Comprehensive immunophenotypic study of acute myeloid leukemia with KMT2A (MLL) rearrangement in adults: A single-institution experience
Sergej Konoplev and Xiaoqiong Wang contributed equally to this work.
Abstract
Background
Acute myeloid leukemia (AML) with KMT2A (MLL) rearrangement is known for monocytic or myelomonocytic differentiation, but the full immunophenotypic spectrum and dynamic changes of the immunophenotype in this genetically defined disease have not been systematically studied.
Methods
We reviewed the immunophenotype, karyotype, and mutations at the time of initial diagnosis and relapse of adults with AML with KMT2A rearrangement in our institution between 2007 and 2020.
Results
We identified 102 patients: 44 men and 58 women with a median age of 52 years (range, 18–87). Forty-three patients were considered to be therapy-related. Twenty-four out of 64 patients relapsed from complete remission after induction therapy, 34 had persistent/progressive disease, and 58 patients died with a median overall survival of 17 months.
We detected five immunophenotypes: immature monocytic (38%); myelomonocytic (22%); myeloblastic (22%); mature monocytic (10%); and acute promyelocytic (APL)-like (8%). By chromosomal breakpoints, we presumed 11 different partners; t(9;11) (p22;q23)/MLLT3-KMT2A was the most common rearrangement (n = 56, 55%), followed by t(6;11) (q27;q23)/AFDN-KMT2A (n = 13,13%). Patients with t(6;11) (q27;q23)/AFDN-KMT2A preferentially showed a myeloblastic phenotype (p = 0.026). Mutations were detected in 39/64 (61%) cases, and RAS pathway (NRAS/KRAS/PTPN11) was involved in 26/64 (41%) cases. None of the APL-like cases had mutations detected. At the time of disease relapse, 10/24 (42%) showed major immunophenotypic change, and 7/10 cases gained additional cytogenetic and/or molecular alterations.
Conclusion
The immunophenotype of AML with KMT2A rearrangement is more diverse than previously recognized, with a substantial subset showing no evidence of monocytic differentiation. Major immunophenotype change is common at the time of relapse.
1 INTRODUCTION
Acute myeloid leukemia (AML) with KMT2A (previously known as MLL) rearrangement is an aggressive form of AML (Liu et al., 2020), and in the literature this neoplasm is most commonly described as exhibiting monocytic or myelomonocytic differentiation (Creutzig et al., 1995; Muñoz et al., 2003; Xie et al., 2020). In fact, the recognition of blasts with a monocytic or monoblastic immunophenotype in the presence of 11q23 translocation is highly suggestive of AML with KMT2A rearrangement, and others have suggested that a monocytic or myelomonocytic phenotype may be used to initiate rapid fluorescence in situ hybridization (FISH) for KMT2A. Other immunophenotype-genotype associations involving KMT2A have been reported to facilitate the differential diagnosis of AML, for example, t(11;17) (q23;q25)/KMT2A-SEPT9 versus acute promyelocytic leukemia (APL) with t(11;17) (q23;q21)/ZBTB16-RARA variant (Balbuena-Merle et al., 2021). Immunophenotype-genotype associations can be helpful in more rapid recognition of genetic abnormalities, such as KMT2A rearrangement, facilitating the assignment of AML patients to the proper frontline treatment protocols.
Recently, the interaction of Menin (MEN1) and MLL (KMT2A) has been shown to be essential in the maintenance of KMT2A fusion-driven gene expression; and small-molecule inhibitors of MEN1/MLL have been shown to have potent anti-leukemia effects leading to the launch of multiple clinical trials (Dzama et al., 2020; Krivtsov et al., 2019). Effective identification of cases with KMT2A rearrangements, especially cryptic rearrangements (Ney Garcia et al., 2017), has therefore become more relevant with evolving therapies. However, in our practice, we have observed a more diverse immunophenotype in cases of AML with KMT2A rearrangement, including a proportion of cases with a non-monocytic phenotype. Furthermore, AML with KMT2A rearrangement has been known for its immunophenotypic/lineage plasticity, and it is one of the most frequent subtypes of acute leukemia with lineage switch post-therapy (Rossi et al., 2012; Yost et al., 2020).
The goal of this study is to understand the full immunophenotypic spectrum of cases of AML with KMT2A rearrangement and to examine if immunophenotypic features of the leukemic blasts can predict the presence of KMT2A rearrangement.
2 MATERIALS AND METHODS
2.1 Patients
We searched the pathology data archives for adult untreated acute myeloid leukemia (AML) patients with KMT2A rearrangement to our institution between January 1, 2007 and December 31, 2020. Patients who initially presented with a relapse or refractory AML with KMT2A rearrangement were excluded due to the concern of major immunophenotype changes and the acquisition of additional molecular genetic changes. Due to the focus of this study, patients with mixed phenotype of acute leukemia or B-lymphoblastic leukemia with KMT2A rearrangement were also excluded from the study. KMT2A rearrangements were confirmed by FISH in all cases. The study was approved by the institutional review board (IRB).
2.2 Morphological assessment and special cytochemistry stains
All cases had routine hematoxylin and eosin (H&E) histologic sections of bone marrow (BM) trephine biopsy specimens and well-prepared Wright–Giemsa-stained aspirate smears. A 500 nucleated cell differential was performed. A myeloperoxidase (MPO) cytochemical stain was performed in most cases, and non-specific esterase (NSE) stain was performed selectively in cases with monocytic differentiation as assessed by morphological evaluation at the time of diagnosis.
2.3 Karyotypic analysis and fluorescence in situ hybridization (FISH) study
Conventional chromosomal analysis was performed on G-banded metaphase cells prepared from unstimulated BM aspirate cultures (24 and 48 hours) using standard techniques. Twenty metaphases were analyzed, and the results were reported using the International System for Human Cytogenetic Nomenclature (ISCN 2013). FISH studies for KMT2A (dual-color break-apart probe, Abbott Molecular/Vysis, Des Plaines, Illinois, USA) rearrangement were performed on freshly harvested BM cells (metaphase or interphase). The positive cut-off for KMT2A rearrangement in our laboratory is 3.6% (Zuo et al., 2017).
2.4 Multicolor flow cytometric immunophenotyping (FCI)
BM aspirate specimens were collected in EDTA anticoagulant tubes and processed within 12 h of collection using a standard lyse/wash technique (PharmLyse™, BD Biosciences, San Diego, CA). A minimum of 200,000 events were acquired on FACSCanto II instruments (BD Biosciences). The acute leukemia panels included essential markers in the diagnosis and characterization of acute leukemias such as CD2, CD3, cytoCD3, CD4, CD5, CD7, CD13, CD14, CD15, CD19, CD33, CD34, CD117, CD123, HLA-DR, CD64, MPO, and TDT. Other markers, including CD22, CD25, CD36, and CD71, were incorporated in relatively recent panels. The CD45 dim-blast region including monocytes based on CD45/side scatter was analyzed.
2.5 Targeted next-generation sequencing
Targeted next-generation sequencing (NGS) studies using panels of genes commonly altered in myeloid neoplasia were performed as part of the clinical workup (28-gene panel or an 81-gene panel) as described previously (Patel et al., 2019; Wang et al., 2016). The 28 gene panel included commonly mutated genes in AML/MDS, including ABL1, ASXL1, TET2, DNMT3A, IDH1, IDH2, NPM1, RUNX1, TP53, GATA2, EXH2, PTPN11, WT1, and NOTCH1, and the 81 gene panel included multiple additional genes, such as ANKRD26, BCOR, PHF6, PIGA, SETBP1, SF3B1, U2AF1, U2AF2, and ZRSR2. NGS data were analyzed using MiSeq Reporter (TruSeq) or SureCall (Haloplex). The Integrative Genomics Viewer (IGV, Broad Institute) was used to visualize read alignment and confirm variant calls. A custom-developed, in-house software package (OncoSeek) was used to annotate sequence variants. Nomenclature of genetic variants was designated following the Human Genome Variation Society recommendations (Richards et al., 2015).
2.6 Statistics
Clinical and laboratory variables were compared among the five immunophenotypic subgroups of AML identified using Fisher's exact tests for categorical variables and Kruskal-Wallis tests for numeric variables. Overall survival (OS) was calculated from the initial diagnosis of AML to the date of expiration or the last known alive date. Survival distributions were analyzed with the Kaplan–Meier method. Statistical analyses were performed using GraphPad Prism 8 and SPSS Statistics 24. Differences between subgroups were considered statistically significant when the p-value was less than 0.05.
3 RESULTS
3.1 Patients
The cohort included 102 patients with AML harboring KMT2A rearrangement: 44 men and 58 women with a median age of 52 years (range, 18–87) (Table 1 and Table S1). Forty-three (42%) patients had received prior chemotherapy with radiation or chemotherapy alone for various malignancies and were considered as therapy-related AML (t-AML). The patients presented with a median white blood count of 8.2 × 109/L (0.5–291.9), hemoglobin 9.5 g/dl (5.5–13.8), platelet count of 42 × 109/L (4–279), absolute monocyte count of 0.2 × 109/L (0–76.8), and circulating blast percentage of 31% (0–97%). The median percentage of blasts in the BM was 70% (20–95%); the median percentage of monocytes in the BM was 4% (3–60%).
| All | Immature monocytic | Mature monocytic | APL-like | Myelo-monocytic | Myelo-blastic | |
|---|---|---|---|---|---|---|
| N = 102 | N = 39 (38%) | N = 10 (10%) | N = 8 (8%) | N = 23 (22%) | N = 22 (22%) | |
| Age, median (range, years) | 52 (18–87) | 52 (18–87) | 57 (33–70) | 54 (26–82) | 52 (18–76) | 47 (18–76) |
| Gender (male, %) | 44/102 (43%) | 21/39 (54%) | 0/10 (0%) | 4/8 (50%) | 10/23 (44%) | 9/22 (41%) |
| PB, median (range) | ||||||
| WBC (×109/L) | 8.2 (0.5–291.9) | 8 (0.5–291.9) | 18.9 (2.3–140.2) | 1.4 (1.0–24.6) | 10.2 (1.1–159.4) | 4.8 (0.6–110.5) |
| Hb (g/dl) | 9.5 (5.5–13.8) | 9.8 (6.9–12.9) | 9.1 (5.5–11.0) | 9.6 (8.2–13.2) | 9.4 (6.7–13.8) | 9.7 (7.1–12.6) |
| Plt (×109/L) | 42 (4–279) | 68 (15–235) | 26 (13–139) | 21 (13–156) | 37 (4–129) | 41 (8–279) |
| Blast (%) | 31 (0–97) | 36 (0–94) | 6 (0–51) | 0 (0–80) | 26 (0–97) | 54 (0–92) |
| Monocyte (%) | 4 (0–76) | 2 (0–33) | 29 (2–58) | 2 (0–11) | 16 (0–76) | 3 (0–28) |
| AMC (×109/L) | 0.2 (0–76.8) | 0.04 (0–15.6) | 4.4 (0.3–29.4) | 0.04 (0–0.7) | 1.1 (0–76.8) | 0.1 (0–1.6) |
| Monocytosis, N (%) | 33/102 (32%) | 10/39 (26%) | 8/10 (80%) | 0/8 (0%) | 12/23 (52%) | 3/22 (14%) |
| BM, median (range) | ||||||
| Blast (%) | 70 (20–95) | 82 (25–95) | 31 (20–55) | 74 (35–89) | 64 (27–88) | 75 (22–91) |
| Monocyte (%) | 4 (0–60) | 1 (0–34) | 21 (2–60) | 3.5 (0–13) | 10 (1–34) | 2 (0–13) |
| Diagnosis, N (%) | ||||||
| t-AML | 43/102 (42%) | 13/39 (33%) | 6/10 (60%) | 4/8 (50%) | 13/23 (57%) | 7/22 (32%) |
| De Novo AML | 59/102 (58%) | 26/39 (67%) | 4/10 (40%) | 4/8 (50%) | 10/23 (43%) | 15/22 (68%) |
| h/o MDS | 15/102 (15%) | 5/39 (13%) | 4/10 (40%) | 0/8 (0%) | 3/23 (13%) | 3/22 (14%) |
| Karyotype, N (%) | ||||||
| Normal | 3/102 (3%) | 1/39 (3%) | 1/10 (10%) | 0/8 (0%) | 0/23 (0%) | 1/22 (4%) |
| Isolated | 61/102 (60%) | 15/39 (38%) | 6/10 (60%) | 7/8 (88%) | 19/23 (83%) | 14/22 (64%) |
| Associated | 31/102 (30%) | 21/39 (54%) | 2/10 (20%) | 1/8 (12%) | 3/23 (13%) | 4/22 (18%) |
| Complex | 7/102 (7%) | 2/39 (5%) | 1/10 (10%) | 0/8 (0%) | 1/23 (4%) | 3/22 (14%) |
| Molecular alterations, N (%) | ||||||
| Any mutations | 39/64 (61%) | 16/21 (76%) | 2/4 (50%) | 0/6 (0%) | 11/17 (65%) | 10/16 (63%) |
| KRAS | 12/64 (19%) | 5/21 (24%) | 1/4 (25%) | 0/6 (0%) | 4/17 (24%) | 2/16 (13%) |
| NRAS | 18/64 (28%) | 7/21 (33%) | 0/4 (0%) | 0/6 (0%) | 5/17 (29%) | 6/16 (38%) |
| PTPN11 | 5/64 (8%) | 1/21 (5%) | 0/4 (0%) | 0/6 (0%) | 3/17 (18%) | 1/16 (6%) |
| ASXL1 | 4/64 (6%) | 3/21 (14%) | 0/4 (0%) | 0/6 (0%) | 1/17 (6%) | 0/16 (0%) |
| FLT3 | 8/64 (13%) | 6/21 (29%) | 0/4 (0%) | 0/6 (0%) | 2/17 (12%) | 0/16 (0%) |
| KMT2A partner, N (%) | ||||||
| MLLT3 | 56/102 (55%) | 22/39 (56%) | 3/10 (30%) | 6/8 (75%) | 16/23 (70%) | 9/22 (41%) |
| AFDN2 | 13/102 (13%) | 4/39 (10%) | 0/10 (0%) | 0/8 (0%) | 4/23 (17%) | 5/22 (23%) |
| ELL | 9/102 (9%) | 1/39 (3%) | 4/10 (40%) | 0/8 (0%) | 2/23 (9%) | 2/22 (9%) |
| MLLT1 | 9/102 (9%) | 5/39 (13%) | 0/10 (0%) | 2/8 (25%) | 1/23 (4%) | 1/22 (4.5%) |
| ABI1 | 3/102 (3%) | 2/39 (5%) | 0/10 (0%) | 0/8 (0%) | 0/23 (0%) | 1/22 (4.5%) |
| Other | 12/102 (12%) | 5/39 (13%) | 3/10 (30%) | 0/8 (0%) | 0/23 (0%) | 4/22 (18%) |
| SCT, N (%) | 47/98 (48%) | 14/37 (38%) | 4/10 (40%) | 5/8 (63%) | 12/22 (55%) | 12/21 (57%) |
| OS, median (mos) | 17 | 9 | 6 | 13 | 30 | 21 |
| Outcome, N (Death, %) | 58/98 (59%) | 22/37 (60%) | 8/10 (80%) | 6/8 (75%) | 10/22 (46%) | 12/21 (57%) |
- Abbreviations: AMC, absolute monocyte count; CR, complete remission; h/o, history of; mos, months; N, number; OS, overall survival; Other, Other partners, besides MLLT3, AFDN2, ELL, MLLT1 and ABI1; SCT, stem cell transplant; t-AML, therapy-related AML.
All patients with complete follow-up information available (n = 98) received chemotherapy, and 47 of these patients (48%) received allogeneic stem cell transplantation (SCT) (Table 1 and Table S1). Sixty-four (65%) patients achieved complete remission (CR) after initial induction therapy, and 34 had persistent/progressive disease. Sixty-one out of 64 patients with CR underwent follow-up BM examination at our hospital, and 24 (38%) patients had relapsed disease. Within a median follow-up of 9.5 months (range, <1–117), 58 (59%) patients died. The median overall survival is 17 months. Therapy-related AML patients had shorter overall survival compared to de novo AML patients (9 mos vs. 23 mos, p = 0.018), but these two groups of patients showed no statistically significant differences in blast immunophenotype, karyotype complexity, KMT2A partner, and molecular alterations.
3.2 Cytogenetics
Karyotyping generated adequate results in all 102 patients; 99 (97%) patients showed abnormalities involving 11q23 rearrangement; whereas three (3%) patients had cryptic 11q23 rearrangement: 2 patients had a normal karyotype and one patient showed 45,XX,der(10;13) (q10;q10) [19]/46,XX [1] (Table 1 and Table S1). 11q23 rearrangement was detected as a sole abnormality in 61 (60%) patients, was associated with one or two additional abnormalities in 31 (30%) patients and was a part of a complex karyotype in 7 (7%) patients.
FISH confirmed KMT2A rearrangement in all cases, involving a median of 92% nuclei examined (range, 9%–100%). Although FISH assessment of partner genes was not performed, partners could be assumed by chromosome banding in references to published data. The most frequent rearrangement was t(9;11) (p22;q23)/MLLT3-KMT2A (n = 56, 55%), followed by t(6;11) (q27;q23)/AFDN-KMT2A (n = 13, 13%); t(11;19) (q23;p13.1)/ELL-MNT2A (n = 9, 9%); t(11;19) (q23;p13.3)/MLLT1-KMT2A (n = 9, 9%); t(10;11) (p11.2;q23)/ABI1-KMT2A (n = 3, 3%); and one each t(2;11) (q31;q23)/AFF1-KMT2A; t(11;15) (q23;q11.2)/ZFYVE19-KMT2A; t(11;17) (q23;q12)/ACACA-KMT2A; t(11;17) (q23;q21)/MLLT6-KMT2A; inv(11) (q21q23)/MAML2-KMT2A; inv(11) (q14q23)/PACALM-KMT2A (Table 1 and Table S1). Partner genes were unknown in six patients, including three patients with a cryptic karyotype (Table S1).
3.3 Mutations by next generation sequencing
Sixty-four patients had 28-gene or 81 gene NGS panel performed. NPM1 mutation was not detected in all these cases. Thirty-nine out of 64 (61%) patients had somatic mutations and the mutations detected in ≥5% patients included NRAS (28%), KRAS (19%), FLT3 (13%), PTPN11 (8%), ASXL1 (6%) (Table 1 and Table S1). Among eight patients with FLT3 mutations, five patients had a D835 point mutation at exon 20 while others had mutations at different exons; no FLT3 internal tandem duplication (ITD) mutation was detected (Table S1). Mutations involving the RAS pathway, NRAS/KRAS/PTPN11 were detected in 26/64 (41%) patients (Table S1).
3.4 Blast immunophenotype
The blast immunophenotype was categorized into five subgroups (Table 2). Group 1, immature monocytic, showed blasts that expressed bright CD64, with substantial loss of CD14, CD13, and CD36; increased expression of CD117, CD15, or CD34 (Figure 1a). Group 2, mature monocytic had an immunophenotype similar to normal mature monocytes, positive for CD64, CD14, CD36, CD4, dimCD15, and negative for CD117 or CD34 (Figure 1b). Group 3, acute promyelocytic leukemia (APL)-like, showed blasts positive for CD117, CD13, CD33, but negative for CD34 and HLA-DR (Figure 1c). Group 4, myelomonocytic, was characterized by myeloblasts and monocytic blasts each represented ≥20% of blast population (Figure 1d), or in some cases blasts expressed both monocytic and granulocytic markers simultaneously (Figure 1e). Lastly, group 5, myeloblastic (AML-myeloblastic), typically showed blasts positive for CD34, CD117, CD13, CD33, and HLA-DR, but the expression of CD34, HLA-DR and MPO could be variable (Figure 1f).
| All | Immature monocytic | Mature monocytic | APL-like | Myelo-monocytic | Myelo-blastic | |
|---|---|---|---|---|---|---|
| N = 102 | N = 39 (38%) | N = 10 (10%) | N = 8 (8%) | N = 23 (22%) | N = 22 (22%) | |
| Special stain, N (positive, %) | ||||||
| MPO | 62/98 (63%) | 9/36 (2%) | 8/10 (80%) | 6/8 (75%) | 22/23 (96%) | 17/21 (81%) |
| NSE | 55/71 (77%) | 26/28 (93%) | 9/9 (100%) | 2/5 (40%) | 17/17 (100%) | 1/12 (8%) |
| Flow cytometry, N (positive, %) | ||||||
| CD45 | 102/102 | 39/39 | 10/10 | 8/8 | 23/23 | 22/22 |
| CD34 | 24/102 (24%) | 1/39 (3%) | 0/10 (0%) | 0/8 (0%) | 7/23 (30%) | 16/22 (73%) |
| CD117 | 72/102 (71%) | 29/39 (74%) | 0/10 (0%) | 8/8 (100%) | 14/23 (61%) | 21/23 (96%) |
| HLA-DR | 87/102 (85%) | 39/39 (100%) | 10/10 (100%) | 1/8 (13%) | 18/23 (78%) | 20/22 (91%) |
| CD33 | 101/102 (99%) | 39/39 (100%) | 10/10 (100%) | 8/8 (100%) | 23/23 (100%) | 21/22 (96%) |
| CD13 | 82/102 (80%) | 24/39 (62%) | 9/10 (90%) | 7/8 (88%) | 22/23 (96%) | 20/22 (91%) |
| CD123 | 78/87 (90%) | 28/32 (88%) | 3/7 (43%) | 7/7 (100%) | 20/21 (95%) | 20/20 (100%) |
| CD4 | 64/84 (76%) | 30/31 (97%) | 7/7 (100%) | 4/7 (57%) | 18/20 (90%) | 5/19 (26%) |
| CD64 | 99/102 (97%) | 39/39 (100%) | 10/10 (100%) | 8/8 (100%) | 22/23 (96%) | 20/22 (91%) |
| CD14 | 36/102 (35%) | 10/39 (27%) | 10/10 (100%) | 0/8 (0%) | 15/23 (65%) | 1/21 (5%) |
| CD56 | 32/102 (31%) | 24/39 (62%) | 4/10 (40%) | 0/8 (0%) | 3/23 (13%) | 1/22 (5%) |
| CD2 | 4/99 (4%) | 1/39 (3%) | 0/9 (0%) | 0/8 (0%) | 2/22 (9%) | 1/21 (5%) |
| CD7 | 3/102 (3%) | 1/39 (3%) | 0/10 (0%) | 0/8 (0%) | 0/23 (0%) | 2/22 (9%) |
| CD19 | 4/102 (4%) | 1/39 (3%) | 1/10 (10%) | 0/8 (0%) | 0/23 (0%) | 2/22 (9%) |
| CD36 | 29/59 (49%) | 14/21 (67%) | 4/4 (100%) | 0/5 (0%) | 9/16 (56%) | 2/13 (15%) |
| CD22 | 3/67 (5%) | 2/24 (8%) | 0/5 (0%) | 0/5 (0%) | 0/16 (0%) | 1/17 (6%) |
| CD25 | 3/65 (5%) | 0/21 (0%) | 0/6 (0%) | 0/5 (0%) | 0/16 (0%) | 3/17 (18%) |
| CD15 | 65/98 (66%) | 36/37 (97%) | 7/10 (70%) | 2/8 (25%) | 15/21 (71%) | 5/22 (23%) |
| CD38 | 92/92 | 35/35 | 10/10 | 7/7 | 20/20 | 20/20 |
| MPO | 50/100 (50%) | 3/39 (8%) | 8/9 (89%) | 4/8 (50%) | 17/22 (77%) | 18/22 (82%) |
| TdT | 10/101 (10%) | 1/39 (3%) | 2/9 (22%) | 1/8 (13%) | 2/23 (9%) | 4/22 (18%) |
- Abbreviations: MPO, myeloperoxidase; NSE, non-specific esterase; Positive, dim to strong stain, all counted as positive.
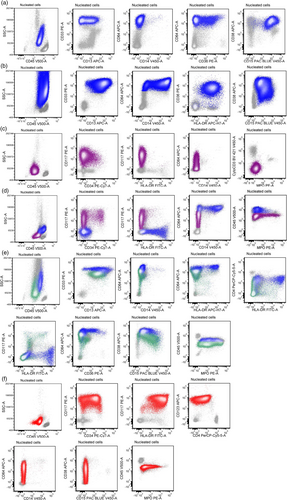
Forty-nine (48%) cases showed monocytic/monoblastic (AMML) immunophenotype. Thirty-nine (38%) exhibited an immature monocytic immunophenotype, which is the most common subtype, and 10 (10%) exhibited a mature monocytic immunophenotype (Table 2 and Table S1). Of the AMML cases, CD56 expression was positive in 28 (57%) cases. Of cases with an immature monocytic phenotype, CD117 (often only in a subset of blasts) was detected in 29 (74%) cases, CD13 was partial to strongly positive in 24 (62%) cases with partial or complete loss of CD13 in 33/39 (85%) cases, and increased CD15 was detected in 21/39 (54%) whereas CD34 was only positive in one patient (3%).
Eight (8%) cases showed an APL-like immunophenotype lacking CD34 as well as absence/dim HLA-DR, but expressing other myeloid markers such as CD13, CD33, and CD117 (Table 2 and Table S1). Unlike classic APL, these cases morphologically lacked abundant cytoplasmic granules, and exhibited lower side scatter. While moderate to bright expression of CD64 was observed in 5/8 cases, other monocytic markers such as CD14 and CD36 were negative in all tested cases. In contrast to true APL with strong and uniform MPO expression, MPO was completely negative in 4 patients, partially positive in small subset of blasts in 3 patients, and uniformly positive in one patient. Non-specific esterase (NSE) was positive in a subset of blasts in 2/5 patients, and negative in 3/5 patients.
Twenty-three (22%) patients showed myelomonocytic immunophenotype. In 21 patients, the blasts contained two distinct populations of myeloblasts and monocytic blasts. The myeloblast immunophenotypes varied in the expression of CD34 and HLA-DR, and in 7 (33%) cases the myeloblasts had an APL-like phenotype with loss of both CD34 and HLA-DR (Figure 1d). The myeloblasts were mostly positive for MPO (n = 22/23) and the monocytic blasts were positive for NSE (n = 17/17). In two cases, monocytic and granulocytic markers were co-expressed on the same blast population, which were strongly positive for both MPO and NSE (Figure 1e).
Twenty-two (22%) patients showed a myeloblastic immunophenotype. An unusual partial or complete loss of either CD34 or HLA-DR expression was observed in 8 (36%) and 8 (36%) patients, respectively. MPO expression was detected in 18 (82%) patient and was negative in 4 (18%) patients, which was further confirmed by cytochemistry stain. NSE cytochemistry stain (n = 12) showed <5% positive cells in all except one case in which blasts were partially positive.
3.5 The correlation between immunophenotype and other laboratory data
APL-like phenotype had low WBC count (1.4 × 109/L, range 1.0–24.6 × 109/L); it was significantly lower than WBC count in patients with immature monocytic phenotype (8 × 109/L, range 0.5–291.9 × 109/L, p = 0.03), mature monocytic phenotype (18.9 × 109/L, range 2.3–140.2 × 109/L, p < 0.01) and myelomonocytic phenotype (10.2 × 109/L, range 1.1–159.4 × 109/L, p < 0.01) (Table 1). The highest median absolute monocyte count was seen in patients with mature monocytic phenotype (4.4 × 109/L, range 0.3–29.4 × 109/L); it was significantly higher than absolute monocyte count in patients with immature monocytic phenotype (0.04 × 109/L, range 0–15.6 × 109/L, p < 0.01), APL-like phenotype (0.04 × 109/L, range 0–0.7 × 109/L, p < 0.01) and myeloblastic phenotype (0.1 × 109/L, range 0–1.6 × 109/L, p < 0.01). The median absolute monocyte count in patients with myelomonocytic phenotype (1.1 × 109/L, range 0–76.8 × 109/L) was significantly higher than absolute monocyte count in patients with immature monocytic phenotype (p < 0.01), APL-like phenotype (p < 0.01) and myeloblastic phenotype (p = 0.014). The difference in absolute monocyte count between patients with mature monocytic and myelomonocytic phenotypes was not statistically significant.
Mature monocytic and APL-like phenotype showed low circulating blasts in peripheral blood (6%, range 0%–51% and 0%, range 0%–80%, respectively), which was significantly lower than the blast percentages in patients with immature monocytic (36%, range 0%–94%, both p < 0.05) and myeloblastic (54%, range 0%–92%, both p < 0.05) phenotype. Mature monocytic phenotype had a lower blast percentage in bone marrow (31%, range 20%–55%), statistically different from all other four groups (p < 0.01).
Table 1 summarizes the relationship between immunophenotype and cytogenetic findings. Mature monocytic phenotype had t(11;19) (q23;p13.1)/ELL-KMT2A as the most common rearrangement (4/10, 40%), followed by t(9;11) (p22;q23)/MLLT3-KMT2A (3/10, 30%), which is statistically different from immature monocytic phenotype, APL-like phenotype and myelomonocytic phenotype, where t(9;11) (p22;q23)/MLLT3-KMT2A was the most common rearrangement. t(6;11) (q27;q23)/AFDN-KMT2A cases more often exhibited typical CD34 + CD117 + CD13 + CD33 + HLA-DR+ myeloblasts (either the entire blast population, 5/22) or including cases mixed with monocytic blasts (9/36).
Mutations involving the RAS pathway, NRAS/KRAS/PTPN11 were detected in 26 patients, including 1 case with a mature monocytic immunophenotype, 11 immature monocytic, 8 myelomonocytic and 6 myeloblastic (Table 1 and Table S1). All 6 cases with an APL-like immunophenotype showed no mutations, which was statistically different from all other four groups (p = 0.014). No statistical differences were present in 5 immunophenotypic subgroups for KRAS, NRAS, PTPN11, ASXL1, or FLT3 mutation.
3.6 AML immunophenotype at relapse
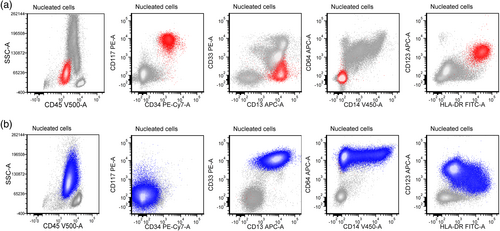
Twenty-four out of 64 patients experienced relapse during the follow-up period and had immunophenotype studies performed at the time of relapse. Major immunophenotypic change was observed in 10 (42%) patients (Table 3). Except for one patient who relapsed with AML with some megakaryocytic markers (by immunohistochemistry), the others had AML with major immunophenotype change between the above five immunophenotypic categories. Figure 2 showed one case with initial myeloblastic phenotype changed to immature monocytic phenotype at relapse (case 6 in Table 3). Karyotype analysis was performed in all 10 patients, and NGS panel was performed in 7 patients at the time of initial diagnosis and/or relapse. Seven out of these 10 patients gained additional cytogenetic and/or genetic alterations at the time of relapse (Table 3).
| Case | Initial IP | Relapsed IP | Karyotype | KMT2A partner | Genetic mutation | Karyotype at relapse | Genetic mutation at relapse |
|---|---|---|---|---|---|---|---|
| 1 | APL-like | Myeloblastic | 46,XX,t(9;11) (p22;q23) [20] | MLLT3 | NA | 46,XX,del(7) (q22q34),t(9;11) (p22;q23) [20] | NA |
| 2 | Immature monocytic | Myeloblastic | 46,XX,t(9;11) (p22;q23) [20] | MLLT3 | NA | 46,XX,t(9;11) (p22;q23) [10] | NA |
| 3 | Immature monocytic | APL-like | 48,XX,t(2;4;11) (p23;q21;q23),del(3) (q21q27), +der(4)t(2;4;11),add(6) (p11.2),del(12) (p12p13), +13,add(19) (p13.3) [20] |
AFF1 | NA | 48,XX,t(2;4;11) (p23;q21;q23),del(3) (q21q27),+der(4)t(2;4;11),add(6) (p11.2),del(12) (p12p13),+13,add(19) (p13.3) [7]/48,idem,del(5) (q31q35) [4]/48 ~ 49,XX,t(2;4;11),(p23;q21;q23),del(3) (q21q27),+der(4)t(2;4;11),add(6) (p11.2),del(7) (q22q34),del(12) (p12p13),+13,add(19) (p13.3)[cp3] | NA |
| 4 | Myelomonocytic | Mature monocytic | 46,XY,t(9;11) (p22;q23) [9]/47,idem,+der(9)t(9;11) [9]/62,idem,-X,-1,+3,-4,+7,-11,-12,-12,+13,+14,-17,-19,-21,-22[1]/46,XY [1] | MLLT3 | None | 47,XY,t(9;11) (p22;q23),+21[16]/48,XY,+der(9)t(9;11) (p22;q23),t(9;11),+16[4] | NRAS |
| 5 | Myelomonocytic | APL-like | 46,XX,t(9;11) (p22;q23) [18]/47,idem,+21[1]/46,XX,del(13) (q12q22) [1] | MLLT3 | None | 46,XX,t(9;11) (p22;q23) [5]/46,XX,t(5;12) (q23;q24.2) [1]/46,XX [14] | None |
| 6 | Myeloblastic | Immature monocytic | 47,XX,+8[13] | MLLT3 | IDH1, JAK2, RUNX1 | 47,XX,+8,t(9;11) (p22;q23) [20] | NA |
| 7 | Mature monocytic | AML with megakaryocytic differentiation | 46,XX,t(9;11) (p22;q23) [14]/50,idem,+13,+19,+19,+21[6] | MLLT3 | None | 53,XX,+3,+8,t(9;11) (p22;q23),+17,+19,+20,+21[5]/46,XY,inv(9) (p12q13)[25] | None |
| 8 | Myelomonocytic | Immature monocytic | 47,XX,+8,t(11;19) (q23;p13.1) [4]//46,XY [16] | ELL | None | 47,XX,+8,t(11;19) (q23;p13.1) [19]/47,idem,del(12) (p13),der(22)t(1;22) (p10;q10) [1] | PTPN11 |
| 9 | Myelomonocytic | APL-like | 46,XX,t(11;19) (q23;p13.3) [20] | MLLT3 | NF1, NRAS, WT1 | 46,XX,t(11;19) (q23;p13.3) [20] | NA |
| 10 | Myeloblastic | APL-like | 46,XY,inv(9) (p12q13),t(9;11) (p22;q23) [16]/45,idem,-Y [4] | MLLT3 | GATA2, PTPN11 | 47,XY,inv(9) (p12q13),t(9;11) (p22;q23),+21[12]//46,XX [8] | None |
- Abbreviations: IP, immunophenotype; NA, not available.
4 DISCUSSION
In this study, we collected one of the largest series of cases of AML with KMT2A rearrangement and conducted detailed immunophenotypic characterization, both at the time of initial diagnosis and at the time of relapse.
In keeping with the general impression and reports from earlier studies (Baer et al., 1998; Swansbury et al., 1998), a significant proportion (71%) of cases showed monocytic differentiation, nearly half of cases presented as acute monocytic/monoblastic leukemia (AMML), and a quarter presented as myelomonocytic AML. Of the cases presenting as AMML, the most cases exhibited an immature monocytic immunophenotype, and about 20% of these cases had a mature monocytic immunophenotype. The immaturity of monocytic blasts manifested as complete or substantial loss of CD14 and CD36, and partial CD117 expression. In addition, we showed that monocyte immaturity was strongly associated with partial or complete loss of CD13 (85%) and elevated CD15 expression (54%). These findings should contribute to the assessment of monocyte maturity, which has been notoriously difficult, both by morphologic examination (Foucar et al., 2020) and flow cytometry immunophenotyping (Matarraz et al., 2017).
In addition, we found that the immunophenotype of AML with KMT2A rearrangement is more diverse than previously recognized. A subset (8%) of cases showed an APL-like immunophenotype with loss of CD34 and HLA-DR, mostly negative for CD15, while expressing CD117, CD13, CD33, and often moderate CD64. In contrast to APL, MPO was either partial or negative in these cases, and blasts had lower side scatter characteristics. Blasts with APL-like immunophenotype were additionally observed in the myeloblast component of the seven cases in myelomonocytic AML subgroup. An APL-like immunophenotype has been described in NPM1-mutated AML that is associated with an improved outcome (Mason et al., 2018); and due to its unique immunophenotype, it has become a feature to raise the suspicion for NPM1-mutated AML at the time of initial diagnosis. We show here that this APL-like immunophenotype is not unique for NPM1+ AML and can be observed in KMT2A-rearranged AML.
In general, the presence of KMT2A rearrangement is associated with a poor clinical outcome. Earlier studies have shown that the outcome of patients with AML associated with t(9;11) (p22;q23)/MLLT3-KMT2A is more favorable than that of patients with other 11q23 abnormalities (Mrózek et al., 1997), especially in patients younger than 60 years of age (Bill et al., 2020). On the other hand, t(6;11) (q27;q23)/AFDN-KTM2A rearrangement is reported to be associated with a poorer outcome (Bill et al., 2020). In our patients, t(6;11) (q27;q23)/AFDN-KTM2A was more frequently associated with a myeloblastic phenotype. Similar to the recently published study (Bill et al., 2020), we showed that somatic mutations detected by NGS in of AML with KMT2A rearrangement mainly involved the RAS pathway (KRAS, NRAS, and PTPN11). Noticeably, cases with an APL-like immunophenotype showed no mutations. Studies have shown that NPM1-mutated AML had similar immunophenotype subtypes. Prognostically, the NPM1-mutated AML with an APL-like immunophenotype had a superior outcome whereas the myeloblastic immunophenotype was associated with an adverse risk (Mason et al., 2018). Due to the fact that over 40% of cases were therapy-related in our study, a confounding factor with an adverse outcome, as well as small number of cases in each immunophenotypical subgroup, there is no statistical difference in overall survival among the five immunophenotype subgroups.
At the time of relapse, the immunophenotype subtype remained the same in 14/24 patients, but showed a major change in 10 patients. The relapse immunophenotypes changed from one subtype to another subtype within the five described immunophenotypes, with an exception for one patient who relapsed with AML showing megakaryoblastic differentiation.
In summary, we show that the immunophenotype of AML with KMT2A rearrangement is more diverse than previously recognized. As reported previously, cases with monocytic or myelomonocytic differentiation are common, but a substantial subset of cases may have a typical myeloblastic immunophenotype; and a minor subset have an immunophenotype mimicking acute promyelocytic leukemia (APL-like). This immunophenotypic diversity may be partly attributable to different partner genes involved or associated somatic mutations. Recognition of the immunophenotypic subtypes and variations of AML with KMT2A rearrangement will be helpful for the initial triage of specimens and future detection of minimal residual disease.
ACKNOWLEDGMENTS
The authors are indebted to Mrs. Sonia Perez for technical assistance with this project.
CONFLICT OF INTEREST
G.C.I. received research funding from Celgene, Kura Oncology, Syndax and Novartis, and received consultancy fees from Novartis and Kura Oncology. Other authors declare no conflict of interest.