The Mitochondrial Genome of the Imperiled Goliath Grouper Epinephelus itajara: Selective Pressures in Protein Coding Genes, Secondary Structure of tRNA Genes, and Phylogenetic Placement
Funding: The authors received no specific funding for this work.
ABSTRACT
The goliath grouper Epinephelus itajara (Perciformes: Epinephelidae) is a large, critically endangered fish distributed across coastal habitats in the western Atlantic Ocean, from Florida to southern Brazil, and with additional populations in the eastern Pacific basin. Conservation concerns for this species stem from historical overfishing, habitat loss, and life-history traits such as slow growth and late sexual maturity. In this study, to aid conservation efforts, we assembled and characterized the complete mitochondrial genome of E. itajara. The mitochondrial genome of Epinephelus itajara is 16,561 bp long and comprises 13 protein-coding genes (PCGs), two ribosomal RNA genes (12S and 16S rRNA), 22 transfer RNA (tRNA) genes, and an 856 bp control region. Gene order is identical to that reported for other congeneric species. The overall A + T content is 56%, and codon usage shows a preference for A + T-rich codons. All PCGs were found to be under purifying selection, with variation in selective pressure among genes; cox1 and nad4 were under the strongest and weakest selection, respectively. Secondary structure analysis of the tRNA genes displayed typical cloverleaf secondary structures, except for trnS1, which lacked a complete D-arm. Comparative analyses between MiTFi and RASP2 revealed that MiTFi provided more accurate predictions of tRNA secondary structures. The control region exhibited a high A + T content (69.9%), multiple microsatellite motifs, and one tandem repeat, along with hairpin secondary structures. These features mirror findings in closely related species. A maximum likelihood phylogenetic analysis based on translated PCGs did not support the monophyletic status of the genus Epinephelus and indicated a sister relationship between Epinephelus itajara and Epinephelus lanceolatus, another large-bodied grouper from the Indo-Pacific Ocean. The newly sequenced mitochondrial genome of Epinephelus itajara provides a new genomic resource that can support future conservation efforts.
1 Introduction
Groupers belonging to the subfamily Epinephelinae (Perciformes: Epinephelidae) can reach large body sizes and exhibit wide mouths with which they create a strong vacuum to draw prey into their mouth, acting as highly effective predators (Sadovy and Eklund 1999; Rimmer and Glamuzina 2019). Among them, the goliath grouper, Epinephelus itajara, is the largest member of the family in the Atlantic Ocean, though larger species exist in other regions such as the giant grouper inhabiting the Indo-Pacific region (Craig et al. 2024). Epinephelus itajara reaches lengths of over 2.4 m and weighs up to 363 kg (Bullock et al. 1992; Morris et al. 2000; Sadovy and Eklund 1999). This massive, mottled brown fish inhabits coastal waters off the Atlantic Ocean, ranging from Florida in the southeastern USA to Paraná in southern Brazil (Froese and Pauly 2024). Additionally, Epinephelus itajara has been reported in the eastern Pacific Ocean, with populations ranging from Costa Rica to Peru. However, genetic evidence suggests that these Pacific populations represent a distinct and separate species from the Atlantic Epinephelus itajara, despite looking alike morphologically (Craig et al. 2009). Small juvenile groupers are commonly found in structurally complex habitats formed by root networks of the red mangrove Rhizophora mangle, which serve as important nurseries by providing shelter and abundant food resources (Koenig et al. 2007). As they mature, they transition to coral reefs, where they have access to a variety of prey. These impressive predators primarily feed on crustaceans such as spiny lobsters, octopuses, shrimp, and crabs, along with other vertebrates such as numerous species of teleost fish and even young sea turtles (Artero et al. 2015). Predators of the goliath grouper include barracudas Sphyraena spp., the king mackerel Scomberomorus cavalla, moray eels (Muraenidae spp.), and sandbar sharks Carcharhinus plumbeus (Bakermans and San Martín 2022). The goliath grouper is a protogynous hermaphrodite (female to male sex changer), exhibits slow growth, and reaches sexual maturity relatively late, around 8 years of age (Murie et al. 2023; Bakermans and San Martín n.d.; Orth 2023). Goliath groupers can live for over 30 years, with the oldest recorded individual attaining 37 years old (Bullock et al. 1992).
Epinephelus itajara faces significant conservation challenges due to historical overfishing and habitat destruction paired with the species' slow growth and late maturity (Koenig et al. 2020), which do not permit their populations to be resilient to intense fishing practices. Goliath groupers are heavily fished, given their large body size and meat quality, which drives a very high market value for this species (Orth 2023). Coastal development and pollution have reduced shelter availability and food sources for this and other fish as well (Koenig et al. 2020). The species' life history traits limit its capacity for population recovery, rendering it highly vulnerable to the combined impacts of excessive fishing pressure and habitat loss. Goliath grouper populations in Florida declined by up to 95% of their population prior to the implementation of anti-fishery protections in 1990, which led to the species being designated as a candidate for listing under the Endangered Species Act in 1991 and later as a species of concern (McClenachan 2009; NOAA Fisheries 2023). Today, the goliath grouper remains classified as ‘Critically Endangered’ by the International Union for Conservation of Nature (IUCN), highlighting the urgent need for comprehensive conservation efforts. These measures include fishing bans, habitat restoration, and broader ecosystem management strategies aimed at ensuring the species' long-term survival (Bertoncini et al. 2018).
Due to the imperiled status of Epinephelus itajara, generating genetic and genomic resources is essential to support conservation strategies focusing on this remarkable species. Unfortunately, there is limited genomic information available for Epinephelus itajara, though several genetic studies have been conducted to support its conservation. Among them, a study of populations from the Northern Brazilian coast that utilized mitochondrial DNA sequences, specifically fragments of the protein-coding gene cytochrome b (cob) and control region, revealed low genetic diversity in the studied populations and greater genetic variability in populations inhabiting well-preserved mangrove forests compared to those present in degraded habitats, highlighting the importance of healthy mangrove ecosystems for the species conservation and fisheries management (Silva-Oliveira et al. 2008). Illegal fishing remains a major threat to the conservation of Epinephelus itajara, jeopardizing efforts to protect and restore its population (Giglio et al. 2014). In a second study, to help prevent grouper mislabeling and curb illegal fishing, species-specific primers were developed for the mitochondrial Cytochrome Oxidase subunit I (cox1) gene for the simultaneous identification of nine different species of the subfamily Epinephelidae from the western Atlantic basin using PCR (Damasceno et al. 2016). In a more recent study, DNA barcoding of the same COI gene was applied to fish market samples along the Brazilian coast, revealing that 77.3% of Epinephelus itajara specimens were illegally commercialized (Almeida et al. 2024). Beyond mitochondrial data, several studies have expanded the nuclear genomic toolkit for the species. Among them, the Florida Fish and Wildlife Research Institute isolated 40 microsatellite markers, developing 29 polymorphic microsatellite loci for both Atlantic and Pacific goliath groupers. These markers were validated using samples from both species and revealed significant genetic differentiation between the Atlantic and Pacific populations. The study above also noted fixed allelic differences and suggested that Epinephelus itajara may show signs of a genetic bottleneck, likely due to historical overharvesting. The developed microsatellite markers provide a valuable resource for future studies of population structure, gene flow, and conservation genetics in these species (Seyoum et al. 2013). Similarly, 12 novel exon-primed intron-crossing (EPIC) nuclear markers were developed for Epinephelus itajara using samples from northern Brazil (Silva-Oliveira et al. 2013). Several of these markers revealed intraspecific polymorphism and demonstrated utility across multiple Epinephelus species. These markers are also valuable tools in population genetic and phylogeographic studies that are beneficial to the conservation of this species. Inter Simple Sequence Repeat (ISSR) nuclear markers can also be used in population genetics and were developed for Epinephelus itajara in Brazil (Benevides et al. 2014). Benevides et al. (2014) found moderate global genetic variation (~50%), but identified a genetically distinct population in Santa Catarina, Brazil, which showed the highest levels of genetic diversity and evidence of historical isolation. Their analyses revealed two evolutionarily significant units (ESUs) within Epinephelus itajara's Atlantic range, suggesting limited gene flow between Santa Catarina, Brazil, and other populations. These findings emphasize the importance of region-specific conservation strategies due to evolutionary distinctiveness. Lastly, a recent development of a simple and cost-effective PCR-based method was utilized to accurately identify Epinephelus itajara using a species-specific primer (16S-RYOP4) targeting a region of the mitochondrial 16S rRNA gene. The method reliably distinguished Epinephelus itajara from other species of Epinephelus and closely related genera by producing a distinctive two-band electrophoresis pattern (423 and 621 bp). This tool is particularly useful for enforcement agencies in regions with high rates of illegal harvesting, as it allows for rapid identification of fishery products even when morphological features are removed (Oliveira et al. 2021).
In this study, we provide a new genomic resource for the goliath grouper; we sequenced the mitochondrial genome of Epinephelus itajara and analyzed it in detail. Following recommendations in Baeza (2022), we analyzed the nucleotide composition of the entire mitochondrial genome, codon usage of and selective pressure in the mitochondrial protein-coding genes (PCGs), examined the secondary structure of the transfer RNA (tRNA) genes, and explored the presence of short tandem repeats and microsatellites within the control region. This genomic resource is expected to support conservation plans for this iconic fish, for instance, biomonitoring of this species using environmental DNA.
2 Methods
The mitochondrial genome of E. itajara was sequenced using DNA extracted from a specimen (USNM: FISH:416759) belonging to the fish collection of the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA. This specimen was collected from the Atlantic coast of Florida, United States (Port Canaveral Main Channel, Brevard County; 28.4100° N, 80.6400° W) at a depth of 2 m. Following Cady et al. (2021) and Skufca and Baeza (2025), muscle tissue was used for extracting genomic DNA (gDNA) using the AutoGenPrep 965 automated DNA extraction robot (AutoGen, Holliston, MA, USA). Then, as in Cady et al. (2021) and Skufca and Baeza (2025), an Illumina shotgun library (paired-end) was prepared utilizing the NEB Ultra II DNA library prep kit (New England Biolabs, Ipswich, MA, USA). Short-read sequencing was achieved using an Illumina MiSeq (Illumina, San Diego, CA, USA) platform (at 2 × 150 cycles). Lastly, the totality of the sequenced reads (n = 7,927,452 pairs) was utilized for the assembly of the mitochondrial genome belonging to the goliath grouper with the pipeline GetOrganelle v. v1.7.6.1 (Jin et al. 2020).
The newly assembled mitochondrial genome was annotated using the program MITOS2 (https://usegalaxy.eu/storage, Donath et al. 2019) as implemented in the platform Galaxy Europe (https://usegalaxy.eu/login/start?redirect=None, Jalili et al. 2020). Next, the software Mega11 (Kumar et al. 2018) was used to estimate the nucleotide composition of the entire mitochondrial genome. The online web server GenomeVx (http://wolfe.ucd.ie/GenomeVx/, Conant and Wolfe 2008) was used to depict the assembled and annotated mitochondrial genome as a circular map.
Codon usage was assessed using the Codon Usage Calculator available on the web server Sequence Manipulation Suite (https://www.bioinformatics.org/sms2/codon_usage.html, Stothard 2000). EZcodon (http://ezmito.unisi.it/ezcodon, Cucini et al. 2021) was also used to calculate the relative synonymous codon usage (RSCU) of the mitochondrial protein-coding genes.
We assessed whether PCGs in the studied mitochondrial genome were subjected to neutral, negative (purifying), or positive (diversifying) selection. To achieve this goal, first, each mitochondrial protein-coding gene was aligned to an orthologous sequence belonging to the congeneric Epinephelus akaara (EU043377.1) using the program MEGA11. Next, we estimated Ka, Ks, and the Ka/Ks ratio using the program KaKs Calculator 3 (Zhang 2022) and the congeneric Epinephelus akaara (EU043377.1) as an outgroup. The non-synonymous substitution rate (Ka), the synonymous substitution rate (Ks), and the ratio of Ka/Ks were used to infer the type of selection acting on a gene. A Ka/Ks ratio < 1 indicates purifying selection, a Ka/Ks ratio = 1 indicates that the locus may be evolving neutrally with respect to selection, and a Ka/Ks ratio > 1 indicates positive selection. For our calculations, we applied the MYN model to account for variability in mutation rate across the length of the studied sequences. The statistical significance of the estimated Ka/Ks ratio is assessed using a p-value from a Fisher test (Zhang 2022).
The secondary structures of all mitochondrial transfer RNA (tRNA) genes were predicted using two different bioinformatic tools, that is, the program MiTFi (Jühling et al. 2012) within the MITOS2 platform and the newly released software RASP2 (http://rasp.zhanglab.net/predstr/, Mu et al. 2025). We compared the secondary structure predicted by the two tools to benchmark the accuracy of RASP2. All secondary structure predictions were visualized using the platform Forna (http://rna.tbi.univie.ac.at/forna/, Kerpedjiev et al. 2015).
In the control region of the studied mitochondrial genome, microsatellites were detected using the web server Microsatellite Repeats Finder (http://insilico.ehu.es/mini_tools/microsatellites/, Bikandi et al. 2004). Tandem repeats were also identified using the program Tandem Repeat Finder (https://tandem.bu.edu/trf/trf.html, Benson 1999). Lastly, the RNAfold Web Server (Lorenz et al. 2011) was used to predict the secondary structure of the control region. RNAfold predicted both a Minimum Free Energy (MFE) secondary structure that represents the CR's most stable conformation with the lowest free energy and a Centroid secondary structure, which evaluates all possible folds and selects the one with the lowest ensemble distance (Lorenz et al. 2011).
2.1 Phylogenetic Placement of the Goliath Grouper Based on Mitochondrial Genomes
We explored the phylogenetic position of the goliath grouper in the genus Epinephelus and examined the monophyletic status of the same genus using the phylogenetic signal provided by mitochondrial PCGs. The phylomitogenomic analysis included the mitochondrial genome of Epinephelus itajara (sequenced as part of BioProject PRJNA720393 and available under SRA accession SRX17832793) along with 54 additional mitochondrial genomes from 40 other Epinephelus species retrieved from GenBank (consulted 25 April 2025). Mitochondrial genomes belonging to the genera Aethaloperca (n = 2 sequences belonging to 1 species), Anyperodon (n = 1 sequence belonging to a single species), Cephalopholis (n = 11 sequences belonging to 10 species), Cromileptes (n = 3 sequences belonging to a single species), Hyporthodus (n = 3 sequences belonging to 3 species), Mycteroperca (n = 3 sequences belonging to 3 species), Triso (n = 1), Plectropomus (n = 6 sequences belonging to 3 species), and Variola (n = 2 sequences belonging to 2 species) belonging to the family Epinephelidae, and other species not belonging to the aforementioned family: Etheostoma (n = 1), Caranx (n = 1), Perca (n = 1), and Toxotes (n = 1), were used as outgroups during the analysis (Table S1).
To start the analysis, we aligned each mitochondrial PCG using the program ClustalW (Thompson 1997) to then remove regions in each alignment that were poorly aligned with the software GBlocks (Castresana 2000; Talavera and Castresana 2007). Next, we selected the best model of character evolution using the program ProtTest (Abascal et al. 2005; Minh et al. 2013). At last, a Maximum Likelihood (ML) phylogenetic analysis was conducted on the PCG-partitioned dataset using the web server IQTREE v. 1.6.10 (http://iqtree.cibiv.univie.ac.at—Nguyen et al. 2015). The robustness of the ML tree topology was evaluated by performing 1000 bootstrap replications of the observed dataset.
3 Results & Discussion
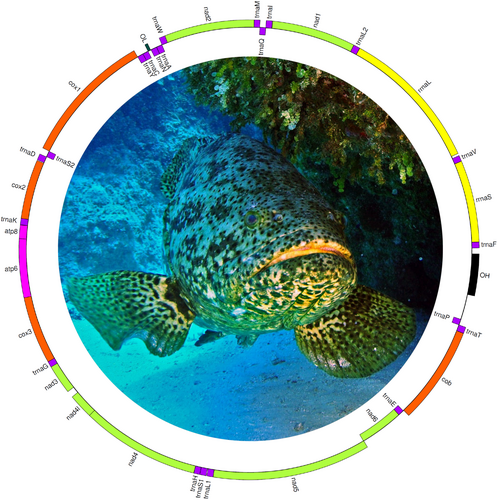
The program GetOrganelle assembled a complete (circular) mitochondrial genome of Epinephelus itajara (GenBank accession number OP056827.1) with an average coverage of 105× per nucleotide. The mitochondrial genome of Epinephelus itajara was 16,561 base pairs (bp) long and contained 13 protein-coding genes, 22 tRNA genes, and 2 rRNA genes. It also includes a non-coding region 856 bp long (Figure 1, Table 1). Genes in the circular mitochondrial genome map were color-coded based on their type and functional classification. Transfer RNA (tRNA) genes are shown in purple, while ribosomal RNA (rRNA) genes (12S and 16S) are highlighted in yellow. The black regions represent the non-coding control region, including the origin of heavy and light strand replication (OH and OL). Green indicates NADH dehydrogenase subunits (nad1–nad6 and nad4L), whereas orange is used for cytochrome oxidase subunits (cox1, cox2, cox3) and cytochrome b (cob). Finally, pink marks the ATP synthase subunits (atp6 and atp8). In congeneric species, the mitochondrial genome length ranges from 16,418 bp in Epinephelus coioides to 16,965 bp in Epinephelus areolatus (Zhuang et al. 2013). In turn, in the family Epinephelidae, mitochondrial genome sizes vary more widely, ranging from 16,504 bp in Cromileptes altivelis to 16,767 bp in Cephalopholis argus (Zhuang et al. 2013). Thus, the mitochondrial genome length in Epinephelus itajara falls within the previously observed range in closely related species. Also, the gene order of Epinephelus itajara is identical to that reported in other congeneric species (e.g., in Epinephelus akaara and Epinephelus areolatus—Zhuang et al. 2013, among others).
| Gene name | Type | Start | Stop | Strand | Length (bp) | Start codon | Stop codon | Anticodon | Continuity |
|---|---|---|---|---|---|---|---|---|---|
| tRNAF | tRNA | 1 | 69 | + | 69 | GAA | 0 | ||
| rRNAS | rRNA | 70 | 1022 | + | 952 | +1 | |||
| tRNAV | tRNA | 1023 | 1092 | + | 69 | TAC | +1 | ||
| rRNAL | rRNA | 1119 | 2797 | + | 1678 | +27 | |||
| tRNAL2 | tRNA | 2798 | 2873 | + | 75 | TAA | +1 | ||
| NaD1 | PCG | 2874 | 3848 | + | 974 | ATG | TAA | +1 | |
| tRNAI | tRNA | 3853 | 3922 | + | 69 | GAT | +5 | ||
| tRNAQ | tRNA | 3922 | 3992 | − | 0 | TTG | 0 | ||
| tRNAM | tRNA | 3993 | 4061 | + | 68 | CAT | +1 | ||
| NaD2 | PCG | 4062 | 5108 | + | 1046 | ATG | TAG | +1 | |
| tRNAW | tRNA | 5107 | 5177 | + | 70 | TCA | −1 | ||
| tRNAA | tRNA | 5179 | 5247 | − | 68 | TGC | +2 | ||
| tRNAN | tRNA | 5248 | 5320 | − | 72 | GTT | +1 | ||
| OL | Replication | 5325 | 5359 | + | 34 | +5 | |||
| tRNAC | tRNA | 5360 | 5427 | − | 67 | GCA | +1 | ||
| tRNAY | tRNA | 5428 | 5498 | − | 70 | GTA | +1 | ||
| COX1 | PCG | 5500 | 7050 | + | 1550 | GTG | TAA | +2 | |
| tRNAS2 | tRNA | 7053 | 7123 | − | 70 | TGA | +3 | ||
| tRNAD | tRNA | 7127 | 7199 | + | 72 | GTC | +4 | ||
| COX2 | PCG | 7208 | 7898 | + | 690 | ATG | T— | +9 | |
| tRNAK | tRNA | 7899 | 7972 | + | 73 | +1 | |||
| ATP8 | PCG | 7974 | 8141 | + | 167 | ATG | TAA | TTT | +2 |
| ATP6 | PCG | 8132 | 8815 | + | 683 | CTG | TAA | −9 | |
| COX3 | PCG | 8815 | 9600 | + | 785 | ATG | TAA | 0 | |
| tRNAG | tRNA | 9600 | 9671 | + | 71 | TCC | 0 | ||
| NaD3 | PCG | 9669 | 10,011 | + | 342 | ATA | T— | −2 | |
| NaD4l | PCG | 10,090 | 10,386 | + | 296 | ATG | TAA | −79 | |
| NaD4 | PCG | 10,380 | 11,760 | + | 1380 | ATG | T— | −6 | |
| tRNAH | tRNA | 11,761 | 11,830 | + | 69 | GTG | +1 | ||
| tRNAS1 | tRNA | 11,831 | 11,904 | + | 73 | GCT | +1 | ||
| tRNAL1 | tRNA | 11,913 | 11,985 | + | 72 | TAG | +9 | ||
| NaD5 | PCG | 11,986 | 13,824 | + | 1838 | ATG | TAA | +1 | |
| NaD6 | PCG | 13,821 | 14,342 | − | 521 | ATG | TAG | −3 | |
| tRNAE | tRNA | 14,343 | 14,412 | − | 69 | TTC | +1 | ||
| COB | PCG | 14,420 | 15,560 | + | 1140 | ATG | T— | +8 | |
| tRNAT | tRNA | 15,561 | 15,634 | + | 73 | TGT | +1 | ||
| tRNAP | tRNA | 15,635 | 15,704 | − | 69 | TGG | +1 | ||
| CR | Non-coding | 15,705 | 16,561 | + | 856 | +1 | |||
| OH | Replication | 16,008 | 16,493 | + | 485 | +304 |
- Note: Arrangement and annotation. Gene name, gene type, start and end position in the mitochondrial genome, position in the leading (+) or lagging (−) strand, and start and stop codons for protein-coding genes are shown.
The mitochondrial genome of Epinephelus itajara exhibits the following nucleotide composition: Adenine (A) = 29.3%, Guanine (G) = 15.2%, Cytosine (C) = 28.8%, and Thymine (T) = 26.7%, resulting in an A + T content of 56%. This AT-rich composition aligns well with values observed in other groupers. In the genus Epinephelus, the A + T content ranges from 55% in Epinephelus amblycephalus to 56% in Epinephelus lanceolatus (Wang et al. 2022; Zhuang et al. 2013). Similarly, in the family Epinephelidae, A + T content remains consistently high, with values ranging from 55.2% in Cephalopholis leopardus to 56.1% in Cephalopholis sonnerati (Wang et al. 2022; Zhuang et al. 2013). In Epinephelus itajara and related species, the AT-rich content of mitochondrial genomes might result from a combination of mutational biases and replication asymmetry (Tamura and Nei 1993; Perna and Kocher 1995; Nikolaou and Almirantis 2005; Uddin et al. 2020; Alvarenga et al. 2024).
In the PCGs of the studied mitochondrial genome, most genes possessed conventional start (ATG) and stop (TAA) codons. However, notable deviations were observed, such as the use of GTG and CTG start codons by cox1 and atp6, respectively, and the presence of incomplete stop codons in cox2, nad3, nad4, and cob. In the mitochondrial genome of Epinephelus itajara, PCGs do not exhibit a proportional codon usage. The most frequently used codons were CTA (Leu), CGA (Arg), AAA (Lys), and CAA (Gln). The least frequently used codons for PCGs, except stop codons, were CGG (Arg), TCG (Ser), ACG (Thr), and GCG (Ala) (Table S2). Furthermore, the RSCU analysis revealed biased usage among synonymous codons in most studied PCGs, with a preference for A + T-rich codons over G + C-rich codons (Figure 2). This non-random, A + T-rich codon usage pattern is consistent with findings in other groupers, including Epinephelus bilobatus, Epinephelus maculatus, and Epinephelus longispinis (He et al. 2024).

In Epinephelus itajara, Ka/Ks values estimated for all 13 mitochondrial PCGs were well below 1, indicating that all these genes are under purifying selective pressure (all p-values < 0.05 in all cases) (Table 2). However, there were qualitative differences among genes. For example, nad4 exhibited a notably higher Ka/Ks ratio (= 0.5669) compared to the other PCGs, suggesting it may be under relatively weaker purifying selection. In turn, cox1 exhibited a much lower Ka/Ks ratio (= 0.0058), indicating that this gene might be under a strong purifying selection. Our results indicate that the strength of selective pressure varies among genes. Selective pressure analyses of mitochondrial PCGs have been conducted in only a few species belonging to the family Epinephelidae. Among them, all mitochondrial PCGs have been shown to experience purifying selection in Corydoras leopardus, Cephalopholis spiloparaea, Epinephelus amblycephalus, and Epinephelus hexagonatus (Wang et al. 2022).
| Gene | Ka | Ks | Ka/Ks | p |
|---|---|---|---|---|
| NaD1 | 0.0249851 | 1.46574 | 0.0170461 | 1.52E-72 |
| NaD2 | 0.0706385 | 0.731155 | 0.0966122 | 5.26E-38 |
| COX1 | 0.0112346 | 1.91669 | 0.00586149 | 1.26E-132 |
| COX2 | 0.0174626 | 1.12656 | 0.0155008 | 1.59E-45 |
| ATP8 | 0.103113 | 0.952473 | 0.108259 | 3.71E-06 |
| ATP6 | 0.0482032 | 1.02193 | 0.0471687 | 3.42E-42 |
| COX3 | 0.0171049 | 0.608101 | 0.0281284 | 1.33E-41 |
| NaD3 | 0.0445345 | 1.17934 | 0.0377622 | 3.00E-22 |
| NaD4l | 0.0384378 | 0.904612 | 0.0424909 | 2.45E-15 |
| NaD4 | 0.345154 | 0.608765 | 0.566973 | 0.252856 |
| NaD5 | 0.0456903 | 1.40191 | 0.0325915 | 2.72E-143 |
| NaD6 | 0.0572596 | 2.17001 | 0.0263869 | 1.52E-34 |
| COB | 0.042934 | 1.15065 | 0.037313 | 6.31E-87 |
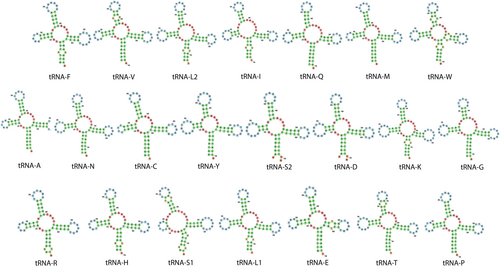
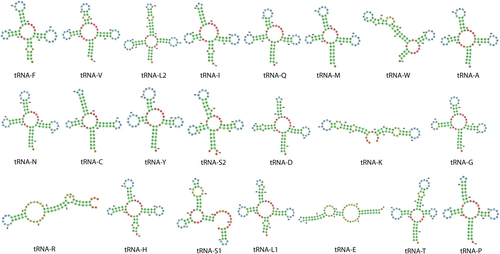
The tRNA genes in the mitochondrial genome of Epinephelus itajara ranged from 67 to 75 bp in length, in tRNAC and tRNAL2, respectively. The secondary structure analysis of the tRNA genes predicted by MiTFi revealed that all these genes exhibited the expected cloverleaf secondary structure except for tRNA-S1, which lacked a complete D arm (Figure 3). This truncation is consistent with observations in the few grouper species where similar analyses have been conducted. For example, Corydoras leopardus and Cephalopholis spiloparaea also exhibit a truncated or absent D arm in their tRNA-S1genes (Wang et al. 2022). Whether the atypical secondary structure of the tRNA-S1gene is a conserved feature in the subfamily Epinephelinae remains to be addressed. Interestingly, major discrepancies were observed between the MiTFi and RASP secondary structure predictions (Figure 4). While MiTFi identified all 22 tRNA genes (except tRNA-S1) with cloverleaf secondary structures, RASP2 only predicted 10 tRNA genes with a cloverleaf secondary structure, suggesting that the latter tool is not as accurate as MiTFi to predict the secondary structure of mitochondrial tRNA genes. We note that MiTFi's predictions align more closely with experimentally validated tRNA structures from related species (Wang et al. 2022).
Both the small and large ribosomal subunit RNAs in the studied mitochondrial genome were located in the leading strand. The 12S rRNA gene is approximately 952 base pairs long and is flanked by the tRNAF gene upstream (5′) and the tRNAV gene downstream (3′). The 16S rRNA gene is 1678 bp long and is flanked by the tRNAV gene upstream (5′) and the tRNAL2 gene downstream (3′). In an earlier study that described the mitochondrial genomes of 22 groupers, the 12S rRNA gene ranged from 952 to 961 bp, and the 16S rRNA gene ranged from 1695 to 1722 bp (Zhuang et al. 2013), in line with our observations. The two ribosomal genes in Epinephelus itajara are A + T-rich, with the small subunit (12S rRNA) containing a 51.42% A + T content and the large subunit (16S rRNA) containing a 54.67% A + T content. This A + T-rich composition is consistent with the results from a comparative analysis of four complete mitochondrial genomes in groupers, which reported A + T contents ranging from 53.0% to 53.6% for the 12S rRNA gene and from 55.4% to 56.9% for the 16S rRNA gene, reflecting a moderate A + T bias typical of mitochondrial rRNA genes in related species (Wang et al. 2022).
The control region (CR) of Epinephelus itajara is 856 bp long and includes the origin of replication of the heavy strand (OH) (Figure 1, Table 1). Notably, the origin of replication for the light strand (OL) was located outside of the CR, flanked by tRNA-P (in the 5′ end) and tRNA-F (3′ end). The studied CR is shorter than those found in other closely related species. For example, E. quoyanus has a CR of 1092 bp while the Red Grouper Plectropomus leopardus has a 1077 bp CR (Peng et al. 2014; Zhu and Yue 2008). Despite differences in length, these CRs share similar nucleotide composition; the control region of E. itajara is A + T-rich (A + T content = 69.9%), a feature commonly observed in mitochondrial CRs across various other closely related species (Zhuang et al. 2013; Peng et al. 2014; Zhu and Yue 2008).
In the mitochondrial CR of Epinephelus itajara, 16 microsatellites were identified, the most common being an A + T dinucleotide motif repeated three times (Table S3). The detected microsatellites were consistently A + T-rich, a trend also observed in related species. For example, in the Orange-Spotted Grouper Epinephelus coioides, microsatellite motifs are predominantly composed of A and T nucleotides (Wang et al. 2011).
A single tandem repeat was identified in the control region of Epinephelus itajara, spanning from position 1 to 205 bp of the studied region and with a period size of 17 bp (repeated 12.1 times). The consensus sequence of this repeat, 5′-AAT TAC ATA TAT GCA TT-3′, is A + T-rich (A + T content = 81%, Table S4). A similar tandem repeat was observed in the congeneric Rock Grouper Epinephelus fasciatomaculosus, where a 141 bp long repeat was found within a 980 bp long control region (Li et al. 2013). The predicted minimum free energy (MFE) and Centroid secondary structure of Epinephelus itajara's mitochondrial control region contained multiple hairpin formations, spanning across the entire region (Figure 5). Studies examining the secondary structure of the CR in other groupers are only a few, but our observations are consistent with findings in other congeneric groupers, including Epinephelus malabaricus, whose CR secondary structure exhibits an array of hairpin structures (Athira et al. 2022). Overall, although studies describing the mitochondrial CR are limited in the genus Epinephelus, in vertebrates, including co-familial species, the control region frequently exhibits microsatellites, short tandem repeats, and hairpin loops (Terencio et al. 2013; Pereira et al. 2008; Cady et al. 2021).

3.1 Phylogenetic Placement of the Goliath Grouper Based on Mitochondrial Genomes
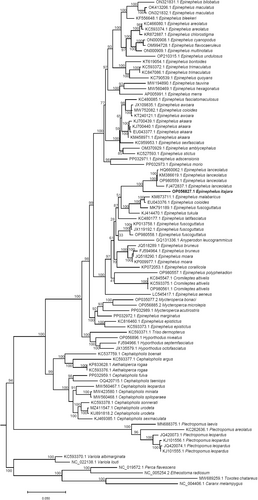
The ML phylogenetic analysis (90 terminal nodes, 3798 amino acids, and 982 informative sites) indicated that the genus Epinephelus was not monophyletic given the position of three genera, Anyperodon (n = 1 species), Cromileptes (n = 3 species), and Mycteroperca (n = 3 species), that clustered deep within a clade comprised of all species of Epinephelus used in our analysis (Figure 6). Within a well-supported clade (bootstrap value [bv] = 98) that included the genera Anyperodon, Cromileptes, Epinephelus, and Mycteroperca, the goliath grouper occupied a derived (late branching) position, being fully supported (bv = 100) as the sister taxon to the Giant Grouper Epinephelus lanceolatus from the Indo-Pacific basin. In turn, Epinephelus itajara and Epinephelus lanceolatus belonged to a poorly supported clade (bootstrap value [bv] = 51) that also included 4 other Indo-Pacific groupers: Epinephelus coioides, Epinephelus fuscoguttatus, Epinephelus malabaricus, and Epinephelus tukula (Figure 6). Additional studies examining the phylogenetic relationships among species in the genus Epinephelus and closely related genera are needed to understand the evolutionary history of this remarkable clade of fish and solve taxonomic classification issues pinpointed by this analysis. Our results are in line with other mitogenome analyses, including a study that recently observed non-monophyly in Epinephelus and noted that Anyperodon and Cromileptes were nested within it (Wang et al. 2022). An older study highlighted the same findings, using 16S rRNA sequences to show that Epinephelus, Cephalopholis, and Mycteroperca do not form distinct, monophyletic groups (Craig et al. 2001). What is new in our analysis is the inclusion of more recently sequenced mitogenomes, such as that of Epinephelus itajara, and a broader sampling of Indo-Pacific species. Together, these data provide a clearer picture of the goliath grouper's evolutionary relationships and further support the need to revise the current classification of Epinephelidae.
4 Conclusion
We analyzed the complete mitochondrial genome of the goliath grouper, Epinephelus itajara, useful for the conservation of this endangered species. The gene order of the newly assembled mitochondrial genome was identical to that reported for other congeneric species. The mitochondrial genome was A + T-rich, codons used more frequently were also A + T-rich, and we provide evidence of purifying selection acting across all protein-coding genes. Secondary structure predictions for tRNA genes indicated that all of them exhibited a cloverleaf secondary structure with the exception of trnS1, which lacked a full D arm, a typical feature observed in other members of the subfamily Epinephelinae. The CR was shorter than those in other closely related species but exhibited typical attributes, including A + T-rich microsatellites, a tandem repeat, and multiple hairpins after prediction of the secondary structure of this region. These findings are consistent with structural features documented in the CR of closely related species. A maximum likelihood phylogenetic analysis supports the utility of PCGs in revealing evolutionary relationships among groupers. This new genomic resource is expected to support conservation efforts in the goliath grouper, Epinephelus itajara.
Author Contributions
Kyla Padgett: data curation (equal), formal analysis (equal), investigation (equal), methodology (equal), project administration (equal), resources (equal), writing – original draft (equal), writing – review and editing (equal). J. Antonio Baeza: data curation (equal), investigation (equal), methodology (equal), project administration (equal), resources (equal), supervision (equal), validation (equal), visualization (equal), writing – original draft (equal), writing – review and editing (equal).
Acknowledgments
The authors are grateful to Dr. Vincent P. Richards (Clemson University) for bioinformatics support and Dr. Katherine Bemis (Smithsonian Institution) for NGS sequencing. Unfortunately, Dr. Katherine Bemis decided not to participate in this study due to heavy time constraints. This study was supported by Creative Inquiry at Clemson University. The authors also thank Daniel DiMichele and Biorepository, National Museum of Natural History, for curating genetic samples; Carrie Craig and the Laboratories of Analytical Biology for support preparing the libraries; Allen Collins and Abigail Reft, NOAA National Systematics Lab and Devon Leopold, Jonah Ventures and for organizing and depositing genetic data on GenBank. Sequencing of the mitochondrial genome analyzed here was provided by a collaborative partnership between the National Systematics Lab, the National Oceanic and Atmospheric Administration, and the National Museum of Natural History, Smithsonian Institution, to develop voucher-based reference libraries for mitochondrial genomes (BioProject: PRJNA720393).
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
Mitochondrial genome assembly data are available online on NCBI GenBank under the accession number OP056827.1.




