Space flight associated changes in astronauts’ plasma-derived small extracellular vesicle microRNA: Biomarker identification
This pilot study suggests relatively short (median 12 days long) low-Earth orbit (LEO) spaceflight induces changes in circulating plasma small extracellular vesicle (sEV) microRNA expression. Normalization of small RNA sequencing (sRNAseq) data and quantitative polymerase chain reaction (qPCR) validation confirmed miR-4732-3p is significantly upregulated up to 3 days post-landing, and enrichment analysis suggests this miRNA is expressed in various central nervous system tissues and hematopoietic cells and may be linked to different organ disorders.
During spaceflight, astronauts are exposed to numerous stressors (i.e. microgravity and ionizing radiation)1 that are linked to various harmful health effects, including cardiovascular (CV), neurodegenerative diseases, and cancers.2, 3 However, the molecular basis of spaceflight-induced pathophysiological changes remains limited. Few studies have suggested LEO spaceflight promotes physiologic stress as noted by increased reactivation of Epstein Barr virus, elevated urine catecholamine and cortisol levels, and increased circulating cell-free mitochondrial DNA and cytokines.1, 4-7 Interestingly, NASA's Twin study also noted unique miRNA signatures (miR-125, miR-16, and let-7a) in peripheral blood mononuclear cells following year-long LEO spaceflight.8 Additionally, emerging evidence shows that sEVs carry cell-specific RNA signatures that can regulate stress responses.9 Thus, changes in sEVs content may serve as potential predictive biomarkers of health and/or subclinical disease. Therefore, we sought to determine whether the spaceflight environment can induce alterations in sEV miRNA content.
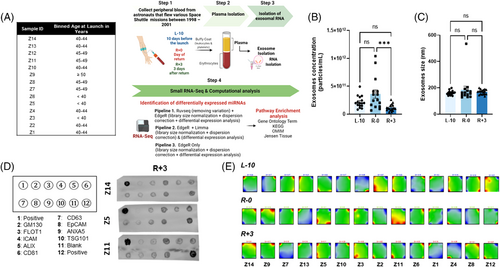
We performed sRNAseq using sEVs isolated from deidentified plasma collected 10 days before launch (L-10), the day of landing (R-0), and 3 days post-landing (R+3) from 14 astronauts who flew various space Shuttle missions between 1998–2001 (Figure 1A). This study was approved by NASA and the Icahn School of Medicine at Mount Sinai's IRBs (STUDY00000075 and HSM19-00367, respectively). Nanosight analysis revealed that the concentration of sEVs was reduced at R+3 but the size remained unchanged (Figure 1B,C). sEV content was further characterized using an exosome-specific antibody array confirming the preponderance of exosomal proteins (Figure 1D). The sRNAseq dataset revealed plasma sEVs were enriched with various RNA species (Figure S1A); however, the current study focused on miRNA. Batch effect assessment of variability of raw miRNA counts showed strong batch effects associated with inter-astronaut variability of miRNA expression (Figure S1B–D). Further, the portrayal of miRNA transcriptome landscapes using self-organizing maps confirmed the considerable interindividual variability at baseline and at both post-flight time points (Figure 1E).
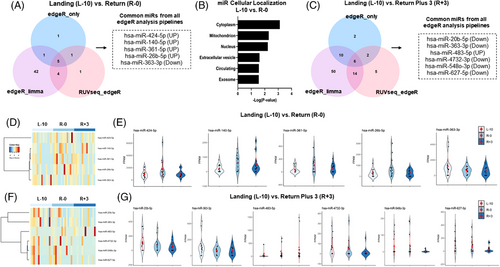
To determine alterations in miRNA expression, we utilized three different normalization methods (edgeR only, edgeR-limma, and RUVseq-edgeR), which identified five overlapping differentially regulated miRNAs at R-0 compared to L-10 (Figure 2A,D,E). Of these, four were upregulated (hsa-miR-424-5p, hsa-miR-140-5p, hsa-miR-361-5p, hsa-miR-26b-5p) and one downregulated (hsa-miR-363-3p). Analyses of predicted subcellular localization revealed that most miRNAs are expressed in the cytoplasm, followed by mitochondria, nucleus, EVs, free in circulation, and exosomes (Figure 2B). Further, we identified six overlapping differentially expressed miRNAs at R+3 compared to L-10, of which five were downregulated (hsa-miR-20b-5p, hsa-miR-363-3p, hsa-miR-4732-3p, hsa-miR-5480-3p, hsa-miR-627-5p) and one was upregulated (hsa-miR-483-5p) (Figure 2C,F,G). We did not detect any significant localization predictions for these six miRNAs.
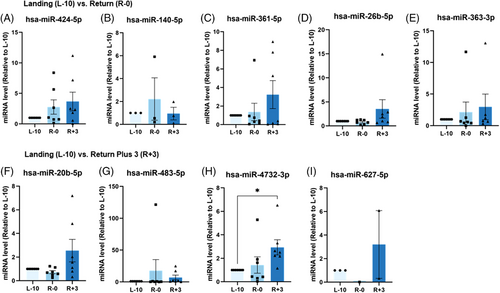
To validate these sRNAseq analysis findings, we assayed the expression of nine miRNAs by qPCR (Figure 3). Only hsa-miR-4732-3p was significantly upregulated at R+3 compared to L-10 (p < 0.05) (Figure 3H). Notwithstanding the substantial interindividual variability, comparing the sRNAseq datasets and qPCR analysis revealed discrepancies between these two approaches. For example, hsa-miR-4732-3p expression was down-regulated at R+3 in sRNAseq compared to qPCR results. Similarly, qPCR analysis for hsa-miR-363-3p, hsa-miR-20b-5p, and hsa-miR-627-5p appeared to be regulated in the opposite direction to sRNAseq data (Figures 2C,F,G and 3E,F,I). Of note, sRNAseq was performed using RNA isolated from sEVs of 14 astronauts, while qPCR analysis could only be performed on seven astronauts due to the exceptionally rare sample availability. Thus, out of an abundance of caution, we narrowed our focus to hsa-miR-4732-3p, which displayed significant regulation at R+3 compared to L-10 by qPCR.
To better understand the potential tissues, biological pathways, and diseases that may be associated with spaceflight-regulated hsa-miR-4732-3p, we performed functional enrichment of the top 20 key hub-nodes for this miR using MiEAA (Figure 4). Network analysis results were plotted with degree centrality and closeness centrality reflecting the node's influence on hsa-miR-4732-3p (Figure 4A). Tissue enrichment analysis using the Jensen Tissues database identified adipose tissue and uterus as the most enriched, whereas the most significant number of target genes were enriched in different tissues of the central nervous system (34%), followed by the immune/lymphatic system (31%) (Figure 4B).
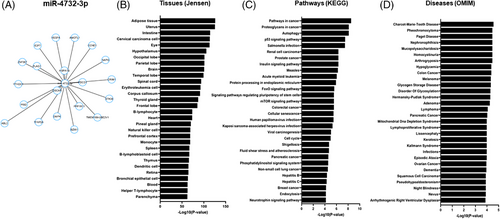
KEGG pathway enrichment analysis revealed that the top 20 pathways identified for hsa-miR-4732-3p are commonly involved in dysregulation of 1) pathways in various cancers, 2) pathogen mediated diseases (Salmonella infection, measles, viral carcinogenesis, human Papillomavirus infection and Hepatitis B and C); 3) homeostatic cell regulation including p53, FoxO and mTOR signalling pathways, cell cycle, cell senescence, autophagy, and pathways regulating pluripotency of stem cells, 4) phosphatidylinositol and insulin signalling pathways and 5) cardiovascular hemodynamics (fluid shear stress and atherosclerosis) (Figure 4C).
Lastly, we extended our analysis to determine potential diseases that hsa-miR-4732-3p may be involved. The OMIM database suggests this miRNA may be involved in various hematopoietic and solid cancers (27%), endocrine/metabolic disorders (27%), and neurodegenerative disorders (17%) (Figure 4D). Other diseases miR-4732-3p may be involved in include musculoskeletal (arthrogryposis, Paget disease), cardiovascular (arrhythmogenic right ventricular dysplasia), and dermatologic disorders (keratosis, nevus), among others (Figure 4D and Data S2).
In conclusion, our data suggest that short LEO spaceflight induced significant changes in plasma-derived sEV miRNA content and identified hsa-miR-4732-3p to be significantly upregulated post-flight. Curiously, NASA's Twin Study showed an increase in carotid intima-media thickness which remained stable 4 days post-flight,3 whereas our bioinformatics analysis revealed hsa-miR-4732-3p involvement in cardiovascular hemodynamics. Further, miR-4732-3p is upregulated in mesenchymal stromal cell-derived sEVs following oxygen-glucose deprivation and contributes to cardioprotection via reduction of apoptosis and levels of reactive oxygen species.10 Further longitudinal studies using samples from a larger astronaut cohort with paired clinical data are warranted to validate the utility of miR-4732-3p as a potential biomarker for monitoring astronauts’ health.
ACKNOWLEDGEMENTS
We would like to thank Stepan Nersisyan Faculty of Biology and Biotechnology, HSE University, Moscow, Russia for his assistance with miRNA sequencing analysis. This work was supported by the Translational Research Institute for Space Health FIP0005 and National Aeronautics and Space Administration grant 80NSSC19K1079 to D.A.G. and by National Aeronautics and Space Administration grant 80NSSC21K0549 to D.A.G. and K.W. This work was also partially supported by American Heart Association Career Development Award 18CDA34110277 and startup funds from the Ohio State University Medical Center to VNSG. This work was supported by American Heart Association Post-Doctoral Fellowship award 915681 to AKR. Computing resources were provided in the frames of the State Target Program of the Government of Armenia “Creating a Cloud Computing Environment for Solving Scientific and Applied Problems” (1-8/TB-21). This work was also partially supported by P01HL134608 to RK. Work was also performed under the auspices of the US Department of Energy by Lawrence Livermore National Laboratory under contract DE-AC52-07NA27344.
CONFLICT OF INTEREST
The authors report no conflict of interest.




