Relationship Between Genetic and Phenotypic Variations in Natural Populations of Perennial and Biennial Sagebrush
Funding: This study was supported by Deutscher Akademischer Austauschdienst, Graduiertenakademie, Technische Universität Dresden, and Bundesministerium für Bildung und Forschung (01LC1820C).
ABSTRACT
Plant responses to environmental heterogeneity depend on life-history traits, which could relate to phenotypical and genetic characteristics. To elucidate this relationship, we examined the variation in population genetics and functional traits of short- and long-lived Artemisia species that are co-occurring in the steppes of Mongolia. Mongolian steppes represent stressful and water-limited habitats, demanding phenotypic modifications in the short term and/or genetic adaptation in the long term. However, detailed knowledge is missing about both plant phenotypic and genetic differentiation, and their interrelationships in temperate grasslands. Here, we investigated 21 populations of the widely distributed subshrub Artemisia frigida and the herbaceous biennial Artemisia scoparia. Genetic variation was assessed with newly developed simple sequence repeats (SSRs) markers. Functional trait data were collected from each individual, and data on environmental variables was collected for each population. We detected significantly higher genetic diversity in the biennial species (HE = 0.86) compared with the perennial (HE = 0.79). For both species, the largest share of genetic variation was partitioned within populations (96%). Population genetic structure in the biennial A. scoparia was weak, while the perennial A. frigida showed some spatial genetic structure, which was impacted by geographical factors, soil nutrients, and precipitation amount. Morphology-related functional traits (i.e., plant height) were predominantly associated with environmental variables rather than with genetic variation, whereas physiology-related trait (i.e., specific leaf area [SLA]) was partly genetically determined.
1 Introduction
It is widely acknowledged that a species’ genetic diversity and its variation are associated with life-history traits, such as life form, breeding system, seed dispersal mechanism, and geographical range (Hamrick and Godt 1996; Nybom and Bartish 2000; Reisch and Bernhardt-Römermann 2014). Species with outcrossing and mixed-mating systems tend to have higher levels of genetic variation than selfing species (Nybom 2004). Short-lived, non-woody, self-compatible, and early-successional species, i.e., annuals/biennials, are characterized by higher genetic variation between populations, but lower genetic variation within populations. In contrast, long-lived, woody, outcrossing, and late-successional species, i.e., many perennials, have higher genetic variation within populations (Reisch and Bernhardt-Römermann 2014). However, comparative studies such as that of Heelemann et al. (2015) found lower within-population variation in perennial Eriocephalus africanus L. than in the annual species Hemimeris racemosa (Houtt.) Merrill. A comparison of perennial and annual wild species of the genus Oryza L. discovered that perennial species had higher population level genetic diversity but less genetic variation among populations than annuals (Zhou et al. 2008).
Even within a species, plant phenotypic variation is often high. Functional trait plasticity related to morphology (e.g., plant height), (eco)physiology (e.g., SLA), and life history (e.g., flowering time and seed traits) was found to be under genetic control in some model plants (Locascio, Lucchin, and Varotto 2009; Hughes, Soppe, and Albani 2019). Several studies detected correlations between phenotypic traits (morphological and functional trait variation) and genetic variation (Waitt and Levin 1998; Karbstein, Tomasello, and Prinz 2019; Csilléry et al. 2020). In particular, Waitt and Levin (1998) presented a meta-study demonstrating a positive correlation between the genetic and phenotypic character traits of 27 species. However, trait variation does not necessarily coincide with genetic variation, especially if the trait is completely plastic (Chevin and Hoffmann 2017). Plasticity, i.e., phenotypic modification, allows for long-term adaptation to the local environment and/or short-term (reversible) responses. However, how genetic diversity and intraspecific functional traits interact at the population level, particularly in natural environments, remains poorly understood.
Artemisia L. (sagebrush) is a large and diverse genus that comprises over 500 taxa of annuals/biennials, perennial herbs, and shrubs or subshrubs distributed across temperate regions of the northern hemisphere (Riggins and Seigler 2012). Many species are clearly wind-pollinated; however, some indication of insect pollination was observed (colorful capitula and sticky pollen; Vallès and McArthur 2001). Artemisia spp. inhabits arid, semi-arid, and mesic environments spanning deserts to tundras, and their range of phenotypic diversity is broad (morphological, (eco)physiological, and reproductive traits), as is their range of ploidy levels (2n = 16 or 18 up to 2n = 144; Sanz et al. 2008). Although the genus offers ample opportunities for comparison, studies on genetic diversity and life history traits are hardly available. Al-Ajmi et al. (2021) compared seven species of Artemisia and found a positive interspecific correlation between similarities in genetic variation among species. However, we do not know of any study that addressed intraspecific variation in traits and genetic structures.
Artemisia frigida Willd. and Artemisia scoparia Waldst. & Kit. are both outbreeding and wind pollinated species (Vallès et al. 2011) with a range of phenotypic variations. In this study, we aimed to test the effects of the environment on genetic variation and genetic structure of the short-lived biennial A. scoparia and the long-lived subshrub A. frigida, which are co-occurring in the steppes of Mongolia. The flora of Mongolia lists 103 native Artemisia species (Baasanmunkh et al. 2022), among which species growing in dry steppes and forest steppes are the most numerous. Mongolia has one of the world's largest steppes, covering 1.2 million km2 and being home to thousands of steppe species (Munkhzul et al. 2021; Baasanmunkh et al. 2022). The continuous plain steppe of Mongolia allows for sufficient genetic exchanges between plant populations, as shown by former studies on the perennial grass Stipa glareosa P.A.Smirn. (Oyundelger et al. 2020) and on A. frigida (Oyundelger et al. 2021, 2023). In these studies, we detected moderate genetic structuring, which was mostly attributed to the differences in climate and edaphic conditions of the local populations rather than the geographical distance. However, the present study covers an even larger area of Mongolia, ranging from the western Altai Mountains to the eastern Mongolian Steppes. Specifically, we aimed to answer the following questions: (i) How do genetic diversity and population structure differ between the two Artemisia species? (ii) Do environmental factors relate to the genetic variation of the species across the Mongolian steppe? (iii) Are functional traits related to genetic diversity and/or abiotic habitat heterogeneity?
2 Materials and Methods
2.1 Study Species: Artemisia frigda and Artemisia scoparia
Perennial prairie-sage (A. frigida) has the largest natural range within its genus, being distributed across the North American prairie and the Eurasian steppe, whereas A. scoparia is a biennial species widely distributed from Central Europe to East Asia. Species’ ranges overlap in Inner Asia and specifically in Mongolia, where they are common steppe plants (Hilbig 1995). They share the same breeding system (outbreeding) and dispersal mechanism (wind), yet differ in their life form (biennial herb vs. perennial subshrub). The perennial A. frigida grows primarily in mountains, hillsides, and ruderal sites in steppes (Tkach et al. 2008). It bears a dense silvery pubescence and has woody ascending stems that are usually strongly branched (Figure 1). The biennial A. scoparia is found in riverbanks, as well as in ruderal sites in steppes and semi-deserts. Its stems are initially pubescent, becoming glabrous and strongly branched in the middle and upper parts (Figure 1). Artemisia frigida and A. scoparia are pioneer plants at grazing disturbed sites and also occur in the early recovery stages of abandoned land that underwent severe soil erosion (Jiao et al. 2013; Wang et al. 2022). Both species have high seed yields with small and light seeds (A. frigida: 0.106 g and A. scoparia: 0.047 g) that are easily propagated by wind and are then buried into soils (Yi et al. 2019).
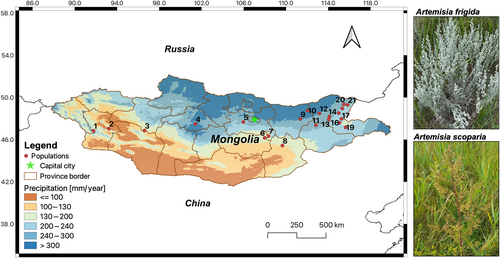
Artemisia scoparia belongs to the subgenus Dracunculus Besser representing the most basal lineage of Artemisia (clade divergence in 17.6 ± 2.1 Mya), while A. frigida is part of the subgenus Absinthium DC. (clade node 6.8 ± 0.8 Mya; Sanz et al. 2011; Hussain et al. 2019). Artemisia frigida comprises diploids (2n = 2x = 16) as well as tetraploids (2n = 4x = 36; Pellicer et al. 2010; Korobkov, Kotseruba, and Probatova 2014). In A. scoparia, mostly diploid cytotypes were observed (2n = 2x = 16 or 18; Pellicer et al. 2010); yet 2n = 4x = 32 or 36 have also been reported from Slovenia, Siberia, and recently from the Western Himalayas (Kawatani 1964; Amelchenko 1979; Gupta, Goyal, and Singh 2014).
2.2 Study Design and Sampling
Sampling was carried out along a broad-scale longitudinal precipitation gradient from western to eastern Mongolia during the summers of 2018 and 2019 (Figure 1). Fresh leaf materials were collected from 21 populations where both species co-occurred. For each population, representative herbarium specimens were deposited at Herbarium Senckenbergianum Görlitz (GLM). As a result, we sampled thirteen eastern (E) populations and four western (W) and four central (C) populations across various steppe vegetation types (Table 1).
| Pop code | Locality and province | Longitude | Latitude | Altitude (m) | MAT (°C) | MAP (mm) | Summer temp. (°C) | Summer prec. (mm) | cvP (%) | Steppe type | Region |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Munkhkhairkhan, Khovd | 91.765 | 46.841 | 1781 | −6.1 | 147 | 8.9 | 87 | 33 | MoS | W |
| 2 | Center of Khovd, Khovd | 93.228 | 47.044 | 1355 | 2.6 | 115 | 19.5 | 77 | 42 | DrS | W |
| 3 | Taishir Soum, Govi-Altai | 96.605 | 46.860 | 2009 | −1.6 | 172 | 14.7 | 105 | 29 | DrS | W |
| 4 | Khotont Soum, Arkhangai | 101.421 | 47.492 | 1608 | −1.3 | 300 | 14.3 | 198 | 24 | MoS | W |
| 5 | Hustai National Park, Tuv | 105.968 | 47.666 | 1264 | 0.9 | 167 | 18.3 | 118 | 24 | MoS | C |
| 6 | Tsagaandelger, Dundgovi | 107.975 | 46.170 | 1280 | 2.2 | 117 | 19.9 | 82 | 42 | DrS | C |
| 7 | Choir, Dundgovi | 108.350 | 46.331 | 1270 | 1.9 | 135 | 19.7 | 92 | 40 | DrS | C |
| 8 | Altanshiree, Dundgovi | 109.660 | 45.438 | 1007 | 3.8 | 126 | 21.9 | 84 | 31 | DeS | C |
| 9 | Batnorov, Khentii | 111.357 | 47.955 | 1078 | 0.5 | 286 | 18.8 | 194 | 25 | DrS | E |
| 10 | Norovlin, Dornod | 112.044 | 48.751 | 1020 | 0.6 | 277 | 18.6 | 192 | 29 | DrS | E |
| 11 | Hulunbuir, Khentii | 112.831 | 47.364 | 1008 | 0.4 | 231 | 18.6 | 164 | 37 | DrS | E |
| 12 | Tsagaan-Ovoo, Dornod | 113.167 | 48.480 | 1009 | 1.4 | 240 | 19.6 | 166 | 34 | DrS | E |
| 13 | Bulgan, Dornod | 114.046 | 47.896 | 961 | 1.5 | 238 | 19.9 | 163 | 46 | DrS | E |
| 14 | Bayantumen, Dornod | 114.109 | 48.190 | 991 | 1.4 | 210 | 19.7 | 142 | 44 | DrS | E |
| 15 | Choibalsan, Dornod | 114.997 | 48.519 | 847 | 1.5 | 209 | 20.3 | 141 | 49 | DrS | E |
| 16 | Matad, Dornod | 115.062 | 47.598 | 761 | 2.1 | 184 | 20.5 | 130 | 52 | DrS | E |
| 17 | Matad, Dornod | 115.250 | 47.971 | 1075 | 1.4 | 198 | 19.9 | 139 | 53 | DrS | E |
| 18 | Choibalsan, Dornod | 115.331 | 48.953 | 909 | 1.0 | 231 | 19.9 | 153 | 46 | DrS | E |
| 19 | Shar-Khudag, Dornod | 115.646 | 47.157 | 1011 | 1.5 | 199 | 19.7 | 144 | 52 | DrS | E |
| 20 | 64n toochig, Dornod | 115.485 | 49.344 | 650 | 1.3 | 232 | 20.4 | 154 | 45 | DrS | E |
| 21 | Otor pasture, Dornod | 115.837 | 49.288 | 821 | 1.1 | 239 | 20.3 | 159 | 44 | DrS | E |
- Abbreviations: C, central; cvP, coefficient of variation of interannual precipitation; DeS, desert steppe; DrS, dry steppe; E, eastern region of Mongolia, coordinates are in WGS84; MAP, mean annual precipitation; MAT, mean annual temperature; MoS, mountain steppe; Summer prec., summer mean annual precipitation; Summer temp., summer mean annual temperature; W, western.
At each site, 15 individuals per species were sampled within a 10 m × 10 m plot. Within these plots, plant community composition and total cover (%) of vascular plants were recorded, and a sample of topsoil (1 – 5 cm depth) with fine plant roots and the humic layer was collected. Soil samples were separated from litter, debris, and after shifting through a 2 mm sieve the following measurements were conducted in the laboratory: pH value, electrical conductivity (EC, as a proxy for salinity), plant available P, N%, organic C%, and C/N ratio. All results refer to oven-dried soil (75°C, 18 h). Moreover, plots were classified into different steppe types according to “The steppe vegetation of Mongolia” (Tuvshintogtokh 2014) based on our sampling location, which was also validated by our field-based plant community composition data.
Three functional traits were measured in the same individuals sampled for molecular data. In the field, “height of inflorescence (HI)” (if plants were flowering), “height of vegetative part (HV),” and leaf area for the trait “SLA” were measured. The HI was determined as the height from ground level to the tip of the highest inflorescence and the HV as the height of one randomly selected vegetative branch per plant. In A. frigida, vegetative and generative shoots differ in length. Thus, both heights were chosen as traits. Artemisia scoparia does not develop sterile shoots, and thus only the HI was applicable. For SLA, two fresh leaves were taken from each individual (30 leaves per site) and scanned using a Conrad P-573 handheld document scanner. Scanned pictures were later analyzed with ImageJ (Abràmoff, Magalhães, and Ram 2004) to determine the leaf area. Leaves were then air-dried for more than a month, and biomass weight was measured with a Mettler Toledo XP6 balance in the laboratory. The SLA was then calculated by dividing leaf area by dry mass (Perez-Harguindeguy et al. 2013). Population-level trait data and their correlation matrices, indicating their independence, are provided in Table S1.
Meteorological data of 20 years (mean annual temperature [MAT], mean annual precipitation [MAP], and mean spring temperature [March–May], mean summer temperature, and mean summer precipitation [June–August] between 1994 and 2013) were retrieved for each locality from the high-resolution CHELSA_V1 dataset, which has the advantage of capture interannual precipitation variation (Karger et al. 2017). The coefficient of interannual variation of annual precipitation (cvP) was estimated based on the retrieved MAP data and was also used as a predictor since cvP is a critical driver of rangeland dynamics (von Wehrden et al. 2012).
2.3 Molecular Analyses and Microsatellite Marker Development
Two randomly selected individuals of each species from two distinct populations were used to develop new SSR markers by applying whole genome sequencing (WGS). A previous study by Oyundelger et al. (2021), gives detailed steps for DNA extraction, library preparation, quality control, and bioinformatics in SSR development. Raw sequencing data were submitted to the NCBI Sequence Read Archive (SRA) and made publicly accessible under BioProject: PRJNA680535.
A total of 20 and 21 SSR markers were then tested for optimization in A. frigida and A. scoparia, respectively, using randomly selected samples from more than ten populations containing 8–16 samples. Furthermore, cross-checking of markers for both species was performed, and ten SSR markers published for A. frigida in the master thesis of Wang (2011) were tested with our samples in parallel. Based on reproducibility and polymorphism, 11 markers were chosen for each species. Detailed information on SSR markers of A. frigida can be found from Oyundelger et al. (2021). Information about species-specific SSR markers for A. scoparia developed for this study are presented in Table 2. Amplifications of a total of 22 SSR markers were performed in a volume of 12.5 μL, and customized PCR reaction mixtures and cycling programs were used (see PCR details from Table S2). Individuals of all 21 populations from both species exhibited a maximum of four alleles per locus, indicating prevailing tetraploidy (see Table S3 for ploidy information).
| No. | Locus | Repeat motif | Primer sequences (5′–3′) | Ta (°C) | Allele size range (bp) | Fluorescent dye | PCR type |
|---|---|---|---|---|---|---|---|
| 1 | Arcs2 | (GT)9 | F: TGTAAAACGACGGCCAGTTCTCCTTTCTGATTCATTGG | 55 | 585–620 | 6 FAM | Multiplex |
| R: CGAGATGAATTTGCGTCAT | |||||||
| 2 | Arsc12 | (TGT)9 | F: TGTAAAACGACGGCCAGTGGACATTTGAATGATGTTCG | 55 | 200–265 | 6 FAM | |
| R: AAGTCTTCCGCCAGCTATA | |||||||
| 3 | Arsc7 | (TG)11 |
F: TGTAAAACGACGGCCAGTTGT CCATCAAGATACCTATGC |
55 | 520–560 | VIC | Multiplex |
| GGTTATCGCCTCTCATTTG | |||||||
| 4 | Arsc11 | (ACA)8 | F: TGTAAAACGACGGCCAGTGAACGGGAAGATTACAAGC | 55 | 130–180 | VIC | |
| R: CACCAATATTACCTGGTGTG | |||||||
| 5 | Arsc18 | (ATG)8 | F: TGTAAAACGACGGCCAGTACACTGGAAAGCTATGTGC | 55 | 610–660 | PET | Multiplex |
| R: CGAGTCACAGTCATGGTC | |||||||
| 6 | Arsc19 | (TGA)8 |
F: TGTAAAACGACGGCCAGTCCT CAAACCTTGAAAGATAGC |
55 | 350–400 | PET | |
| R: CCGTATGAGTTAAGCAATCAG | |||||||
| 7 | Arsc17 | (TGA)8 | F: TGTAAAACGACGGCCAGTAATGGATTATGTTGATAGCCA | 55 | 135–160 | 6 FAM | Singleplex |
| R: CAAGTTCCGTTGACTCG | |||||||
| 8 | Arsc14 | (ATA)8 |
F: TGTAAAACGACGGCCAGTATG CACATAATATCCGAGC |
55 | 270–325 | VIC | Singleplex |
| R: GTGCTGAGACCGAATGC | |||||||
| 9 | Arsc20 | (ACA)14 |
F: TGTAAAACGACGGCCAGTGAC ACCCATAGACAGGAGC |
55 | ~500 | NED | Singleplex |
| R: GTCAGCTCGAAGCTTTCC | |||||||
| 10 | Arsc21 | (TGT)8 |
F: TGTAAAACGACGGCCAGTTGC CTTTGCAACAATTAAC |
55 | 110–128 | NED | Singleplex |
| R: GCTGCAAACATTACGTAAGC | |||||||
| 11 | Ch468 | NA |
F: TGTAAAACGACGGCCAGTTAG GGTTGCAGAAGATAAAC |
55 | 160–236 | PET | Singleplex |
| R: GCTTCTTCACTTCCTACTAAAG |
- Note: Details on SSR markers for Artemisia frigida can be found in Oyundelger et al. (2021).
2.4 Statical Analyses
2.4.1 Analysis of Genetic Diversity and Population Structure
To compare the genetic diversity within each species, we employed two programs, which allowed handling of microsatellite data for polyploids and species with mixed ploidy: GenoDive v.3.04 (Meirmans 2020) and the R-package Polysat v. 1.7 (Clark and Jasieniuk 2011) in R v.4.0.3 (R Core Team 2020). Estimators of genetic diversity comprised allelic diversity (AD), percentage of polymorphic loci (PPL), observed heterozygosity (HO), expected heterozygosity (HE), and inbreeding coefficient (GIS), all of which were calculated using GenoDive. Bruvo distances were computed with the R-package Polysat v.1.7 (Bruvo et al. 2004). Using the R-package vegan (Oksanen et al. 2007), we calculated the mean Bruvo distance among individuals for any given population (hereafter ‘Bruvo index’; see detail in Oyundelger et al. 2021), which was then used as a surrogate for genetic diversity (see Table S4 for the genetic diversity indices). A paired T-test was used to determine the significance of the difference in genetic diversity indices between two species.
Coefficients of genetic differentiation (FST and GST) were estimated using Polysat (Table S5). Population genetic structure was further analyzed with principal coordinate analysis (PCoA) using population-wise FST distance using the R-package ape (Paradis and Schliep 2019). In order to reveal environmental variables that were significantly associated with population genetic structure of the species, environmental and vegetation variables were fitted post hoc on the ordination using vegan, and plots were visualized with ggplot2 (Wickham 2011).
To examine the partitioning of genetic variation between and within populations, analysis of molecular variance (AMOVA; Excoffier, Smouse, and Quattro 1992) was performed in R-package poppr (Kamvar, Tabima, and Grünwald 2014) based on the individual-level Bruvo distance matrix estimated with Polysat.
2.4.2 Relationship Between Genetic and Spatial Distances
To assess the overall relationship between genetic and spatial distances, Mantel tests between genetic distance (linearized population level pairwise FST (FST/(1 − FST))) and geographic distances (Euclidean distances) were computed through 10000 randomizations using the R-package vegan (Oksanen et al. 2007). Further Mantel tests were then conducted between genetic distances and (a) climatic differences (Euclidean distance of centred and standardized climatic variables); (b) distance of soil indicator variables (Euclidean distance of centred and standardized variables), and (c) differences in plant community composition (Bray-Curtis's distance based on log-transformed species' cover).
2.4.3 Relationships of Functional Trait Variation With Genetic and Environmental Patterns
We estimated population-level means and coefficients of variation (CV) for trait variables, the latter as the ratio of standard deviation to mean. We checked collinearity among traits (mean and CV) with Pearson's coefficient (Table S1) using the R-package corrplot (Wei et al. 2021). As correlation coefficient values (r) of the mean and CVs were below ~|0.7|, we did not exclude particular functional traits.
To assess whether functional traits are related to environmental heterogeneity and genetic diversity, we fitted linear models (Dobson and Barnett 2018) with mean and CV of traits as the dependent variables. We again used corrplot for an exploratory analysis of associations among measures of genetic diversity. As a result, HE was chosen as the main response variable, as it had the highest correlation and depends less on population history (e.g., bottlenecks) compared to the other indices (Rosenberg 2004; Szczecińska et al. 2016). For the predictors, we first checked correlations among environmental variables to select representative variables based on their importance and independencies (r < |0.7|; see Table S6 for the data and their correlations). As a result: MAP, MAT, and cvP for climate; altitude for topography, and soil C/N ratio for soil nutrient contents were initially used as predictors for the models.
All predictors were first scaled to zero mean—unit variance (z-scores) to make effect sizes comparable. The response variable: cvIH of A. frigida was log-transformed due to its non-normal distribution; other response variables (cv and means) were in normal distribution, and thus no transformation was done. We then conducted model simplification by dropping the least relevant variables from linear models until a null model with intercept only. Models were compared using ANOVA, the summary was used to estimate significance and to choose the most parsimonious models. Lastly, plotting was used to check residuals of the models for possible deviations from normality and reasonable distribution of variances.
3 Results
3.1 Comparison of Genetic Diversity Between the Perennial and Biennial Artemisia
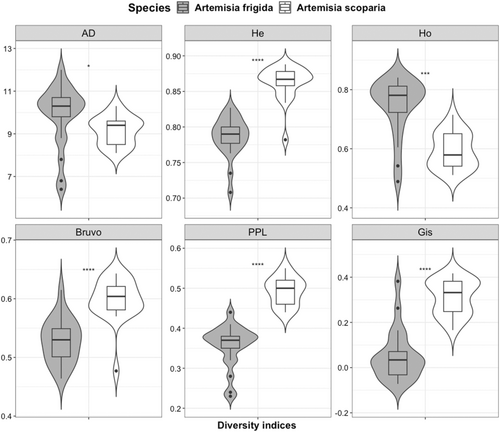
The overall polymorphic information content (PIC) of newly developed species-specific SSR markers was high (PIC = 0.77 and 0.84) for both A. frigida and A. scoparia. Paired T-test revealed that proxies of genetic diversity differed between two the Artemisia species (Figure 2). Specifically, HE, Bruvo, PPL, and GIS of the biennial A. scoparia were significantly higher than in the perennial A. frigida. In contrast, AD and HO were larger in the perennial than the annual species, yet with lower significance. Details for estimators of genetic diversity are presented in Table S4.
3.2 Population Genetic Variation and Relationship With Environmental Variables
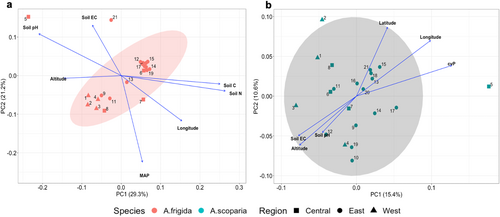
Coefficients of genetic differentiation of the two species across 21 populations were low overall, suggesting that isolation is at most moderate over the distances considered here. However, population differentiation of A. frigida was slightly more pronounced (Global FST = 0.078 and Global GST = 0.071) than of A. scoparia (Global FST = 0.064 and Global GST = 0.055). The most genetically distant population was population 5 (Hustai National Park) in both species (dissimilarity data provided in Table S5). Analysis of molecular variance showed that in both species, the highest genetic variation resided between individuals, while genetic variation partitioned among regions was slightly higher in A. frigida than A. scoparia (0.97% and 0.83%, respectively; Table 3). The ordination plots suggested that there was no pronounced genetic differentiation among steppe types and regions of Mongolia (individual level PCoA in Table S7), although A. frigida exhibited some population level genetic structure (Figure 3). In the PCoA ordination of A. frigida, the first two axes explained about 50% of the genetic variation, and some structuring of eastern vs. western populations mixed with central populations was discernible. According to post hoc fitting of predictor variables, longitude, altitude, mean annual precipitation (MAP), soil carbon, nitrogen, pH, and soil electrical conductivity (EC) showed a significant association with genetic structure (Figure 3a). In total, 26% of the total genetic variation was explained by the first two axes in the populations of A. scoparia, representing more continuous patterns among populations. Main structures along axis 1 and 2 were significantly correlated with altitude, soil pH, and EC together with longitude, latitude, and coefficient of variation of interannual precipitation (cvP), with western populations being in the upper left (Figure 3b). The ordinations demonstrated that soil pH and EC, as well as soil C and N, exhibit covariance, as proven by their high correlations (r = 0.78 and r = 0.99; Table S6). Results of post hoc fitting predictor variables on the PCoA are provided in the Table S8.
| Source of variance | df | Sum sq | Variance component | % Total | Φ statistic |
|---|---|---|---|---|---|
| Artemisia frigida | |||||
| Between regions | 2 | 12.56 | 0.028 | 0.97*** | 0.037 |
| Between populations | 18 | 72.03 | 0.080 | 2.69*** | 0.027 |
| Within populations | 283 | 806.44 | 2.850 | 96.34*** | 0.009 |
| Total | 303 | 891.03 | 2.958 | ||
| Artemisia scoparia | |||||
| Between regions | 2 | 2.21 | 0.004 | 0.83*** | 0.035 |
| Between populations | 18 | 11.25 | 0.013 | 2.69*** | 0.027 |
| Within populations | 282 | 128.68 | 0.456 | 96.47*** | 0.008 |
| Total | 302 | 142.14 | 0.473 | ||
- Abbreviations: % total, percentage of variation; df, degrees of freedom; Sum Sq, sum of square.
- *** p ≤ 0.0001.
The Mantel tests on association between genetic structures (linearized FST—FST/(1 − FST)) with various environmental variable distances revealed an overall negligible relationship with the genetic distances in both species (Table S9). Geographic distance and the distance of soil nutrient values in particular showed a significant but weak correlation with the genetic distance of A. frigida (r2 = 0.05*** and r2 = 0.02*). In contrast, A. scoparia did not exhibit an isolation by distance effect, while a weak correlation with climatic distance was observed.
3.3 Associations of Functional Traits With Genetic and Environmental Variations
Results of linear models showed that means as well as coefficients of variation of functional traits in A. frigida were associated with climatic and geographic variables, whereas in A. scoparia genetic diversity and soil nutrients had a significant relationship with SLA (Table 4). In the perennial A. frigida, altitude was positively associated with the physiology-related trait (mean SLA), while variations of morphology-related traits, cvHI and cvVH, were significantly affected by MAT, MAP, and cvP. In the biennial A. scoparia, genetic diversity showed an association with mean SLA and soil nutrient contents with the variation of SLA. With the exception of altitude and cvP, all significant associations were negative (scatter plots with linear regression lines of the significant models are provided Tables S10 and S11).
| Functional traits | Predictor | Estimate | Std. error | Pr(>|t|) | |
|---|---|---|---|---|---|
| A. frigida | Mean SLA | (Intercept) | 0.15 | 0.006 | < 0.001*** |
| Altitude | 0.02 | 0.006 | 0.012* | ||
| CV height of inflorescence | (Intercept) | 1.29 | 0.022 | < 0.001*** | |
| MAP | −0.09 | 0.023 | 0.002** | ||
| MAT | −0.08 | 0.025 | 0.005** | ||
| cvP | 0.07 | 0.025 | 0.01** | ||
| CV height of vegetative part | (Intercept) | 31.78 | 1.652 | < 0.001*** | |
| MAT | −3.86 | 1.693 | 0.034* | ||
| A. scoparia | Mean SLA | (Intercept) | 0.15 | 0.007 | < 0.001*** |
| H E | −0.03 | 0.007 | 0.001*** | ||
| CV SLA | (Intercept) | 1.61 | 0.037 | < 0.001*** | |
| Soil C/N | −9.06 | 3.602 | 0.021* |
- Note: Pr(>|t|) – significance p-value. Significance codes: ***p ≤ 0.001; **p < 0.01; *p ≤ 0.05.
4 Discussion
4.1 Population Genetic Diversity and Differentiation of Artemisia frigida and Artemisia scoapria
Life form and breeding system of plants are known to have a major influence on species' genetic diversity and population genetic structure (see Nybom and Bartish 2000; Reisch and Bernhardt-Römermann 2014; De Kort et al. 2021). Our chosen Artemisia species both have a wide range of distribution, are wind/water dispersed, outcrossing, and showed prevailing tetraploid cytotypes, making a direct comparison of diversity indices possible. Population-level mean values of the genetic diversity in both Artemisia species were higher (A. frigida: HE = 0.79 and A. scoapria: HE = 0.86) than in the review of Nybom (2004) for similar life history traits. The genetic diversity was significantly higher in the biennial A. scoparia than in the perennial species, according to four of the six diversity indices (HE, GIS, Bruvo, and PPL; Figure 2). This is in line with the study of Balfourier, Charmet, and Ravel (1998), who compared outcrossing annual and perennial ryegrass (Lolium L.) species. Probably, the effective population size and recombination rate are higher in the biennial than in the perennial. In short-lived species, recombination rate is higher as a result of their shorter life cycles and smaller genome/lower DNA content (Brazier and Glémin 2022), which may lead to a higher level of genetic diversity. Indeed, Garcia et al. (2004) reported that genome size of A. scoparia was the smallest (1C = 1.77 pg) within the studied species, while the genome size of A. frigida was 2.63 pg. Furthermore, in A. frigida, a smaller number of plants may participate in reproduction, as it is often subject to intensive grazing in natural and permanent pastures, and some individuals may survive vegetatively over several seasons. However, this observation is in contrast to some review studies that compared the genetic diversity of different life forms, utilizing allozyme and RAPD markers (see Hamrick and Godt 1990, 1996; Nybom and Bartish 2000; Nybom 2004) and AFLP markers (Balfourier, Charmet, and Ravel 1998; Reisch and Bernhardt-Römermann 2014). Nonetheless, individual life history traits, as well as genetic markers and diversity indices utilized, affect estimates of population genetic diversity, making the direct comparisons among studies somewhat questionable.
Patterns of genetic variation in the two species did not differ much, with spatial differences (among regions) explaining about 1% of the genetic variation, while barely 2%–3% variation resided among populations, and the highest variation (more than 95%) was explained by within-population variations (Table 3). Yet, the populations of the perennial A. frigida represented some structure illustrated in the PCoA, having fuzzy eastern and western clusters associated with altitude, longitude, amount of precipitation, and soil salinity (Figure 3a). Patterns in the biennial species were more continuous and impacted by geographical factors, like longitude, latitude, and altitude, as well as the coefficient of interannual precipitation variation (Figure 3b). Population 5 (Hustai NP) is a geographically central population that, however, represented the greatest genetic distance from others in both species (see PCoA; Figure 3 and Table S5 for differentiation matrices). This pattern has been seen in our former studies (see Oyundelger et al. 2021, 2023) and is now supported by the analysis of a second species, indicating this region has a distinct regime of gene flow and/or population connectivity, most likely due to its proximity to the local livestock trade center where animals from all over the country are brought in and may carry seeds.
Only few studies have compared the genetic variation of herbaceous species with different life forms (perennial vs. annual) in the same spatial context (Balfourier, Charmet, and Ravel 1998; Zhou et al. 2008; Heelemann et al. 2015), but their findings were contradictory: Zhou et al. (2008) found the highest molecular variation among populations in the annual (78%) than the perennial wild rice species (52%). While Balfourier, Charmet, and Ravel (1998) and Heelemann et al. (2015) reported that most of the total genetic variation was accounted for within populations in perennial (91%) and annual ryegrass (90%); and wild rosemary species (perennial: 89% and annual: 87%), respectively. Our result was in line with the latter, as within population variations were as high as 96% in both species. Furthermore, genetic variation between populations of the perennial was only marginally higher than that of annual species; yet both were comparably low. The low level of genetic variation between populations and regions, as well as weak correlations between genetic differences with environmental distances, indicates considerable historical and current gene flow between populations, supporting our former studies (Oyundelger et al. 2021, 2023).
4.2 Associations of Functional Traits With Genetic and Environmental Variations
Mean values as well as variations of morphology- (IH and VH) and (eco)physiology- (SLA) related traits were predominantly associated with environmental variables rather than with genetic variation (Table 4). This indicates that the traits showed substantial plasticity in response to environmental differences, as demonstrated by a number of other studies (see Gratani 2014; Chevin and Hoffmann 2017; Matesanz and Ramírez-Valiente 2019). Specifically, climate (MAP, MAT, and cvP) was found to be the most important factor influencing the morphological trait variations of the perennial Artemisia. This, of course, indicates the importance of climatic conditions for plant growth, as has been previously shown for plant species occurrence and abundance in the Mongolian steppe (von Wehrden and Wesche 2007; von Wehrden et al. 2010). In A. frigida, morphological differentiation is probably promoted by site-dependent microhabitat differences, primarily in temperature and water availability. Morphological differences become even more pronounced, particularly due to the harsh climate in steppes (MAT: min (−6.1) to max +3.8°C) with overall limited water availability (MAP: min 117 mm to max 300 mm), as demonstrated by our linear model (Table 4). Phenotypic differences were pronounced between sites/populations, whereas genetic differentiation was less evident (Global FST = 0.064). This is in line with a large body of literature showing plant phenotypic trait responses and genetic differentiation patterns varying highly in abiotic and biotic environmental conditions (Odat, Jetschke, and Hellwig 2004; Bucher et al. 2016; König et al. 2018), and plant trait differentiations being even enhanced in extreme environments (Chevin and Hoffmann 2017; Karbstein, Tomasello, and Prinz 2019).
SLA relates to photosynthesis, relative growth rate, and stress tolerance (Perez-Harguindeguy et al. 2013) and is known to be subject to substantial plasticity (Pan et al. 2013; Stotz et al. 2022) as well as being partly under genetic control (Knight and Ackerly 2003; Scheepens, Frei, and Stöcklin 2010). In our study, mean SLA was significantly associated with altitude in A. frigida and with genetic diversity in A. scoparia. Soil nutrient availability also had a significant impact on the variation of the SLA in A. scoparia, supporting the common observations, as we detected the effect of both environment and genetics on SLA (Table 4). Significant relationships of the mean and cvSLA with environmental variables were observed in other studies. For instance, Woodward (1983) noted a negative association between altitude and SLA in Festuca L. and Carex L. species, which was explained by an underlying relationship between altitude and temperature. Yulin et al. (2005) detected an increasing SLA in habitats with higher amounts of soil nutrients (total nitrogen and organic carbon) in Artemisia halodendron Turcz. ex Besser, as soil nutrient stress is a major limiting factor for plant growth. A global study has shown a positive association between soil fertility and SLA, whereas negative relationships exist between soil C/N ratio and SLA (Ordoñez et al. 2009), supporting our findings. Furthermore, genetic effects on SLA variance were observed in Campanula L. (Scheepens, Frei, and Stöcklin 2010), which were attributed to selection-induced adaptations. The same may hold true for our observation that genetically less diverse populations represented a larger mean SLA, as a result of local adaptation. Yet, this negative association might be rather an artifact attributed to the (natural outlier) population 5 (Hustai NP), where the lowest population-level diversity (HE = 0.78) and the largest mean SLA (0.24 mm/mg) were detected (see relationship in Table S10).
5 Conclusion
Understanding plant adaptation—both in terms of morphological and genetic aspects—to environmental heterogeneity has been a focal point of many studies. However, steppe plants have rarely been investigated, and no comparative studies of species with different life-history traits have been conducted to date. Our findings demonstrated that genetic diversity in both species was relatively high (A. frigida: HE = 0.79 and A. scoparia: HE = 0.86), and their genetic variation and functional trait characteristics were significantly affected by geographical factors and soil nutrient contents. Surprisingly, climatic factors exhibited a relatively limited impact, and when there was an effect, it was primarily associated with the amount and variation of precipitation. This aligns with the overarching observation in Mongolia that precipitation serves as the primary limiting factor for plant growth, occurrence, and abundance. Thus, plants in these areas require significant adaptations to thrive in the water-limiting habitats while retaining sufficient genetic diversity.
Author Contributions
Khurelpurev Oyundelger: data curation (equal), formal analysis (lead), investigation (lead), methodology (lead), project administration (supporting), validation (equal), visualization (lead), writing – original draft (lead), writing – review and editing (lead). Lisa Großmann: formal analysis (supporting), investigation (supporting), methodology (supporting), visualization (supporting), writing – review and editing (supporting). Veit Herklotz: data curation (supporting), investigation (equal), methodology (equal), software (supporting), writing – review and editing (supporting). Dörte Harpke: data curation (supporting), investigation (equal), methodology (equal), software (lead), writing – review and editing (supporting). Oyuntsetseg Batlai: data curation (supporting), methodology (supporting), resources (equal), writing – review and editing (equal). Karsten Wesche: conceptualization (lead), data curation (equal), formal analysis (equal), funding acquisition (lead), investigation (equal), methodology (equal), project administration (equal), resources (lead), supervision (lead), validation (lead), writing – review and editing (supporting). Christiane M. Ritz: conceptualization (lead), data curation (equal), funding acquisition (lead), investigation (equal), methodology (equal), project administration (lead), resources (lead), supervision (lead), validation (equal), writing – review and editing (supporting).
Acknowledgments
The authors would like to thank the German Ministry for Science and Education (BMBF, No. 01LC1820C—MoreStep project in the BioTip scheme) and Summer School Mongolia, financed by the German Academic Exchange Service (DAAD) for their support for the consecutive field works. A special thank to the students, who helped with the sampling and soil sample preparation, and D. Davaasuren for his persistent commitment as a great field driver. The sample collection was carried out as a part of a cooperation agreement between the National University of Mongolia (Faculty of Biology) and the Museum of Natural History in Görlitz, Germany. We are also grateful to O. Munkhzul for organizing sample shipment to Germany and to Y. Jäschke for providing soil nutrient data. We thank our technical assistants M. Schwager and B. Schlitt (Museum of Natural History Görlitz) for their support during laboratory work, as well as our colleagues of the Senckenberg Biodiversity and Climate Research Institute (SBiK-F) for their technical support. Oyundelger was funded by a scholarship program for the promotion of early-career female scientists of Technische Universität (TU) Dresden. Open Access funding enabled and organized by Projekt DEAL.
Conflicts of Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Khurelpurev Oyundelger reports financial support was provided by TU Dresden. Karsten Wesche reports travel was provided by German Federal Ministry of Education and Research. Christiane Ritz reports travel was provided by German Academic Exchange Service.
Open Research
Data Availability Statement
WGS raw sequencing data are available in the NCBI Sequence Read Archive (SRA) under BioProject PRJNA680535. Further dataset generated and analyzed during the current study are provided in the Supplement material tables. The data that support the findings of this study are available on request from the corresponding author.




