The Updated Genome of the Burying Beetle Nicrophorus vespilloides, a Model Species for Evolutionary and Genetic Studies of Parental Care
Funding: This work was supported by Agricultural Research Service, 58-6080-9-006; Division of Integrative Organismal Systems, DBI-2217859, IOS-1354358.
ABSTRACT
Understanding the evolution of social behavior requires establishing links between genomes and social phenotypes. High quality genomic resources from a diverse set of social species are required for both broad scale comparative genomic analyses and targeted functional genomic experiments and are therefore crucial for this goal. Here, we report on an updated genome for the burying beetle Nicrophorus vespilloides, an evolutionary and genomic model species for social behavior and parental care. The new assembly used PacBio sequencing reads and long read assemblers. This version of the genome greatly improves the continuity of the assembly and added new annotations, particularly lncRNA's. These updates will allow this resource to continue to be useful for newer functional genomic techniques. This improved assembly will also keep N. vespilloides a valuable comparative genomic resource. Updating genomic resources will continue to allow the field to make discoveries about the evolution of complex phenotypes, such as parental care.
1 Introduction
A complete understanding of phenotypes can only be achieved by investigating broad scale evolutionary forces and the genomic substrate upon which those forces act (Hofmann et al. 2014). Social behavior presents a particular challenge because of its complexity and flexibility, and how multiple levels of biology are integrated to produce social traits (Boake et al. 2002; O'Connell and Hofmann 2011). Tackling this complexity requires behavioral models be chosen broadly, developed with high quality genomic resources, and studies to find core mechanisms controlling social behavior (Phelps et al. 2010; Hofmann et al. 2014). To that end, we developed genomic resources for the burying beetle Nicrophorus vespilloides, a beetle with elaborate parental care (Cunningham et al. 2015). Here, we report a significantly improved and updated N. vespilloides genome produced using PacBio data and long read assemblers.
The burying beetle genus Nicrophorus is an ecological, evolutionary, and molecular model of complex social behavior and parental care (Eggert and Muüller 1997; Scott 1998; Cunningham 2020; Potticary et al. 2024). Beetles of this group will find small vertebrate carcasses, bury them, protect them from decomposition, and feed offspring directly through regurgitation of partially digested flesh (Figure 1a). Nicrophorus vespilloides is the most studied of the group, including at the molecular level. The role of neuropeptides, neurotransmitters, and immunity during the parenting of N. vespilloides has been examined with many genomic (Sun et al. 2020; Lewis et al. 2018; Sarkies et al. 2024), transcriptomic (Parker et al. 2015; Palmer et al. 2016; Jacobs et al. 2016; Benowitz et al. 2017; Cunningham et al. 2019), epigenetic (Cunningham et al. 2019; Lewis et al. 2020; Sarkies et al. 2024), and proteomic tools (Cunningham et al. 2017). All these studies have begun unraveling the complex transcriptional regulation of the parental care of this species. However, the next step for more refined and cell-type specific functional genomic studies is an improved genomic resource (Pool et al. 2023; Hoedjes et al. 2024).
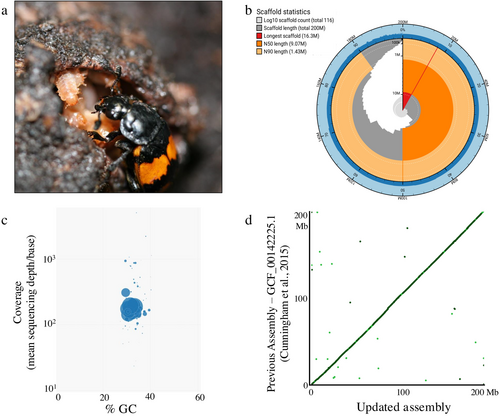
This resource is intended to further facilitate and improve molecular studies of this species' social behavior and parental care. This new assembly and annotation will allow N. vespilloides to continue to serve as a molecular model of social behavior as functional genomics techniques, such as single cell RNA-sequencing (scRNA-seq) or Assay of Transposon Accessible Chromatin using sequencing (ATAC-seq) continue to improve and require ever higher quality of genomic resources. It will also provide another high-quality genomic resource for comparative studies of beetles and insects.
2 Results and Discussion
2.1 Genome Assembly
After assembly and contamination screening, we produced an assembly of 116 scaffolds with a cumulative length of 199.97 Mb (Figure 1b,c; Table 1). This represents 97.8% of the genome's predicted size of 204 Mb based on a flow cytometry estimate (Cunningham et al. 2015). The previous assembly was 195 Mb in 4,664 scaffolds (Cunningham et al. 2015; Table 1). The new assembly increases the scaffold L/N50 to 8/9.1 Mb from 344/122 kb, while estimating the same GC content—31.8 vs. 31.9%. This represents a large improvement in the continuity of the assembly while keeping most of the previous sequence (Figure 1d). BUSCO assessed the assembly as having a near complete compliment of conserved genes (Table 1). The repeat content of the genome increased to 17.11%, compared to the previous 12.85%. The top three classifications of repeats were unclassified (9.59%), DNA elements (4.58%), and simple repeats (2.36%). The difference is likely attributable to more repetitive parts of the genome being assembled with longer reads.
| Genome | Previous assembly | Updated assembly | ||
|---|---|---|---|---|
| Scaffold number | 4650 | 116 | ||
| Contig number | 8129 | 232 | ||
| Assembly length | 195 | 199.974 Mb | ||
| Scaffold L/N50 | 344/122.407 kb | 8/9.069 Mb | ||
| Contig L/N50 | 623/67.958 kb | 13/4.349 Mb | ||
| Max scaffold length | 1.795 Mb | 16.295 Mb | ||
| % Scaffolds > 50 kb | 74.2 | 99.6 | ||
| % GC | 31.9 | 31.9 | ||
| % Repeat | 12.9 | 17.1 | ||
| BUSCO | 99.1% | 99.5% | ||
| Complete | Single Copy | 97.8% | 98.0% | |
| Complete | Duplicated | 1.3% | 1.5% | |
| Fragmented | 0.5% | 0.2% | ||
| Missing | 0.4% | 0.3% |
| Annotation | Previous annotation | Updated annotation | ||
|---|---|---|---|---|
| Gene models | 13,761 | 16,229 | ||
| Protein coding | 12,642 | 12,838 | ||
| lncRNAs | 983 | 2593 | ||
| Pseudogenes | 136 | 130 | ||
| tRNAs | 676 | 668 | ||
| BUSCO | 98.6% | 98.7% | ||
| Complete | Single Copy | 97.4% | 97.2% | |
| Complete | Duplicated | 1.2% | 1.5% | |
| Fragmented | 0.5% | 0.6% | ||
| Missing | 0.9% | 0.5% |
- Abbreviation: BUSCO, Benchmarking Universal Single-Copy Orthologue.
2.2 Genome Annotation
The previous genome annotation was 98.5% present in the new assembly (protein-coding, tRNA's, pseudogene, lncRNA's, and miscellaneous non-coding gene models). There were 2,973 genomic loci that did not overlap gene models from the previous annotations produced by StringTie from the mapped RNA-seq reads. Of these, 1,871 returned BLAST hits to an insect species and were retained. Gene models that annotated to known protein-coding (n = 216) or long non-coding RNAs (n = 16) were retained. Gene models that did not annotate to know protein-coding genes or long non-coding RNAs were analyzed with CPC2 as either putative protein-coding (n = 161) or putative long non-coding RNA (n = 1,478) gene models. All of these novel gene models returned best BLAST hits to beetles species. This genome annotation produced a BUSCO assessment of 98.7% complete (single copy—97.2%, duplicated—1.5%), 0.6% fragmented, and 05% missing gene compliment using the most expressed transcript from every annotated locus. The finished genome annotation contains 12,838 protein-coding and 2,593 long non-coding RNA gene models (Table 1). All of these metrics are in line with the previous assembly and other insect genomes generally (Thomas et al. 2020), and give confidence that the new assembly and annotation are not lacking any significant biological information.
3 Conclusions
Understanding social behavior requires broad taxonomic sampling and resources to interrogate both the evolutionary forces acting upon it and its genetic underpinnings. Here, we presented an updated genome assembly and annotation for Nicrophorus vespilloides, a beetle that has elaborate parental care. The updated genome greatly improved the continuity of the assembly while retaining the near-complete gene complement of the original assembly. This resource maintains valuable taxonomic representation for comparative studies and more refined cell-type specific functional genomics studies moving forward.
4 Materials and Methods
4.1 Genome Assembly & Annotation
4.1.1 Sequencing
We assembled 14.53 Gb of PacBio's RS II P5-C4 CLR sequencing reads prepared at The University of Maryland's Institute for Genomic Sciences using sequencing from two SMRT cells run with a 14.4 kb and one SMRT cell run with a 15.3 kb long insert PacBio libraries (~71.2× coverage total). The first PacBio SMRT cell was also used to scaffold an Illumina assembly to produce the previous assembly (31% of the PacBio data; Cunningham et al. 2015). All data was extracted, processed, and prepared for assembly as described in Cunningham et al. (2015). This produced 7.1 Gb of error-corrected reads using CANU (~34.8× coverage; v2.2; Koren et al. 2017).
4.1.2 Assembly
Non-default parameter values for each bioinformatic step can be found in Data S1 or mentioned here inline if we used a web-based service. We used FLYE (v 2.9.2; Kolmogorov et al. 2019) and CANU (v 2.2; Koren et al. 2017) to generate draft assemblies. These each were scaffolded with LRScaf (v 1.1.11; Qin et al. 2019) and merged with quickmerge (v0.3; Chakraborty et al. 2016) following the program's suggested protocol. We purged haplotigs with purge_dups (v 1.2.5; Guan et al. 2020) at each step of the assembly. The assembly was then scanned for contamination with blobtools (v 1.1.1; Laetsch and Blaxter 2017) using BLAST+ (v 2.14.1; Camacho et al. 2009) against NCBI nt database (downloaded 2024-05-02). Scaffolds were retained if they annotated to Arthropoda. The assembly was then polished with PILON (v1.24; Walker et al. 2014) for seven rounds using the Illumina genomic reads used for the primary assembly of Cunningham et al. (2019; NCBI SRA SRX1058748) after adapter trimming using cutadapt (v4.5; Martin 2011). We also assessed each step for BUSCO completeness (v 5.5.0; Manni et al. 2021) using the Endopterygota (v.odb10) and continuity using BBMap (v 39.01; Bushnell 2014) to ensure each was producing a higher quality assembly. We used Blobtk (v0.5.3; Challis 2017) to produce the Snail plot, blobtools (v1.1.1; Laetsch and Blaxter 2017) was used to produce the Blob plot, and D-Genies (1.5.0; Cabanettes and Klopp 2018; non-default parameter: sorted, hide noise selected, 67% value of filter small matches) was used to produce the dotplot of the previous and current assembly.
4.1.3 Repeats
We used RepeatModeler (v2.0.4; Flynn et al. 2020) to identify and classify the repetitive elements of the assembled genome. RepeatMasker (v4.1.4; Smit, Hubley, and Green 2013–2015) was then used to mask the genome and produce repetitive element content estimates.
4.1.4 Annotation
We used Liftoff (v 1.6.3; Shumate and Salzberg 2021) to map the current NCBI RefSeq Annotation (Accession Nicrophorus vespilloides Annotation Release 100) of the N. vespilloides genome (NCBI Accession GCF_001412225.1) to the new assembly. In addition, we produced new gene annotations from RNA-seq data. We used FLASH (v 2.2.0; Magoc and Salzberg 2011), HISAT2 (v 2.2.1; Kim et al. 2019), and StringTie (v 2.2.1; Pertea et al. 2015) to assemble new gene models using RNA-seq data after quality control with cutadapt from Parker et al. (2015), Palmer et al. (2016), Jacobs et al. (2016), Benowitz et al. (2017), and Cunningham et al. (2019). The gene models generated were compared to the annotation that was mapped from the previous annotation. Any genomic loci that were identified that were not overlapping with any gene model from the previous annotation that recovered BLAST hits from NCBI's nt database (−evalue 1e-30) that annotated to an insect species was retained. If the gene model was annotated to a known protein-coding or long non-coding RNA, it was annotated as such. The remaining gene models were analyzed with CPC2's webserver using default settings (vBeta; Kang et al. 2017). We assessed the completeness of the final annotation using BUSCO with the Endopterygota (v.odb10) dataset using the highest expressed isoform from each locus measured by TPM.
Author Contributions
Christopher B. Cunningham: conceptualization (lead), data curation (lead), formal analysis (lead), investigation (lead), methodology (lead), project administration (lead), visualization (lead), writing – original draft (lead), writing – review and editing (lead). Kyle M. Benowitz: conceptualization (supporting), formal analysis (supporting), investigation (supporting), methodology (supporting), visualization (supporting), writing – review and editing (equal). Allen J. Moore: conceptualization (equal), funding acquisition (equal), methodology (equal), supervision (equal), writing – review and editing (equal).
Acknowledgements
We would like to thank Elizabeth McKinney for help with the preparation of genomic DNA. This work was supported by a National Science Foundation grant (IOS-1354358) to AJM. The work was also partially supported by a USDA-ARS Non-Assistance Cooperative Agreement (#58-6080-9-006) to AJM and a National Science Foundation grant (DBI-2217859) to KMB. We would also like to thank Rahia Mashoodh and other reviewers for critical feedback that improved this manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
Open Research
Data Availability Statement
The raw and processed data that support the findings of this study are openly available in under NCBI BioProject PRJNA1123269. This genomic resource is also available at the USDA's National Agricultural Library i5k Workspace.




