Comprehensive genomic and digital pathology profiling of tobacco-chewer female oral cancer patients simultaneously with integration of single-cell datasets identifies clinically actionable patient subgroups
Arnab Ghosh and Siddharth Singh contributed equally to this work.
Dear Editor,
Genomic alteration landscape of oral squamous cell carcinoma (OSCC) among female patients remains understudied,1, 2 although certain regions – southeast Asia, including India – show high burden of OSCC among females.3 Through generation of whole exome sequencing (WES) and copy-number-array from paired tumour and paired normal tissues, and integrative analysis with digital pathology image from same tumour sections, this study identified somatic alterations associated with differential immune infiltration profiles of these tumours (Figure S1). Further integration of large-scale single-cell transcriptomics data reveals novel epithelial and immune interactions for this immune suppression which led to identification of two therapeutically actionable subgroups within these patients.
WES analysis from 38 female OSCC patients (Table S1) identified 5141 somatic mutations (of which 3846 were non-synonymous) with a median of 105.5 mutations per patients with a range of 8–564 (Figure 1A and Table S2). Ten genes, CASP8 (mutated in 57.89% patients), TP53 (55.26%), FAT1 (42.11%), NOTCH1 (31.58%), CDKN2A (23.68%), HLA-B (21.05%), HRAS (21.05%), EPHA2 (15.79%), PIK3CA (15.79%) and ARID2 (13.16%), were found to be significantly mutated (q < .1) (Table 1 and Figures 1B and S2). We also detected somatic mutations in KMT2B, HLA-A, PTEN and FBXW7 genes which are previously known to be associated with head and neck cancer. Based on somatic mutational landscape of the driver genes, patients could be stratified into four broad groups (Figure 1B). We showed CASP8 mutations alter pro-caspase-8 and active caspase-8 levels in tumour lysates as evident from western blot assays (Figure S3). We did not find any significant (p = .345) differences in frequency of TP53 somatic mutations in these female patients (55.26%) as compared with male patients in ICGC-India OSCC cohort (60.92%),2 but the frequency was significantly (p = .0001) lower than the HPV(−) male patients in TCGA-HNSC cohort (84.93%) (Table 1). TP53 mutated patients had significantly (p = .035) lower recurrence-free survival (Figure S4). In contrast, prevalence of CASP8, NOTCH1, CDKN2A, HRAS, HLA-B and EPHA2 mutations were significantly higher (p < .026) in these female patients than male patients in ICGC-India OSCC cohort1, 2 (Table 1). Oncogenic hotspot somatic mutations in TP53, PIK3CA or HRAS genes were identified in 28.95% of the patients (Figure 1B). These oncogenic somatic mutations occurred significantly (p = .0331) more frequently in tumours with CASP8 mutations (45.45%) than among the remaining (12.5%). The somatic mutations in this cohort were found to be contributed by 12 frequently detected (≥10% tumours) somatic-mutational-signatures linked to tumour aging, smoking, APOBEC hyperactivity, UV and defective DNA repair (Figure 1C).
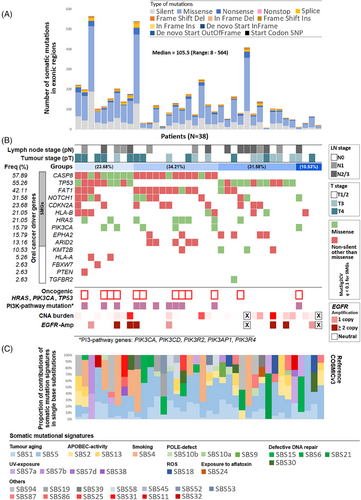
| SMG | dbGENVOC | p value | p value | |||
|---|---|---|---|---|---|---|
| Rank | Gene | Female centric OSCC cohort | ICGC-India | (Fisher's exact test, one-sided) | TCGA HNSCC | (Fisher's exact test, one-sided) |
| Female (N = 38) | Male (N = 87) | HPV(−) Male (N = 292) | ||||
| 1 | TP53 | 21 (55.26%) | 53 (60.92%) | .3454 | 248 (84.93%) | .0001 |
| 2 | CDKN2A | 9 (23.68%) | 5 (5.75%) | .0059 | 73 (25.00%) | .5196 |
| 3 | HLA-B | 8 (21.05%) | 6 (6.90%) | .026 | 11 (3.77%) | .0004 |
| 4 | CASP8 | 22 (57.89%) | 33 (37.93%) | .0307 | 28 (9.59%) | .0001 |
| 5 | HRAS | 8 (21.05%) | 7 (8.05%) | .0429 | 17 (5.82%) | .0037 |
| 6 | EPHA2 | 6 (15.79%) | 2 (2.30%) | .0098 | 13 (4.45%) | .0138 |
| 7 | FAT1 | 16 (42.11%) | 23 (26.44%) | .0644 | 65 (22.26%) | .0087 |
| 8 | ARID2 | 5 (13.16%) | 10 (11.49%) | .502 | 9 (3.08%) | .0143 |
| 9 | NOTCH1 | 12 (31.58%) | 9 (10.34%) | .0049 | 50 (17.12%) | .0319 |
| 10 | PIK3CA | 6 (15.79%) | 12 (13.79%) | .4832 | 40 (13.70%) | .4414 |
- Note: 6 significantly mutated genes (SMG) showing statistically significant difference in mutation frequency in the female OSCC cohort as compared to male OSCC cohort in India are highlighted.
The median copy-number-alteration burden was 0.09% ranging between 0.0016 and 23.5%, with significant (q < .1) amplification of 3p, 8p, 8q, 19p and 19q, and deletion of 3q, 8q, 9p, 9q and 14q chromosome arms (Table S3). Significant focal amplification of ALDH1L1 (56.5%) and EGFR (17.4%) (Figure 1B) were identified. Alongside, focal amplification of KCNJ11, and deletions of CDK7, CDKN2A, BRAF and FAT1 genes were identified.
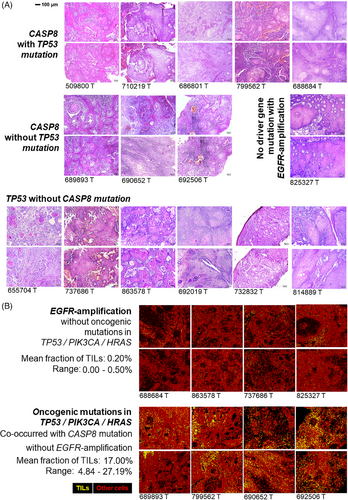
Somatic alteration patterns are often shown to influence immune infiltration in solid tumours. In this study, we have utilised whole slide imaging (WSI) data generated (20×-magnification) from haematoxylin–eosin-stained (H&E) tumour tissue sections for a subset of patients (Figure 2A) to infer immune infiltration. Recently developed deep-learning model4 was utilised to quantify tumour infiltrating leukocytes (TILs) (Table S4 and Figure S5) from WSIs, followed by validation using an orthogonal model (Figure S6) at cell level (Figure 2B). From these WSIs, a median TILs infiltration of 4.84% with a range between 0 and 27.19% was detected for these tumours. Based on the distribution of the proportion of TILs, the patients could be stratified in two groups – (a) low-TIL (with 0–0.5% TIL) and (b) high-TIL (with 4.84–27.19% TIL) (Table S4). The somatic mutational burden in these tumours were not correlated with the proportion of TILs. We undertook an unbiased approach to correlate somatic alteration status (mutation or copy number alterations) with TIL infiltration and found – tumours with low-TIL are significantly (p = .007) more enriched with EGFR copy number amplification (71.4%) as compared with tumours with high-TIL (0%). The average fraction of TILs in EGFR-amplified tumours was 0.17% ranging between 0 and 0.5%, while the remaining tumours had an average of 12.5% TILs ranging between 0.14 and 27.19% (Figure and Table S4). In contrast, CASP8-mutated tumours (enriched for co-occurrence of other oncogenic-mutations) showed significantly (p = .026) higher infiltration of TILs (median = 9.98%), than the rest (0.17%) (Figure 2B and Table S4). These findings were validated in TCGA-HNSC cohort (Figure S7A).
As EGFR expression was positively correlated with its genomic-copy-number in TCGA-HNSC cohort (Figure S7B), to further elucidate the plausible mechanism of EGFR driven immune suppression, we have created and analysed an integrated gene expression atlas of 66,809 single cells from 55 oral cavity tumours (Figure 3A–E). We show that tumour epithelial cells with high EGFR expression featured – (a) significant down-regulation of (a.1) antigen-processing and presentation (HLA-A, HLA-B, HLA-DRA, B2M, etc.), (a.2) immune-stimulating genes (CXCL17, LCN2 and SAA1), and (b) significant up-regulation of (b.1) pro-angiogenic factors VEGFB and FGFBP1, (b.2) glutathione biosynthesis related gene GPX2, and its interacting partners AKR1C1, AKR1C2 and AKR1C3 (Figure 3F). High EGFR expressing tumour cells were shown to have elevated epithelial-to-immune interactions through (1) PTN (interacting with NCL on immune cells), (2) PLAU (interacting with PLAUR on macrophages), (3) ADM (interacting with CALCRL on pDCs) and (4) MDK (interacting with ITGA4 and ITGB1 on macrophages) (Figure 3G,H) – potentially facilitating pro-tumourigenic microenvironment. Whereas interactions necessary of T-cell activation, like MIF (interacting with CXCR4, CD74 and CD44), PPIA (interacting with BSG) and CXCL16 (interacting with CXCR6 on CD8+ T-cells) were down-regulated in high EGFR expressing tumour cells. Moreover, we show that these EGFR-high tumour cells overexpressed LGALS3, CD59, laminins (LAMB1 and LAMB3), PROS1 and so on that induced LAG3 and CD44 expression in the associated CD8+ T cells (Figure 3I–K) featuring an exhaustion state. Recently EGFR-amplification was shown to decrease immune-infiltration in lung5 and gastroesophageal-adenocarcinoma.6 Detailed methodology and results are available as Supporting Information.
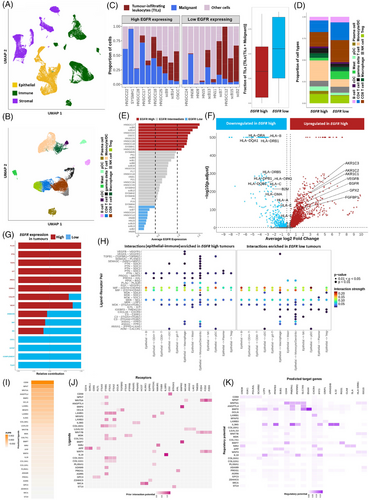
This study provides novel insights into EGFR-amplification mediated immune suppression in oral tumours, which was present in 22.22% of tumours from female OSCC patients. We identified key epithelial–immune interactions in high EGFR expressing tumours such as LGALS3–LAG3, PLAU–PLAUR, MDK–(ITGA4+ITGB1) which can be therapeutically targeted with available inhibitors such as relatlimumab,7 upamostat8 and natalizumab9 alongside currently prescribed anti-EGFR cetuximab10 to increase immune infiltration within tumour for potential prognostic benefit. Additionally, the results suggest tumours with oncogenic somatic mutations (found in 31.58% female OSCC patients) that showed comparatively high immune infiltration may benefit from a combination alpelisib or tipifarnib (targeting PIK3CA or HRAS)11 and adjuvant immune-checkpoint inhibitors.
AUTHOR CONTRIBUTIONS
N. K. B., T. K. K. and P. P. M. conceptualised the project. T. K. K., S. S., K. S. G. and A. Z. M. coordinated patient recruitment, biospecimen collection. A. M. coordinated sequence data generation through the Indian national genomics core. A. G., S. C., C. D. and N. K. B. performed NGS data analysis. T. R. M., A. G. and N. K. B. integrated publicly available scRNAseq data and interpreted. S. S., S. V. B. and T. K. K. generated histopathology and western blot data. A. G. and N. K. B. integrated genomic and digital pathology data. A. G., S. S., T. R. M., S. C., C. D., P. P. M., T. K. K. and N. K. B. wrote the manuscript.
ACKNOWLEDGEMENTS
N. K. B., T. K. K., P. P. M. and A. M. acknowledge Department of Biotechnology, Government of India for funding via its Virtual National Oral Cancer Institute (VNOCI) Grant. P. P. M. acknowledges National Science Chair Grant. A. G. acknowledges ICMR-SRF fellowship (GENOMICS-BMS/2021-10619) and BRIC-RCB (RCB/NIBMG-PhD/2022-23/A/394/1002). S. S. acknowledges the Integrated Ph.D. program fellowship of Jawaharlal Nehru Centre for Advanced Scientific Research (JNCASR) and SVB acknowledges CSIR-UGC fellowship. T. R. M. acknowledges ICMR-JRF fellowship [3/1/3/JRF-2022/HRD-07 (19654)] and BRIC-RCB (RCB/NIBMG-Int-PhD/2020/1003). S. C. acknowledges DBT fellowship (DBT/2019/NIBMG/1225) and BRIC-RCB (RCB/NIBMG-PhD/2019/1011). T. K. K. is a recipient of the J C Bose National Fellowship. The software pipelines were developed with funding from NSM grant, Meity to N. K. B.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interest.
FUNDING INFORMATION
This study was funded by Department of Biotechnology, Government of India via the Virtual National Oral Cancer Institute (VNOCI) Grant No. BT/PR17576/MED/30/1690/2016.
ETHICS STATEMENT
This study was approved by the ethics committee of Sri Devaraj Urs Academy of Higher Education and Research (Comprising Sri Devaraj Urs Medical College), Kolar, India [No. SDUAHER/KLR/R&D /81 /2018-19]. Data and bio-specimens were collected with voluntary and informed consent.
Open Research
DATA AVAILABILITY STATEMENT
Whole exome sequence data have been deposited to Indian Biological Data Center (IBDC) and European Nucleotide Archive (ENA). It can be accessed through study accession – IBDC (INDA-CA): INCARP000295 and ENA: PRJEB74692.




