and M307 (5′-CGGACTTCACCCGGGTGTGGGGAGCA-3′ and 5′-GGTGCTCCCCACACCCGGGTGAAGT-3′); (4) the putative Pax-5-binding site CD23-3 was obtained by annealing the oligonucleotide pair M308 and M309 (5′-GTGGTATGATTCAGTGTGCAGTAACAGTGG TTC-3′ and 5′-GTGAACCACTGTTACTGCACACTGAATCATA-3′); (5) CD23-1mu1 (mutated nucleotides are underlined) was obtained by annealing the oligonucleotide pair M310 and M311 (5′-GGGTGTGGGAAGTGCCAGTATAAGTTGTG-3′ and 5′-ACGCACAACTTATACTGGCACTTCCCAC-3′); (6) CD23-1mu2 (mutated nucleotides are underlined) was obtained by annealing the oligonucleotide pair M265 and M266 (5′-GTGTGGGGAGAACCAGTAGAGGCCATGCGTG-3′ and 5′-CACGCATGGCCTCTACTGGTTCTCCCCA-3′).
4.6 Transfection/infection assays
Recombinant retroviral particles were generated using the pHIT packaging system as described by Soneoka et al. 47.
Briefly, 293T cells were transiently cotransfected using the standard calcium phosphate method with 5 μg of each of the packaging vectors pHIT456 (which codes for the amphotropic env protein) and 5–7 μg of the retroviral construct pEGZ/MCS and EGZ/Pax-5. Sixteen hours later the transfection solution was replaced by DMEM. Viral supernatants were harvested 48 h later and filtered (0.45 μm). Polybrene (Sigma) was added to a final concentration of 10 μg/ml. U-937 cells were mixed with the retroviral supernatant and centrifuged for 3 h at 1000×g. Thereafter cells were replated in RPMI. Transduced cells were selected with Zeocin (250 μg/ml).
The cell line selected for experiments stained positive for EGZ in 97% of the cells, as revealed by FACS analysis.
4.7 RNA preparation and RNase protection assay
U-937 cells (5x106) were maintained in RPMI 1640 medium (Gibco), supplemented with 10% FCS and antibiotics and were stimulated with IL-4 (50 ng/ml), PMA (3 ng/ml) or both. After 48 h the cells were lysed in 1 ml TRIZOL reagent (Gibco) and total RNA was prepared following the manufacturer's instructions.
The hybridization probe (CD23a RNA) was generated by in vitro transcription (MAXIscriptTM T7/T3 -Ambion) using EcoRI linearized pBS-23A vector as a template. The probes give rise to a 299 bp band for CD23a and a 184 bp band for CD23b. Between 40 and 50 μg of total RNA were used in a HybSpeedTM RPA Kit (Ambion) according to the manufacturer's instructions, except that hybridization was extended overnight. Results were assessed by autoradiography. In addition to the CD23 probe, a second probe for β-actin was included in each experiment as an internal control. This probe resulted in a double band of around 130 bp.
Acknowledgements
We thank Drs. Stefan Klein-Hessling and Anne-Sophie Rouzière for helpful discussion. We also thank Prof. Anneliese Schimpl for critically reviewing the manuscript and Kathrin Zehe for her expert technical assistance. We are grateful to Dr. J. Altschmied for kindly providing the pLuc+ vector, to Dr. E. Pfitzner for providing the pXM-STAT6 vector and toProf W. Sebald for the gift of IL-4. This work was supported by "Interdisziplinäres Zentrum für Klinische Forschung" (BMBF, 01 KS 9603) and the Graduate College 520 "Immunmodulation" (DFG).
- WILEY-VCH
- WILEY-VCH
- WILEY-VCH
- WILEY-VCH
- WILEY-VCH




