Loss of the IR region in conifer plastomes: Changes in the selection pressure and substitution rate of protein-coding genes
Funding information
This work was funded by the National Natural Science Foundation of China (31670200, 31770587, 31872670, and 32071781).
Abstract
Plastid genomes (plastomes) have a quadripartite structure, but some species have drastically reduced or lost inverted repeat (IR) regions. IR regions are important for genome stability and the evolution rate. In the evolutionary process of gymnosperms, the typical IRs of conifers were lost, possibly affecting the evolutionary rate and selection pressure of genomic protein-coding genes. In this study, we selected 78 gymnosperm species (51 genera, 13 families) for evolutionary analysis. The selection pressure analysis results showed that negative selection effects were detected in all 50 common genes. Among them, six genes in conifers had higher ω values than non-conifers, and 12 genes had lower ω values. The evolutionary rate analysis results showed that 9 of 50 common genes differed between conifers and non-conifers. It is more obvious that in non-conifers, the rates of psbA (trst, trsv, ratio, dN, dS, and ω) were 2.6- to 3.1-fold of conifers. In conifers, trsv, ratio, dN, dS, and ω of ycf2 were 1.2- to 3.6-fold of non-conifers. In addition, the evolution rate of ycf2 in the IR was significantly reduced. psbA is undergoing dynamic change, with an abnormally high evolution rate as a small portion of it enters the IR region. Although conifers have lost the typical IR regions, we detected no change in the substitution rate or selection pressure of most protein-coding genes due to gene function, plant habitat, or newly acquired IRs.
1 INTRODUCTION
The plastid genomes (plastomes) of most land plants have a highly conserved quadripartite structure, consisting of a large single-copy (LSC) region and a small single-copy (SSC) region separated by a pair of inverted repeat (IR) regions (Wicke et al., 2011). Due to selective pressure on photosynthesis-related elements, plastids have highly conserved gene content and order (Ruhlman & Jansen, 2014). And due to their highly conservation, a large copy number, lack of recombination, and uniparental inheritance, plastomes have been used to evaluate phylogeographic relationships, phylogeographic histories, and evolutionary events (Barrett et al., 2016; Julian et al., 2017; Moore et al., 2006; Pacheco et al., 2019; Shaw et al., 2014; Wang et al., 2020).
Inverted repeat regions are important in replication initiation (Heinhorst & Cannon, 1993), genomic stability (Maréchal & Brisson, 2010; Palmer & Thompson, 1982), and gene conservation (Palmer & Thompson, 1982; Wolfe et al., 1987). IR changes alter the size of the plastid genome (Kwon et al., 2020). From green algae to angiosperms, IR regions typically contain at least four rRNAs and five tRNAs (Mower & Vickrey, 2018). However, the IR region, as a hot spot of structural rearrangement, often contracts, expands, or is lost. In related species, the boundary of the IR region changes little, resulting in the gain or loss of a small number of genes (Downie & Jansen, 2015; Li et al., 2016; Ping, Li, et al., 2021; Wicke et al., 2014). The IR region of Pelargonium, Psilotum, and Trochodendraceae has undergone large-scale expansion, gaining a large number of genes from the SC region (Chumley et al., 2006; Grewe et al., 2013; Sun et al., 2013). The IR region is absent in the plastomes of some plant lineages, such as prasinophyceans (Lemieux et al., 2014), trebouxiophyceans (Turmel et al., 2015), streptophytes (Lemieux et al., 2016), Ulvophyceae (Cai et al., 2017), and Euglenaceae (Karnkowska et al., 2018) among algae; Pinaceae, Cupressophytes (Li, Gao, et al., 2016; Wu, Lin, et al., 2011; Wu, Wang, et al., 2011), and Taxaceae (Zhang et al., 2014) among gymnosperms; and the putranjivoid clade of Malpighiales (Jin et al., 2020), Tahina spectabilis (Arecaceae) (Barrett et al., 2016), legumes (Palmer & Thompson, 1981, 1982), Carnegiea gigantea (Cactaceae) (Sanderson et al., 2015), and Erodium species (Geraniaceae) (Blazier et al., 2016; Guisinger et al., 2011; Ruhlman et al., 2017) among angiosperms.
The sequence substitution rate in the IR region differs from that in the SC region. In some legumes with missing IR regions, the synonymous substitution rate of genes entering the SC region from the IR region is similar to that of those already in the SC region (Perry & Wolfe, 2002). The IR region has a low substitution rate in Cycas (Wu & Chaw, 2015). Zhu et al. (2016) found that the synonymous substitution rate of genes in the SC region in vascular plants (angiosperms, gymnosperms, and ferns) is 3.7-fold that of genes in the IR region; after the transfer of genes between the SC and IR regions, the substitution rate becomes consistent. Some genes in the IR region have high synonymous substitution rates in some species (Pelargonium, Plantago, and Silene). In ferns, psbA, rps7, 3'-rps12, and ycf2 showed a reduced substitution rate and increased GC content after entering the IR region (Li, Kuo, et al., 2016). Recently, Ping, Feng, et al. (2021), Ping, Li, et al. (2021) reported that the substitution rate of 3'-rps12 in the IR region is lower than that of 5'-rps12 in the SC region in ferns and gymnosperms.
The IR region has important effects on genome stability and the evolutionary rate (Maréchal & Brisson, 2010; Palmer & Thompson, 1982; Wolfe et al., 1987). During gymnosperm evolution, conifers lost the IR region. Araucaria cunninghamii (Araucariaceae) is native to southeastern coastal areas of Oceania and is the main afforestation tree species in tropical and subtropical regions. It is one of the top five park tree species worldwide and an important garden ornamental plant, and it is cultivated widely in China. Callitropsis funebris (Cupressaceae) is endemic to China and distributed widely; it grows rapidly and has a wide range of uses and strong adaptability. Its phylogenetic classification is a focus of research (Zheng & Fu, 1978). As these two species are important representatives of their genera, we sequenced their plastomes. In this study, we selected 78 gymnosperms and 2 Polypodiales (outgroup) and analyzed selection pressure and the evolutionary rate using the maximum likelihood method. We investigated whether the selection pressure and evolutionary rate of protein-coding genes in conifers differed according to the presence of the IR region; whether the selection pressure and evolutionary rate of genes that enter the IR region differ from those that enter the SC region; and the heterogeneity of gymnosperm evolutionary rates.
2 MATERIALS AND METHODS
2.1 Sequencing and sequence preparation
Fresh leaves of Callitropsis funebris (E113°35’, N23°15’) and Araucaria cunninghamii (E113°200’, N23°90’) were taken from the campus of South China Agricultural University. Specimens were stored in Herbarium of the College of Life Sciences, SCAU (specimen no.: PJY-NYS1910 and PJY-BM1910). Plastid genomic DNA was extracted using the DNASECure Plant Genomic DNA Extraction Kit (Tiangen) and double-ended sequencing was performed on Illumina HiSeq2500 platform. Clean reads filtered by Trimmomatic V0.32 (Bolger et al., 2014) were spliced and assembled in Velvet V1.2.03 (Zerbino & Birney, 2008). The genes were predicted by the DOGMA Program (Wyman et al., 2004) and the plastome was mapped using the online site OGDRAW v1.3 (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html) after gene annotation (Greiner et al., 2019). Finally, the sequence information was submitted on the Banklt Platform (https://www.ncbi.nlm.nih.gov/WebSub/), and Genbank accession no. is MT227812 and MT227813, respectively.
In addition, 76 gymnosperms and 2 Polypodiales (as outgroups) were downloaded from the NCBI database. A total of 78 gymnosperms cover 13 families and 51 genera (Table 1). Genious prime 2020 (Kearse et al., 2012) software was used to extract 50 protein-coding genes. And performed sequence alignment and correction through the ClustalW (codons) module in MEGA X (Kumar et al., 2018).
| Order | Family | Species name | GenBank accession no. | Species name | GenBank accession no. |
|---|---|---|---|---|---|
| Cycadales | Cycadaceae | Cycas revoluta | NC_020319 | Cycas szechuanensis | NC_042668 |
| Cycas panzhihuaensis | NC_031413 | Cycas taitungensis | NC_009618 | ||
| Boweniaceae | Bowenia serrulata | NC_026036 | |||
| Zamiaceae | Stangeria eriopus | NC_026041 | Zamia furfuracea | NC_026040 | |
| Ceratozamia hildae | NC_026037 | Encephalartos lehmannii | NC_027514 | ||
| Dioon spinulosum | NC_027512 | Lepidozamia peroffskyana | NC_027513 | ||
| Macrozamia mountperriensis | NC_027511 | ||||
| Ginkgoales | Ginkgoaceae | Ginkgo biloba | NC_016986 | ||
| Gnetales | Gnetaceae | Gnetum montanum | NC_021438 | Gnetum ula | NC_028734 |
| Gnetum parvifolium | NC_011942 | Gnetum gnemon | NC_026301 | ||
| Ephedrales | Ephedraceae | Ephedra equisetina | NC_011954 | Ephedra intermedia | NC_044772 |
| Ephedra foeminea | NC_029347 | Ephedra sinica | NC_044773 | ||
| Welwitschiales | Welwitschiaceae | Welwitschia mirabilis | EU342371 | ||
| Cupressales | Cupressaceae | Cryptomeria japonica | NC_010548 | Cupressus tonkinensis | NC_039562 |
| Taiwania cryptomerioides | NC_016065 | Cupressus gigantea | NC_028155 | ||
| Taiwania flousiana | NC_021441 | Cupressus sempervirens | NC_026296 | ||
| Cunninghamia lanceolata | NC_021437 | Callitropsis funebris | MT227813 | ||
| Juniperus monosperma | NC_024022 | Callitropsis nootkatensis | KP099642 | ||
| Juniperus recurva | NC_042763 | Callitropsis vietnamensis | KX832629 | ||
| Taxodium distichum | NC_034941 | Hesperocyparis lusitanica | MH121051 | ||
| Taxodium mucronatum | NC_045277 | Chamaecyparis formosensis | NC_034943 | ||
| Calocedrus formosana | NC_023121 | Chamaecyparis hodginsii | NC_036996 | ||
| Glyptostrobus pensilis | NC_031354 | Thuja occidentalis | NC_042177 | ||
| Metasequoia glyptostroboides | NC_027423 | Thuja sutchuenensis | NC_042176 | ||
| Callitris rhomboidea | NC_034940 | ||||
| Taxaceae | Cephalotaxus sinensis | MF977938 | Taxus fuana | NC_038099 | |
| Cephalotaxus oliveri | NC_021110 | Pseudotaxus chienii | NC_041503 | ||
| Amentotaxus argotaenia | NC_027581 | Torreya fargesii | NC_029398 | ||
| Amentotaxus formosana | NC_024945 | ||||
| Sciadopityaceae | Sciadopitys verticillata | NC_029734 | |||
| Pinales | Pinaceae | Cedrus deodara | NC_014575 | Picea neoveitchii | NC_043913 |
| Pinus massoniana | MF564195 | Keteleeria davidiana | NC_011930 | ||
| Pinus yunnanensis | NC_043856 | Tsuga chinensis | NC_030630 | ||
| Pseudolarix amabilis | NC_030631 | Larix sibirica | NC_036811 | ||
| Pseudotsuga sinensis | NC_016064 | Larix decidua | NC_016058 | ||
| Abies fargesii | NC_042775 | Abies fanjingshanensis | NC_042777 | ||
| Araucariales | Podocarpaceae | Nageia nagi | NC_023120 | Retrophyllum piresii | NC_024827 |
| Podocarpus lambertii | NC_023805 | Dacrydium elatum | NC_045880 | ||
| Dacrycarpus imbricatus | NC_034942 | Manoao colensoi | NC_044893 | ||
| Araucariaceae | Agathis dammara | NC_023119 | Araucaria heterophylla | NC_026450 | |
| Wollemia nobilis | KP259800 | Araucaria araucana | NC_045394 | ||
| Araucaria cunninghamii | MT227812 | Araucaria bidwillii | NC_045395 | ||
| Araucaria angustifolia | NC_039155 | ||||
| Polypodiales | Polypodiaceae | Lepisorus clathratus | NC_035739 | ||
| Athyriaceae | Athyrium anisopterum | NC_035738 |
2.2 Analysis of phylogenetic relationships
We constructed the phylogenetic relationships based on the common gene dataset (the two Polypodiales as the outgroup). MEGA X and PAUP4.0 (Swofford, 2002) were used to construct the NJ (neighbor-joining) tree and MP (maximum-parsimony) tree, respectively. RaxmlGUI2 software built ML (maximum-likelihood) tree with GTRGAMMAI substitution model and 1,000 bootstrap (Stamatakis, 2014). BI (Bayesian inference) tree was built with MrBayes 3.2.6 software (Huelsenbeck & Ronquist, 2001). We reconstructed a background tree combined with the published phylogenetic relationship to analyze selection pressure and evolution rate, finally.
2.3 Analysis of selection pressure
Codeml program in PAML4.9 was used to analyze the selection pressure (Yang, 2004). The M0 (one-ration) model assumes that each branch has the same value of ω under the Branch model. The Model 2 (two-ratio) assumes the foreground branch and the background branch have different ω values. The likelihood ratio test of M0 and Model 2 can be used to detect the difference of selection pressure between the foreground branch and the background branch.
2.4 Analysis of evolutionary rate
We used HyPhy 2.2.4 software (Pond et al., 2005) to calculate the evolution rate, which is based on the maximum likelihood method and in the context of the phylogenetic tree. The transition rate (trst), transversion rate (trsv), and trsv/trst (ratio) of each branch were calculated under the nucleotide HKY85 substitution model with the local parameter. Similarly, the synonymous (dS) and non-synonymous (dN) substitution rates and dN/dS (ω) of each branch were calculated under the codon MG94×HKY85×3_4 substitution model with the rate Het. parameter and 4 rate classes. IBM SPSS v22.0 (IBM Corporation, 2014) software was used to perform the Mann–Whitney U test and Spearman's rank correlations test on related parameters.
3 RESULTS
3.1 Plastome characteristics
The 76 gymnosperms were divided into two types according to the presence of a typical IR region: 56 gymnosperms lacked a typical IR region (SC-56; conifer) and 22 gymnosperms had a typical IR region (gnetophytes, cycads, and Ginkgo biloba; IR-22; non-conifer). For SC-56 (Appendix S1), the genome size ranged from 117,720 bp (K. davidiana) to 146,723 bp (A. heterophylla), with an average of 130,858 bp. The GC content ranged from 34.3% (T. sutchuenensis) to 39.1% (C. davidiana), with an average of 36.2%.
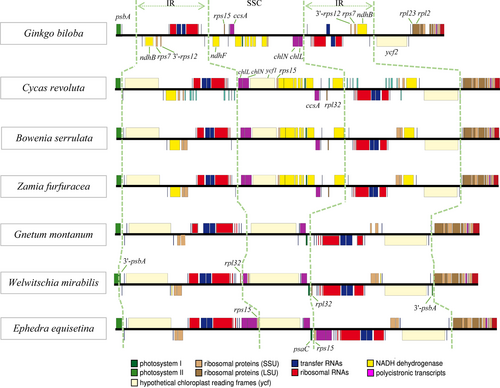
For IR-22 (Table 2), the genome size ranged from 109,518 bp (Ephedra equisetina) to 166,341 bp (Macrozamia mountperriensis), with an average of 142,888 bp. The GC content ranged from 36.6% to 40.1%, with an average of 38.7%. The size of the IR region ranged from 17,732 bp (G. biloba) to 26,137 bp (Ceratozamia hildae). Compared with other groups, the plastome of the gnetophytes was the smallest, and the proportion of SSC region was also the smallest (7.2–9.3%), but the proportion of the IR region was largest (16%–18.9%). The three groups all contained the protein-coding genes rps7 and 3'-rps12 in the IR region, and ndhB was present in G. biloba and cycads. All plants, except G. biloba, possessed ycf2. Individually, 3'-psbA was present in Welwitschia mirabilis, Gnetum parvifolium, and Gnetum gnemon, rpl32 was detected in W. mirabilis, and rps15 was detected in E. equisetina (Figure 1).
| Species name | Genome size/bp | GC/% | LSC/bp | LSC percentage | SSC/bp | SSC percentage | IR/bp | IR percentage | Number of genes in the IR region | |
|---|---|---|---|---|---|---|---|---|---|---|
| CDS | tRNA | |||||||||
| Ginkgo biloba | 156,988 | 39.6 | 99,254 | 63.2% | 22,267 | 14.2% | 17,732 | 11.3% | 3 | 6 |
| Cycas revoluta | 162,489 | 39.4 | 88,978 | 54.8% | 23,379 | 14.4% | 25,066 | 15.4% | 4 | 6 |
| Cycas panzhihuaensis | 162,470 | 39.4 | 88,932 | 54.7% | 23,448 | 14.4% | 25,045 | 15.4% | 4 | 7 |
| Cycas szechuanensis | 162,083 | 39.4 | 88,970 | 54.9% | 23,107 | 14.3% | 25,003 | 15.4% | 4 | 7 |
| Cycas taitungensis | 163,403 | 39.5 | 90,216 | 55.2% | 23,039 | 14.1% | 25,074 | 15.3% | 4 | 7 |
| Bowenia serrulata | 165,695 | 39.9 | 90,733 | 54.8% | 23,156 | 14.0% | 25,903 | 15.6% | 4 | 7 |
| Stangeria eriopus | 163,548 | 39.5 | 89,850 | 54.9% | 23,006 | 14.1% | 25,346 | 15.5% | 4 | 7 |
| Ceratozamia hildae | 165,734 | 39.7 | 90,487 | 54.6% | 22,973 | 13.9% | 26,137 | 15.8% | 4 | 7 |
| Dioon spinulosum | 161,815 | 40.1 | 88,754 | 54.8% | 23,355 | 14.4% | 24,853 | 15.4% | 4 | 7 |
| Zamia furfuracea | 164,953 | 39.7 | 90,441 | 54.8% | 23,228 | 14.1% | 25,642 | 15.5% | 4 | 7 |
| Encephalartos lehmannii | 165,822 | 39.9 | 90,724 | 54.7% | 23,302 | 14.1% | 25,898 | 15.6% | 4 | 7 |
| Lepidozamia peroffskyana | 165,939 | 39.9 | 90,804 | 54.7% | 23,299 | 14.0% | 25,981 | 15.7% | 4 | 7 |
| Macrozamia mountperriensis | 166,341 | 39.8 | 91,171 | 54.8% | 23,334 | 14.0% | 25,918 | 15.6% | 4 | 7 |
| Gnetum montanum | 115,019 | 38.2 | 66,139 | 57.5% | 9,494 | 8.3% | 19,693 | 17.1% | 3 | 8 |
| Gnetum parvifolium | 114,914 | 38.2 | 66,095 | 57.5% | 9,559 | 8.3% | 19,630 | 17.1% | 4 | 8 |
| Gnetum ula | 113,249 | 38.5 | 64,914 | 57.3% | 8,791 | 7.8% | 19,772 | 17.5% | 3 | 8 |
| Gnetum gnemon | 115,022 | 38.2 | 66,591 | 57.9% | 8,329 | 7.2% | 20,051 | 17.4% | 5 | 8 |
| Ephedra equisetina | 109,518 | 36.6 | 59,936 | 54.7% | 8,078 | 7.4% | 20,752 | 18.9% | 6 | 8 |
| Ephedra foeminea | 109,584 | 36.7 | 60,708 | 55.4% | 8,078 | 7.4% | 20,399 | 18.6% | 6 | 8 |
| Ephedra intermedia | 109,667 | 36.6 | 59,936 | 54.7% | 8,247 | 7.5% | 20,742 | 18.9% | 6 | 8 |
| Ephedra sinica | 109,550 | 36.7 | 59,961 | 54.7% | 8,103 | 7.4% | 20,743 | 18.9% | 6 | 8 |
| Welwitschia mirabilis | 119,726 | 36.7 | 68,556 | 57.3% | 11,156 | 9.3% | 20,007 | 16.7% | 5 | 8 |
Rank sum tests indicated no significant differences in genome size (p = .202) and significant differences in GC content (p < .01) between IR-22 and SC-56.
3.2 Phylogenetic analysis
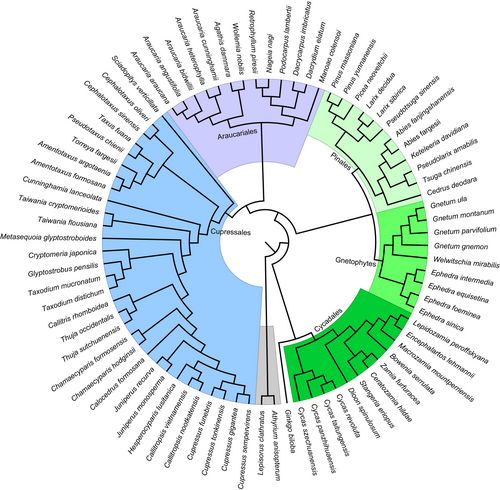
The 50 protein-coding genes comprised 30 photosynthetic system genes, 15 genetic systems, and 5 other genes (Table 3). The constructed phylogenetic results (Appendix S2) showed that the phylogenetic relationship constructed by the NJ method was the clearest, and each group formed a monophyletic branch. Using the BI and ML methods, we found that all groups, except the Pinales, formed a monophyletic branch. In the MP tree, the relationship between Cupressales and Araucariales was unclear.
| Gene type | Gene name |
|---|---|
| Genes for photosynthesis | |
| Photosystem | psaA psaB psaC psaI psaJ |
| Photosystem II | psbA psbB psbC psbD psbE psbF psbH psbI |
| psbJ psbK psbL psbM psbN psbT | |
| Genetic system genes | |
| Cytochrome | petA petB petD petG petL petN |
| ATP Synthase | atpB atpE atpF atpI |
| RubiscoCO large subunit | rbcL |
| Ribosomal Proteins (LSU) | rpl14 rpl20 rpl33 rpl36 |
| Ribosomal Proteins (SSU) | rps2 rps4 rps7 rps8 rps11 rps18 rps19 |
| RNA Polymerase | rpoA rpoB rpoC1 rpoC2 |
| Other genes | |
| Envelop membrane protein | cemA |
| C-type cytochrome synthesis | ccsA |
| Hypothetical chloroplast reading frames | ycf2 ycf3 ycf4 |
The NJ (bootstrap value = 100) and MP (bootstrap value = 50) trees supported gnetophytes as the basic group of gymnosperms, and the MP (bootstrap value = 100) and BI (posterior probability = 1) trees supported gnetophytes and P. neoveitchii as sister groups. Based on previous reports, we accepted the Gnepine hypothesis, and performed a manual adjustment to obtain the phylogenetic tree for selection pressure and evolution rate analysis (Figure 2).
3.3 Evolutionary analysis based on the presence of a typical IR region
With IR-22 as the foreground branch and SC-56 as the background branch, likelihood ratio tests of M0 and Model 2 yielded 18 genes with significant differences (p < .05). That is, these 18 genes experience different selection pressure in IR-22 and SC-56. Among them, six genes had an ω value in IR-22 that were lower than SC-56, and the rest were higher (Table 4).
| Gene | lnL M0 | lnL Model 2 | 2 ΔƖ | P-value | ω foreground | ω background | ω foreground/ ω background |
|---|---|---|---|---|---|---|---|
| atpF | −6580.533 | −6587.056 | 13.046 | 0 | 0.367 | 0.195 | 1.882 |
| ccsA | −11747.691 | −11749.697 | 4.012 | .045 | 0.268 | 0.346 | 0.775 |
| cemA | −10261.516 | −10266.192 | 9.352 | .002 | 0.418 | 0.272 | 1.537 |
| petD | −3345.317 | −3348.100 | 5.566 | .018 | 0.066 | 0.117 | 0.564 |
| psaA | −16296.555 | −16299.080 | 5.050 | .025 | 0.078 | 0.057 | 1.368 |
| psaJ | −1520.374 | −1522.340 | 3.932 | .047 | 0.269 | 0.132 | 2.038 |
| psbA | −6124.730 | −6133.245 | 17.029 | 0 | 0.026 | 0.071 | 0.366 |
| psbC | −9768.612 | −9773.699 | 10.173 | .001 | 0.042 | 0.073 | 0.575 |
| psbE | −1676.534 | −1681.768 | 10.468 | .001 | 0.058 | 0.183 | 0.317 |
| psbJ | −1229.013 | −1231.721 | 5.416 | .020 | 0.133 | 0.366 | 0.363 |
| psbM | −1381.099 | −1384.893 | 7.588 | .006 | 0.479 | 0.132 | 3.629 |
| rpoB | −38294.175 | −38327.819 | 67.289 | 0 | 0.286 | 0.156 | 1.833 |
| rpoC1 | −26817.747 | −26840.736 | 45.978 | 0 | 0.357 | 0.191 | 1.869 |
| rpoC2 | −50272.622 | −50279.519 | 13.795 | 0 | 0.372 | 0.294 | 1.265 |
| rps2 | −8506.349 | −8508.885 | 5.071 | .024 | 0.281 | 0.205 | 1.371 |
| rps11 | −4972.071 | −4979.642 | 15.142 | 0 | 0.339 | 0.152 | 2.230 |
| rps18 | −5208.180 | −5212.761 | 9.163 | .002 | 0.352 | 0.160 | 2.200 |
| ycf2 | −142683.719 | −142688.968 | 10.499 | .001 | 0.768 | 0.635 | 1.209 |
Note
- Foreground is IR-22 and background is SC-56; Bold font indicates that ωforeground is less than ωbackground.
We analyzed and tested the evolutionary rates of the 50 selected common genes (Appendix S3). Among these, we found that the related parameters of nine genes were significantly different between IR-22 and SC-56 (p < .05; Figure 3). Compared with SC-56, the genes with a higher evolution rates in IR-22 were trst (.003 vs. .001), trsv (.026 vs. .010), ratio (.082 vs. .031), dN (.007 vs. .001), dS (.070 vs. .025), and ω (.100 vs. .032) of psbA (2.6- to 4.2-fold that of the SC-56); trst (.052 vs. .021), dN (.033 vs. .014), dS (.112 vs. .051), and ω (.631 vs. .226) of rps8 (2.3- to 2.4-fold); dN (.010 vs. .002) of psbE (5-fold); and ω (.064 vs. .002) of petG (32-fold). Compared with IR-22, the genes with higher evolution rates in the SC-56 were trsv (.033 vs. .009), ratio (.504 vs. .145), dN (.063 vs. .022), dS (.068 vs. .024), and ω (.904 vs. .024) of ycf2 (1.2- to 3.6-fold those of IR-22); trsv (.008 vs. .005) of rps4 (1.8-fold); and ratios of rpoC1 (.224 vs. .087), rbcL (.234 vs. .077), and cemA (.183 vs. .064) (2.6-, 3-, and 2.8-fold, respectively). Spearman's rank correlation test results showed that the evolutionary rate of genes other than dS of rps8 (Spearman's rho = .157, p = .169) and ω of psbA (Spearman's rho = .212, p = .063) was significantly correlated (p < .05) with the IR region (Appendix S4).
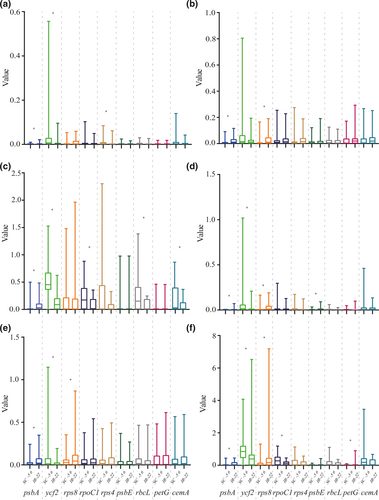
The evolutionary rate of psbA (Appendix S5) was abnormally high in Cycas taitungensis and G. biloba and in Cycas spp. By contrast, the species evolutionary rate of SC-56 was low. For rps8 (Appendix S6), evolutionary rates were high in G. biloba, W. mirabilis, and cycads; the values for most other species branches in SC-56 were 0, except C. rhomboidea and S. verticillata. For rps4 (Appendix S7A), the transversion rate of IR-22 was low, and most values were 0. All species branches in SC-56 had high transversion rates, with those of C. rhomboidea and S. verticillata being abnormally high. For psbE (Appendix S7B), the dN of IR-22 was high (especially in W. mirabilis and Dioon spinulosum) and was 0 for most species in SC-56 (n = 49). For ycf2 (Appendix S8), substitution rates were high in most branches of SC-56, especially C. rhomboidea and S. verticillata. In IR-22, the value of the other branches was 0, except in W. mirabilis and G. biloba.
3.4 Evolutionary analysis based on genes entering the IR region
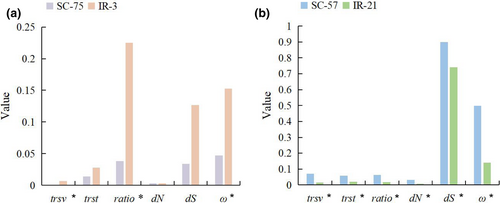
The IR regions of 22 species contained the protein-coding genes 3’-rps12 and rps7 (Figure 1). In W. mirabilis, G. parvifolium, and G. gnemon, part of the psbA (3’-psbA) entered the IR region; thus, it was divided into two categories (IR-3 and SC-75). In G. biloba, ycf2 has been left from the IR region; thus, it was divided into two categories (IR-21 and SC-57).
Likelihood ratio test results revealed that the selection pressure differed among categories. For psbA, ωIR-3 was 0.03474, with IR-3 as the foreground branch, and ωSC-75 was 0.0001, with SC-75 as the background branch. For ycf2, ωIR-21 was 0.76124, with IR-21 as the foreground branch, and ωSC-57 was 0.67832, with SC-57 as the background branch. The trsv, ratio, and ω values of psbA were significantly higher (Ptrsv = .048, Pratio = .003, and Pω = .027) in the IR region than in the SC region, and the evolutionary rate of ycf2 was significantly lower (p < .05) in the IR region than in the SC region (Figure 4).
4 DISCUSSIONS
4.1 Changes in genome size
Evolution has decreased the gymnosperm genome size. Conifers and non-conifers have genomes of similar sizes, which is related to the large compact genome of gnetophytes (115,022–109,518 bp). This compact genome may increase the gnetophyte survival rate in harsh and competitive environments (Wu et al., 2009). After the removal of gnetophytes, a significant difference was detected between groups (p < .01). The shrinkage of the conifer plastome is a result mainly of the loss of the IR region (IRb in Pinaceae and IRa in cupressophytes). In addition, the reduction in the cupressophyte plastome size may be caused by intergenic shrinkage resulting from the mutational burden (Wu & Chaw, 2014). Pinaceae and cupressophytes have lost their typical IRs during evolution, but have acquired one or more short, novel IRs, some of which also exhibit recombination to generate genomic structural diversity (Guo et al., 2014).
Compared with that of ginkgo, the IR region of other non-conifers has expanded. Although the IR region in gnetophytes is smaller than that in cycads, it accounts for a large proportion of the genome and contains more genes. Gnetophytes lost the ndh gene and non-coding sequence, resulting in a compact, more economical genome (Wu et al., 2009). In Pinaceae, the IR comprises only the full trnI-CAU gene and, in most species, a portion of the psbA gene (Lin et al., 2010; Wu, Lin, et al., 2011).
The GC content was increased in non-conifers, which may be related to a GC bias in gene conversion in the IR region. Indeed, the IR region of cycads contains more A/T to GC substitutions (Wu & Chaw, 2015).
4.2 Phylogenetic location of gnetophytes
The four phylogenetic trees constructed using a tandem dataset of 50 common protein-coding genes were not consistent, and the application of different algorithms yielded different topological structures. Compared with those produced by NJ and MP, the phylogenetic relationships constructed by ML and BI are more accurate (Hall, 2005). The ML and BI trees showed that gnetophytes and P. neoveitchi were sister groups with a high degree of support. Although Pinaceae was a non-monophyletic group, it was closest to the gnetophytes, supporting the Gnepine hypothesis.
The phylogenetic position of the gnetophytes has been debated (Mathews, 2009; Palmer et al., 2004). Burleigh and Mathews (2004) examined 13 genes in gymnosperms (five plastid, four nuclear, and four mitochondrial) and almost 19 000 nucleotides, providing evidence for the Gnepine hypothesis. Wu and Chaw (2014) analyzed the rearrangement of DNA in the plastome of gymnosperms, and the results supported the Gnepine hypothesis. Li et al. (2017) analyzed single-copy genes from the genome and transcriptome, and the phylogenetic relationships constructed from their different datasets supported two topological structures: a sister relationship between gnetophytes and other gymnosperms and gnetophytes as a sister group to Pinaceae. Ran et al. (2018) constructed a phylogenetic tree with 1308 loci based on transcriptome data, which also supported the Gnepine hypothesis. Ping, Feng, et al. (2021) constructed ML and BI trees using rbcL and matK, which supported the classification of gnetophytes and Pinaceae as sister groups.
4.3 Heterogeneity of evolutionary rates
The change in evolutionary rate was independent of the presence of the IR region, but rather indicates an increased substitution rate of gnetophytes (especially W. mirabilis) and a lower evolutionary rate of conifers. The evolutionary rate of four genes was increased in non-conifers. The increased substitution rates of rps8, psbE, and psbA were related to the low evolutionary rate of conifers. The petG value was abnormally high. The dN or dS value for most species branches was 0, resulting in false-positive results. By contrast, the substitution rates of rps4 and ycf2 were decreased in non-conifers, which is related to their generally low evolutionary rate. The evolutionary rate of most genes in W. mirabilis was high. The phylogenetic tree constructed using 51 common genes showed that the gnetophytes, especially W. mirabilis and Ephedra sinica, had high evolutionary rates. McCoy et al. (2008) reported that about 75% of plastid protein-coding genes showed high substitution rates in W. mirabilis. The substitution rates of most protein-coding genes did not differ significantly between conifers and non-conifers, which may be related to the newly acquired short IR region of conifers (Hirao et al., 2008; Wu & Chaw, 2014; Yi et al., 2013). In Cephalotaxus oliveri, the 544-bp IR (repeated trnQ-UUG gene) was inferred to have recombination activity (Yi et al., 2013).
Gnetophytes have a higher substitution rate than do other gymnosperms (Ran et al., 2018; Wang et al., 2015; Wu et al., 2009), possibly because of higher mutation rates, changes in selective pressure, increased fixation of mutations by genetic drift, and biological characteristics (Fry & Wernegreen, 2005; Lanfear et al., 2010). Wu et al. (2009) proposed that the high frequency of AT-rich codons in gnetophytes leads to a higher substitution rate. Wang et al. (2015) proposed that the generation time and plant height underlie the increased substitution rate of gnetophytes (Lanfear et al., 2010, 2013). Ran et al. (2018) found that gnetophytes and angiosperms have similar rates of molecular evolution, which are higher than those of other gymnosperms, suggesting that gnetophytes and angiosperms experienced similar selection pressure during their evolutionary histories. As conifers are typically taller than non-conifers, the long-term rate of mitosis in the apical meristem is slower, the frequency of DNA replication is lower, and the accumulated errors per unit time are reduced, resulting in low mutation and substitution rates (Lanfear et al., 2013).
4.4 Negative selection pressure
Genes in the IR region experience strong negative selection (Ping, Feng, et al., 2021; Ping, Li, et al., 2021). The IR region is important for genome structural stability, and genomes lacking an IR region may experience different natural selection effects. However, the selection pressure of most genes did not differ significantly, indicating that they are not affected by the IR region. When ω < 1, smaller ω values reflect stronger negative selection pressure (Yang, 2006). The selection pressure of 18 genes differed between conifers and non-conifers, and their ω values were <1 (Table 4). Compared with conifers, six genes (four PSII genes, one cytochrome gene, and one c-type cytochrome synthesis gene) showed lower ω values in non-conifers, and 12 genes (three PSI genes, one PSII gene, six genetic system genes, and two other genes) showed higher ω values in non-conifers. The lower ω values of these genes indicates the greater the negative selection effect, which may be more conducive to their function in various groups. The differences in selection pressure between conifers and non-conifers may be related to plant height; non-conifers are typically shorter than conifers. Efficient photosynthesis is required to obtain enough sunlight, necessitating negative selection pressure on the genes encoding the photosynthetic machinery. However, some such genes in conifers experienced negative selection pressure, which may be related to their structure or function or to habitat differences. The negative selection exerted by the habitats of gnetophytes leads to gene-specific reductions in dN/dS in their plastomes and genomes (Wang et al., 2015).
4.5 Reduced substitution rate in the IR region
Genes in the IR region tend to have a low substitution rate (Li, Kuo, et al., 2016; Li, Gao, et al., 2016; Ping, Feng, et al., 2021; Ping, Li, et al., 2021). In this study, ycf2 in the IR region showed a reduced substitution rate (Figure 4) but weak selection pressure (ωIR-21 = 0.76124, ωSC-57 = 0.67832), indicating the lack of correlation between these factors. However, Lin et al. (2012) showed that the substitution rate of ycf2 in the SC region of G. biloba was not significantly higher than that in the IR region in seven other species. These discrepancies in results are attributable to differences in sample size. In gymnosperms, 3'-rps12 (exons 2–3) shows a reduced substitution rate and negative selection pressure when present in the IR region (Ping, Feng, et al., 2021).
The substitution rate and selection pressure of rps7 in the IR region did not change, indicating high stability during gymnosperm evolution. The other gene, psbA, encodes the D1 protein in the PSII core complex. In ferns, psbA, ycf2, rps7, and 3'-rps12 (exons 2–3) showed reduced substitution rates when present in the IR region (Li, Kuo, et al., 2016; Li, Gao, et al., 2016). psbA only partially enters the IR region in three gymnosperm species and shows increased substitution rates (Figure 4) and weak selection pressure. These characteristics differ from those of other genes; the most reasonable explanation is that this gene is undergoing dynamic changes in gymnosperms, resulting in changes in its substitution rate and selection pressure.
IR regions may play an important role in the maintenance of genomic stability (Maréchal & Brisson, 2010), such as by replication initiation (Heinhorst & Cannon, 1993), genome stabilization (Palmer & Thompson, 1982), and gene conservation (Palmer & Thompson, 1982; Wolfe et al., 1987). IR loss is related to structural recombination. In some taxa lacking an IR region, plastids underwent significant structural changes, including gene and intron loss, multiple inversion, and translocation and duplication of plastome segments (Jin et al., 2020; Hirao et al., 2008; Mower & Vickrey, 2018). Sequences in the IR region often have a reduced substitution rate, which is related to the region's characteristics. Due to its double-copy nature, the frequency of gene conversion is higher, and some mutation sites are repaired, resulting in a low substitution rate (Birky & Walsh, 1992; Khakhlova & Bock, 2006).
ACKNOWLEDGEMENTS
We thank the National Natural Science Foundation for its support (31670200, 31770587, 31872670, and 32071781).
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTION
Jingyao Ping: Conceptualization (equal); Data curation (equal); Formal analysis (equal); Investigation (equal); Methodology (equal); Writing – original draft (equal). Jing Hao: Data curation (equal); Formal analysis (equal); Methodology (equal). Jinye Li: Data curation (equal); Formal analysis (equal); Resources (equal). Yiqing Yang: Formal analysis (equal); Methodology (equal). Yingjuan Su: Funding acquisition (equal); Writing – original draft (equal); Writing – review & editing (equal). Ting Wang: Funding acquisition (equal); Writing – original draft (equal); Writing – review & editing (equal).
Open Research
DATA AVAILABILITY STATEMENT
Data source is NCBI database:
https://www.ncbi.nlm.nih.gov/nuccore/NC_020319
https://www.ncbi.nlm.nih.gov/nuccore/NC_031413
https://www.ncbi.nlm.nih.gov/nuccore/NC_042668
https://www.ncbi.nlm.nih.gov/nuccore/NC_009618
https://www.ncbi.nlm.nih.gov/nuccore/NC_026036
https://www.ncbi.nlm.nih.gov/nuccore/NC_026041
https://www.ncbi.nlm.nih.gov/nuccore/NC_026037
https://www.ncbi.nlm.nih.gov/nuccore/NC_027512
https://www.ncbi.nlm.nih.gov/nuccore/NC_026040
https://www.ncbi.nlm.nih.gov/nuccore/NC_027514
https://www.ncbi.nlm.nih.gov/nuccore/NC_027513
https://www.ncbi.nlm.nih.gov/nuccore/NC_027511
https://www.ncbi.nlm.nih.gov/nuccore/NC_016986
https://www.ncbi.nlm.nih.gov/nuccore/NC_021438
https://www.ncbi.nlm.nih.gov/nuccore/NC_011942
https://www.ncbi.nlm.nih.gov/nuccore/NC_028734
https://www.ncbi.nlm.nih.gov/nuccore/NC_026301
https://www.ncbi.nlm.nih.gov/nuccore/NC_011954
https://www.ncbi.nlm.nih.gov/nuccore/NC_029347
https://www.ncbi.nlm.nih.gov/nuccore/NC_044772
https://www.ncbi.nlm.nih.gov/nuccore/NC_044773
https://www.ncbi.nlm.nih.gov/nuccore/EU342371
https://www.ncbi.nlm.nih.gov/nuccore/NC_010548
https://www.ncbi.nlm.nih.gov/nuccore/NC_016065
https://www.ncbi.nlm.nih.gov/nuccore/NC_021441
https://www.ncbi.nlm.nih.gov/nuccore/NC_021437
https://www.ncbi.nlm.nih.gov/nuccore/NC_024022
https://www.ncbi.nlm.nih.gov/nuccore/NC_042763
https://www.ncbi.nlm.nih.gov/nuccore/NC_034941
https://www.ncbi.nlm.nih.gov/nuccore/NC_045277
https://www.ncbi.nlm.nih.gov/nuccore/NC_023121
https://www.ncbi.nlm.nih.gov/nuccore/NC_039562
https://www.ncbi.nlm.nih.gov/nuccore/NC_028155
https://www.ncbi.nlm.nih.gov/nuccore/NC_026296
https://www.ncbi.nlm.nih.gov/nuccore/MT227813
https://www.ncbi.nlm.nih.gov/nuccore/KP099642
https://www.ncbi.nlm.nih.gov/nuccore/KX832629
https://www.ncbi.nlm.nih.gov/nuccore/MH121051
https://www.ncbi.nlm.nih.gov/nuccore/NC_034943
https://www.ncbi.nlm.nih.gov/nuccore/NC_036996
https://www.ncbi.nlm.nih.gov/nuccore/NC_031354
https://www.ncbi.nlm.nih.gov/nuccore/NC_027423
https://www.ncbi.nlm.nih.gov/nuccore/NC_042176
https://www.ncbi.nlm.nih.gov/nuccore/NC_042177
https://www.ncbi.nlm.nih.gov/nuccore/NC_034940
https://www.ncbi.nlm.nih.gov/nuccore/MF977938
https://www.ncbi.nlm.nih.gov/nuccore/NC_021110
https://www.ncbi.nlm.nih.gov/nuccore/NC_027581
https://www.ncbi.nlm.nih.gov/nuccore/NC_024945
https://www.ncbi.nlm.nih.gov/nuccore/NC_038099
https://www.ncbi.nlm.nih.gov/nuccore/NC_041503
https://www.ncbi.nlm.nih.gov/nuccore/NC_029398
https://www.ncbi.nlm.nih.gov/nuccore/NC_029734
https://www.ncbi.nlm.nih.gov/nuccore/NC_014575
https://www.ncbi.nlm.nih.gov/nuccore/MF564195
https://www.ncbi.nlm.nih.gov/nuccore/NC_043856
https://www.ncbi.nlm.nih.gov/nuccore/NC_030631
https://www.ncbi.nlm.nih.gov/nuccore/NC_016064
https://www.ncbi.nlm.nih.gov/nuccore/NC_043913
https://www.ncbi.nlm.nih.gov/nuccore/NC_011930
https://www.ncbi.nlm.nih.gov/nuccore/NC_030630
https://www.ncbi.nlm.nih.gov/nuccore/NC_036811
https://www.ncbi.nlm.nih.gov/nuccore/NC_016058
https://www.ncbi.nlm.nih.gov/nuccore/NC_042775
https://www.ncbi.nlm.nih.gov/nuccore/NC_042777
https://www.ncbi.nlm.nih.gov/nuccore/NC_023120
https://www.ncbi.nlm.nih.gov/nuccore/NC_023805
https://www.ncbi.nlm.nih.gov/nuccore/NC_034942
https://www.ncbi.nlm.nih.gov/nuccore/NC_024827
https://www.ncbi.nlm.nih.gov/nuccore/NC_045880
https://www.ncbi.nlm.nih.gov/nuccore/NC_044893
https://www.ncbi.nlm.nih.gov/nuccore/NC_023119
https://www.ncbi.nlm.nih.gov/nuccore/KP259800
https://www.ncbi.nlm.nih.gov/nuccore/MT227812
https://www.ncbi.nlm.nih.gov/nuccore/NC_039155
https://www.ncbi.nlm.nih.gov/nuccore/NC_026450
https://www.ncbi.nlm.nih.gov/nuccore/NC_045394
https://www.ncbi.nlm.nih.gov/nuccore/NC_045395




