Cytoskeletal regulation of mitochondrial movements in myoblasts
Monitoring Editor: Joseph Sanger
Abstract
Mitochondria are distributed in the cell to match the energy demands, and it is their interaction with the cytoskeleton that controls their movement and displacement. Our purpose was to determine which cytoskeletal components are primarily responsible for mitochondrial movement in muscle cells. Live-cell imaging was used to visualize mitochondrial dynamics in myoblasts. Destabilization of microtubules (MT) reduced the total path length and average speed traveled by mitochondria by 64–74%, whereas actin disruption only reduced these variables by 37-40%. Downregulation of the microtubule motor proteins, Kif5B and dynein, by siRNA resulted in decreases in the average speed of mitochondrial movements, by 30 to 40%. We observed a reduction in the average speed of mitochondrial movements (by 22 to 48%) under high calcium conditions. This attenuation in the presence of calcium was negated in cells pre-treated with siRNA targeted to the microtubule motor protein adaptor, Milton, suggesting that Milton is involved in mediating mitochondrial arrest in the presence of high calcium within muscle cells. Thus, we have demonstrated that, in myoblasts, mitochondria primarily move along microtubules tracks with the aid of the motor proteins Kif5B and dynein, in a manner which is inhibited by calcium. These observations will eventually help us understand organelle movements in more complex muscle systems, such as mature myotubes subjected to elevated calcium levels and contractile activity. © 2014 Wiley Periodicals, Inc.
Introduction
Mitochondria are vital organelles, critical for energy supply and cell survival. Such functional versatility is paralleled by the structural complexity of the organelle. Mitochondria can compensate for alterations in energy requirements by adjusting their size and distribution within skeletal muscle [De Vos et al., 2005]. It is their interaction with the cytoskeleton that is used to control their position, movement and anchorage within cells [Tanaka et al., 1998]. The three cytoskeletal elements, actin, microtubules and intermediate filaments act to vary the degree of scaffolding along which mitochondria are capable of moving.
The distribution of mitochondria to specific sites within the cell has consistently been linked to microtubules and actin. In neurons and cardiac myocytes, microtubules are the primary tracks governing mitochondrial movements [Morris and Hollenbeck, 1995; Yi et al., 2004], while in Saccharomyces cerevisiae the positioning of mitochondria is reliant on actin-based transportation [Simon et al., 1997]. Intracellular transport of cargo, such as mitochondria, to specific locations within the cytoplasm involves molecular motors that bind directly to the cargo and the cytoskeletal track. Dynein and kinesins motor proteins use microtubules to transport cargo throughout the cell. In neurons, movement along microtubules occurs either towards the cell body using cytoplasmic dynein, or away, using kinesins. There are 14 kinesin superfamilies encoded by the human and mouse genomes, consisting of more than 45 members [Hirokawa et al., 2009]. In neurons, Kinesin-1 and Kinesin-3 have been identified to regulate mitochondrial movements [Nangaku et al., 1994; Pilling et al., 2006]. Kinesin-1, also known as conventional kinesin, consists of three distinct members, Kif5A, Kif5B and Kif5C. Kif5A and Kif5C are neuron-specific, whereas Kif5B is ubiquitously expressed [Aizawa et al., 1992; Argyropoulos et al., 2009]. Inhibition of Kinesin-1 in Drosophila halts mitochondrial movement in motor axons [Pilling et al., 2006], and in non-neuronal cells, whereby deletion of Kif5B alters mitochondrial positioning [Tanaka et al., 1998; Wu et al., 1998]. The movement of motor proteins along microtubules takes advantage of the dynamic instability of the tubulin polymer. Growth of microtubules allows for the continued expansion of the cytoskeleton, which motor proteins use as tracks. Conversely, the ability of microtubules to shrink is thought to be important for morphological changes in the cell, and in the reorientation of microtubules during the locomotion of organelles [Mitchison and Kirschner, 1984].
In muscle, calcium (Ca2+) is a key regulator of contractile activity, but it is also an important signaling ion. Ca2+ has a role in the regulation of mitochondrial respiration, and it is involved in the opening of the mitochondrial permeability transition pore (mtPTP) [Crompton and Costi, 1988]. This dissipates the electrochemical proton gradient, thereby halting ATP production by oxidative phosphorylation. Calcium influx has also been found to inhibit mitochondrial movement in neurons and in cardiac myocytes [Yi et al., 2004; Macaskill et al., 2009; Wang and Schwarz, 2009]. The ability of mitochondria to respond to intracellular signals such as Ca2+, is mediated by motor protein/adaptor complexes. The core of this complex is composed of the Kinesin-1 motor along with two mitochondrial-specific adaptors, mitochondrial Rho GTPase (Miro) and Milton [Wang and Schwarz, 2009]. Milton binds directly to Kinesin-1 [Smith et al., 2006], whereas Miro serves as an anchor linking Milton and Kinesin-1 to mitochondria [Glater et al., 2006]. Elevated calcium binds to the EF-hands of Miro and induces a conformational change in the protein, resulting in the dissociation of the complex from microtubules and arresting mitochondrial movements [Macaskill et al., 2009; Saotome et al., 2008; Wang and Schwarz, 2009].
It is now well-known that mitochondria are quite dynamic within certain cell types, however, how the distribution of these organelles is controlled and regulated in myoblasts has not been established. Thus, our purpose was to investigate the cytoskeletal components governing mitochondrial movements within myoblasts, and the relationship of these movements to changes in cytosolic calcium signaling. We hypothesize that microtubules and their associated motor proteins are the main cytoskeletal filament proteins involved in mitochondrial dynamics within myoblasts. Moreover, we believe that mitochondrial motility would be regulated by changes in intracellular calcium, and that adaptor proteins would be involved in the process.
Results
Microtubules are the Primary Filaments Used by Mitochondria During Trafficking
To elucidate which cytoskeletal elements are primarily responsible for mitochondrial motility within myoblasts, we disturbed microtubules and actin, and observed their effects on organelle movement. We examined the co-localization of actin (Fig. 1A) and mitochondria (Fig. 1B) in control cells (Fig. 1C), and compared them to myoblasts exposed to the actin disrupting agent cytochalasin D (Fig. 1D). Exposure to cytochalasin D resulted in the drastic rearrangement of actin filaments, such that they appeared disjoined and no longer extended throughout the length of the cell. Actin disruptions reduced the total path length and average speed of mitochondria by 37 and 40%, respectively (Figs. 1I and 1J).
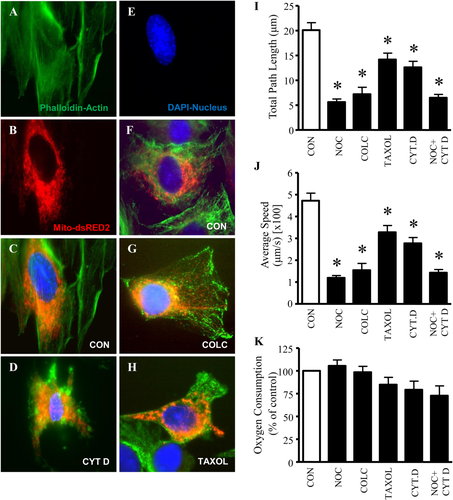
Microtubules are the primary filaments used by mitochondria during trafficking. A: Myoblasts stained for actin (phalloidin, green), B: transfected with Mito-dsRED2 to visualize mitochondria (red), or E: stained with DAPI for nuclei (blue) alone. C: Merged images are overlays of actin (phalloidin, green), Mito-dsRED2 (mitochondria, red) and DAPI (nuclei, blue). Non-treated cells exhibited normal morphology of actin filaments. D: After 1 hr of treatment with cytochalasin D (CYT D), myoblasts presented cells with actin disruptions (phalloidin, green). F: Additionally, untreated myoblasts display normal microtubule (GFP-tubulin, green), mitochondria (Mito-dsRED2, red) and nuclei (DAPI, blue) organization. Myoblasts pre-treated with the microtubule disrupting agents. G: Colchicine (COLC) and H: TAXOL for 1 hr exhibited alterations in the microtubule structures (GFP-tubulin, green). COLC and Taxol were successful at disrupting microtubules, as indicated by the images above. I: Quantification of the total path length and J: the average speed traveled by mitochondria over the 5 min capturing time. Movement of mitochondria was determine from an average of n = 36–72 mitochondria from nine separate cells. K: Oxygen consumption was assessed in myoblasts with microtubule and/or actin disruptions using the microtubule disruptors Nocodazole (NOC), COLC, Taxol, and/or the actin disrupting agent CYT D, along with a control treatment (CON, DMSO). Values are expressed as averages ± SEM, N = 6. *Significantly different from CON, P < 0.05.
Next, we then examined the effects of microtubule disruptions on mitochondrial dynamics. Cells stained with FITC-conjugated α-tubulin antibodies and treated with colchicine or taxol, displayed alterations in the structure of microtubules compared to untreated myoblasts (Figs. 1F–H). The destabilize agent, colchicine, resulted in enhanced depolymerization of microtubules (Fig. 1G). Conversely, the stabilizer, taxol is known to block microtubule disassembly. As depicted in Fig. 1H, taxol preventing normal polymerization of the filaments.
In response to depolymerization of microtubules using nocodazole and colchicine, the total path length of mitochondria was reduced by 64–72% (Fig. 1I), similar reductions were observed in the average speed traveled by the organelles (68–74%, Fig. 1J). In addition, the stabilization of microtubules by taxol also significantly altered mitochondrial motility, albeit to a lesser degree (Figs. 1I and 1J). To deduce the effects that microtubule and actin disruption would have in combination, we co-incubated myoblasts with nocodazole and cytochalasin D. Changes in mitochondrial motility were similar to the results observed with nocodazole treatment alone. We then ascertained whether disruptions in the cytoskeleton and mitochondrial movement had any effect on oxygen utilization. No significant effects on oxygen consumption were observed (Fig. 1K).
Treatment With Microtubule Disrupting Drugs Arrested Mitochondria to a Greater Extent Than Actin Depolymerization
Given the effects that cytoskeletal disruption had on mitochondrial speed, it was of interest to examine whether this reduction was due to slower movements, or to an overall greater percentage of time that mitochondria remained stationary. By transfecting mito-dsRED2 to visualize mitochondria (Fig. 2A), we determined the percentage of time that mitochondria spent in motion. To analyze the movement characteristics of individual mitochondria, kymographs were generated from the live-cell imaging videos to document the dynamic properties of the organelles. We determined that mitochondria remained in constant motion, and frequently changed their direction (Fig. 2B), as reflected by horizontal deviations in the kymograph tracings. The addition of microtubule disrupting agents reduced the motility of mitochondria, as depicted by decreased horizontal deviations in the kymograph tracings over the 5 min recording period with the addition of colchicine, nocodazole, or taxol. Interestingly, the kymograph data generated from treatment with cytochalasin D was comparable to control conditions. Quantification of organelle motility revealed that alterations in microtubules increased the percentage of time that mitochondria were stopped by 1.6- to 2.7-fold (Fig. 2C). However, changes to the structure of actin filaments using by cytochalasin D treatment had no significant effect on the amount of time mitochondria were in motion in these myoblasts.
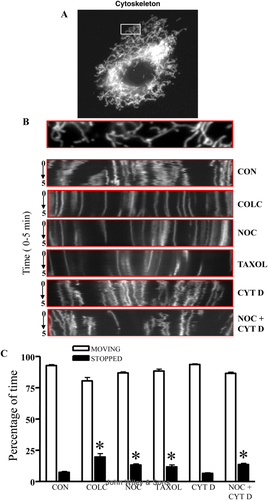
Treatment with microtubule disrupting drugs arrested mitochondria to a greater extent than actin depolymerization. A: Mitochondrial movement in a representative myoblast transfected with mito-DsRed2. B: The first frame in a live-cell imaging series is displayed above with representative kymographs generated from the movies from control (CON, DMSO) Nocodazole (NOC), Colchicine (COLC), Taxol, Cytochalasin D (CYT D), or NOC + CTY D treatments. Time (0–5 min) progresses from the top to bottom in the kymograph. Whereas, the x-axis represents mitochondrial position. Vertical white lines correspond to stationary mitochondria, and horizontal deviations (vibrations from the vertical) depict moving mitochondria. C: The percentage of time that mitochondria spent in motion as a result of microtubule and actin disruptions was determined by monitoring the position of the organelle throughout the capturing time. Mitochondria were considered in motion if movement was greater than 0.01 μm in the 2 sec of capturing time. Movement of mitochondria was determined and averaged from n = 36–72 mitochondria from nine separate cells. Error bars represent mean ± SEM, *P < 0.05 vs. CON.
Knockdown of Microtubule Motor Proteins Attenuates Mitochondrial Dynamics
For mitochondria to bind to cytoskeletal elements, motor proteins are required. The transfection of siRNA probes against Kif5B and dynein in C2C12 myoblasts resulted in significant reductions in their protein levels by 59 and 40%, respectively (Figs. 3A and 3B). The reduction in Kif5B was specific, and did not alter levels of mitochondrial biogenesis markers (Fig. 3A) or mitochondrial morphology proteins Mfn2, Opa1, Fis1, or Drp1 (unpublished observations). Reductions in both Kif5B and dynein resulted in similar significant decreases in the average speed of mitochondrial movements, by 30–40%, as well as in the total path length travelled by the organelles (decreased by 28–37%, Fig. 3C and 3D).
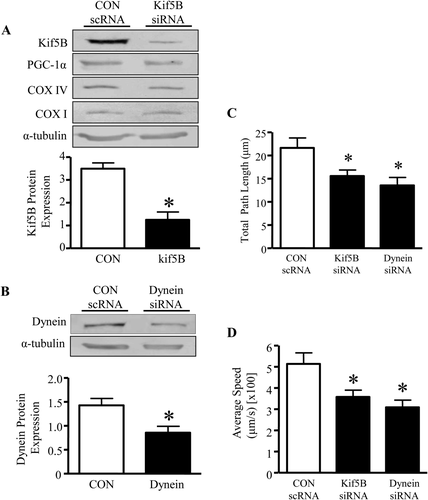
Knockdown of microtubule motor proteins attenuates mitochondrial dynamics. A: Representative Western blots of markers of mitochondrial content in myoblasts treated with either scrambled control (scRNA CON) or siRNA for Kif5B (above). Quantification of protein expression of Kif5B siRNA and scRNA control (below). B: Representative Western blots of dynein treated with an antisense probe and its respective scRNA control (above), along with graphical quantification (below). α-tubulin was unchanged between the conditions and was used as a loading control. Experiments were repeated for five separate trials. C: The total path length and D: the average speed traveled by mitochondria was assessed in scRNA CON and siRNA treatment for Kif5B and dynein. Movement of mitochondria was averaged from n = 20–40 mitochondria from 5 separate cells, *P < 0.05 vs. CON scRNA.
Cytosolic Calcium Regulates Mitochondrial Movement in Myoblasts
Calcium is an important signaling molecule in skeletal muscle, and as such we were interesting in examining its effects on mitochondrial motility. The Ca2+ ionophore, ionomycin, triggered an increase in Ca2+ levels that was suppressed by the addition of EGTA, as assessed by Fura2-AM ratiometric analysis (Figs. 4A and 4B). The rising phase of the cytosolic Ca2+ spike was accompanied by decreases in organelle motility in myoblasts (Figs. 4B and 4C). The total path length and average speed traveled by mitochondria were attenuated by 42 and 48%, respectively. The addition of EGTA permitted the recovery of mitochondrial movement, which coincided with reductions in the amount of cytosolic Ca2+, (Figs. 4A–4C). Similar effects were observed with the incubation of thapsigargin, a SERCA inhibiting agent. The addition of thapsigargin significantly reduced the total path length and average speed of mitochondria by 19 and 22%, respectively (Figs. 4E and 4F). Mitochondrial motility and Ca2+ levels returned to resting levels with the addition of the Ca2+ chelator BAPTA-AM (unpublished observations).
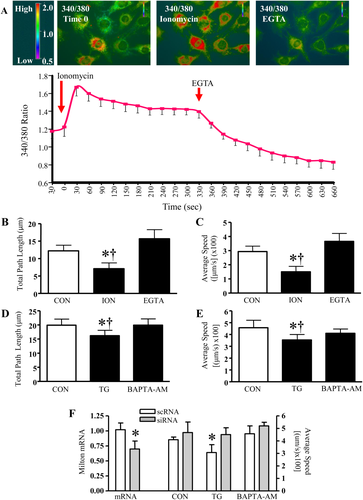
Cytosolic calcium regulates mitochondrial movement in myoblasts. A: Intracellular calcium levels were assessed in Fura2-AM loaded C2C12 cells. Fura2 fluorescence was measured using excitation of Ca2+-bound (340 nm, red) and the Ca2+-free form (380 nm, green). Thus, elevations evoked by the addition of Ionomycin (ION) and Thapsigargin (TG) appear as increases in the red component. Elevated calcium concentrations were achieved through the addition of ionomycin and reduced by EGTA. B, C: Increased calcium attenuated mitochondrial movement, and organelle transport was recovered by the addition of EGTA. Endogenous calcium levels were elevated with Thapsigargin treatment and suppressed with BAPTA-AM. D, E: High calcium concentrations inhibited the total path length and the average velocity traveled by the organelles. (n = 6–8, *P < 0.05 vs CON). F: Milton mRNA levels were assessed in myoblasts treated with 100 nM of scRNA or siRNA specific for Milton (left). The average speed traveled by mitochondria was determined in scRNA and siRNA transfected C2C12 cells, treated with Thapsigargin and BAPTA-AM (right).
The mechanisms by which Ca2+ exerts its effects are still under investigation. Milton is thought to be involved, since in neurons, Milton links mitochondria to Kif5B motor proteins, allowing mitochondria to move along microtubules in a Ca2+-dependent manner [Macaskill et al., 2009; Wang and Schwarz, 2009]. Thus, we reduced the level of Milton in myoblasts and observed its influence under conditions of elevated Ca2+ in the presence of thapsigargin (Fig. 4G). The decrease in mitochondrial speed of movement observed with thapsigargin treatment alone was negated in cells pre-treated with Milton siRNA, suggesting that Milton is involved in mediating mitochondrial arrest in the presence of Ca2+.
Discussion
In order to efficiently supply the cell with energy, mitochondria must be transported to areas of high energy demands. In neurons, mitochondria are delivered to areas where they required, and retained at sites where energy demand is high, such as growth cones, nodes of Ranvier and synapses [Fabricius et al., 1993; Morris and Hollenbeck 1993; Bindokas et al., 1998]. Within neurons, areas of high activity possess greater densities of mitochondria [King and Stocco 1996; Nguyen et al., 1997]. The movement of mitochondria to specific sites within the cell is dependent on a cytoskeleton-based transportation system. Moreover, environmental and physiological changes are thought to influence the positioning of organelles within the cells. Thus, our purpose was to determine which cytoskeletal components were primarily responsible for mitochondrial movement within myoblasts, and to investigate the effects of calcium on mitochondrial motility. To do so, we utilized live-cell imaging techniques to monitor the dynamics of these organelles.
To address the influence of the cytoskeleton on mitochondrial motility we assessed (1) the total path length, (2) average speed, and (3) percentage of time that organelles remained stationary. Our results indicate that microtubule disruption resulted in greater impediments in mitochondrial motility as compared to conditions in which actin polymerization was disturbed in myoblasts. Administration of microtubule depolymerizing or stabilizing agents resulted in similar effects on mitochondrial motility, yet they varied in the degree of their effects. The presence of taxol enhanced the stability of microtubules, rendering them more susceptible to undergoing post-translational modifications, such as acetylation, detyrosination, and polyglutamylation [Hammond et al., 2010]. These post-translational modifications may explain the disparity between the effects of taxol and nocodazole or colchicine treatments
Since kinesin and dynein motors drive the movement of mitochondria along microtubules, we speculated that the downregulation of these components would alter organelle transport. By reducing the levels of Kif5B, a member of the Kinesin-1 family, we observed decreased mitochondrial motility. It has previously been reported that the deletion of similar kinesin-1 family members in fungus and mouse neurons, led to altered mitochondrial distribution [Tanaka et al., 1998; Wu et al., 1998]. Analyses in the Drosophila nervous system support the concept that the primary motors for mitochondrial movement are Kinesin-1, in coordination with dynein, which moves mitochondria in the opposite direction [Pilling, 2006].
Calcium influx has been found to inhibit mitochondrial movement in neurons and cardiac myocytes using motor protein/adapter complexes [Yi et al., 2004; Wang and Schwarz 2009]. However, the influences of Ca2+ within skeletal muscle, as well as the mechanism by which Ca2+ exerts its effects are still under investigation. Our results indicate that elevations in Ca2+ exert a negative effect on mitochondrial movement, and cause an inhibition of motility. Linker proteins coordinate the movement of mitochondria with environmental signals, and are necessary for proper localization of the organelle. In neurons, HEK293 and COS-7 cells, Milton acts as a mitochondrial-specific linker that redistributes the organelle to appropriate regions within the cell, in response to increases in Ca2+ [Stowers et al., 2002; Smith et al., 2006; Wang and Schwarz, 2009]. Mutations in the protein result in mitochondrial trafficking defects, while other organelles retain their normal distribution [Stowers et al., 2002]. In this context, it is interesting that we observed Milton to also have a vital role in mitochondrial movement when Ca2+ levels were elevated in myoblast cells. Inhibition of Milton disrupted Ca2+-induced mitochondrial transport. Thus, Milton may mediate associations between Kinesin-1 and mitochondria in muscle cells.
We next wanted to examine the effect that chronic changes in motility might have on mitochondrial content. Argyropoulos et al. previously found that inhibition of the motor protein, Kif5B, reduced cytochrome b mRNA [Argyropoulos et al., 2009]. Therefore, we speculated that reductions in Kif5B would alter the expression of proteins involved in mitochondrial biogenesis. We observed no change when we examined the prominent regulator of mitochondrial biogenesis, PGC-1α, as well as the mitochondrially- and nuclear-encoded proteins COXI and COXIV, respectively. Therefore, our data are not consistent with a role for changes in motor protein expression on mitochondrial biogenesis.
In summary, mitochondria primarily move along microtubules tracks. In myoblasts, mitochondria can pause, change their direction, and their velocity of movement. We demonstrated that organelle displacement within myoblasts is regulated by Ca2+ signals, which inhibit mitochondrial movements in a Milton-dependent manner. By understanding mitochondrial movements and the signaling events that regulate their dynamics, we can elucidate the underlying basis for organelle interactions, leading to the formation of the mitochondrial reticulum and the distribution of energy in muscle cells.
Materials and Methods
Overview of the Experimental Strategy
To elucidate which cytoskeletal component is primarily responsible for mitochondrial movement within myoblasts, we initially began by using a pharmacological approach to disrupt actin and microtubules. Subsequently, we sought to establish which motor proteins were involved in mitochondrial dynamics by using siRNA technology. Finally, since calcium is an important regulator of mitochondrial function, we sought to determine the relationship between mitochondrial motility and calcium signaling in myoblasts.
Cell Culture and Transfections
C2C12 murine skeletal muscle cells were grown in DMEM supplemented with 10% fetal bovine serum and 1% Penicillin/Streptomycin (Growth Media, GM). Cells were cultured in custom made glass bottom culture plates and maintained at 37°C in 5% CO2. Upon reaching a confluence of approximately 60%, mito-DsRED2 was transfected into myoblasts using Lipofectamine 2000 (Life Technologies) to visualize mitochondria. Mitochondrial movement was observed in C2C12 cells using an expression vector that encodes a fusion of red fluorescence protein and the mitochondrial targeting sequence from cytochrome c oxidase subunit VIII (mito-dsRED2).
Treatments
Forty-eight hours following transfection with mito-DsRed2, myoblasts were treated with microtubule and/or actin disrupting drugs, or a vehicle control (DMSO; Sigma-Aldrich). Microtubules were depolymerized using nocodazole (20μM; Toronto Research Chemicals) and colchicine (20 μM; Sigma-Aldrich), while Taxol (2 μM; Sigma-Aldrich) was used for stabilization. Actin was disrupted using cytochalasin D (2 μM; Enzo Life Sciences). Pre-incubation with these agents was 1 hr prior to imaging.
In order to study the regulation of mitochondrial dynamics by calcium, C2C12 cells were treated with either ionomycin (1 μM, Sigma-Aldrich), a Ca2+ ionophore, or thapsigargin (1 μM, Enzo Life Science), an inhibitor of the sarcoplasmic reticulum Ca2+-ATPase. Ca2+ levels were suppressed using the chelators EGTA (5 mM, Sigma-Aldrich) and BAPTA-AM (2 μM, Calbiochem).
Mitochondrial Movement Analysis
Mitochondria were visualized using an inverted Nikon Eclipse TE2000-U fluorescent microscope equipped with 100×/1.5 oil objective lens, with a custom designed chamber designed to maintain a constant temperature of 37°C with 5% CO2. Mitochondrial dynamics were captured at 2 sec intervals for a total time of 5 mins using real-time imaging. Analyses were performed using the NIS Element AR 3.1 software, and was limited to regions of interest in the periphery of the cells, where individual mitochondria were readily resolved and could be used for analysis, while avoiding the confounding influence of high density mitochondrial networks.
Immunocytochemistry of Actin and Microtubules
For immunocytochemistry, the transfected cells were washed 3× with PBS at room temperature. The cells were then fixed with 3.7% formaldehyde for 7 min and washed with 3× with PBS. Cells were permeabilized with 0.1% Triton X-100 at 4°C for 15 min and washed 3× with PBS. Subsequently, cells were stained with a FITC conjugated α-tubulin primary antibody (Abcam), or Acti-stain 488 phalloidin (Cytoskeleton Inc) for 2 hrs or 30 mins, respectively. The cells were then washed 3× with PBS and incubated with DAPI (Sigma-Aldrich) for 5 mins to stain the DNA. After washing twice with PBS, the media was replaced with GM without phenol red.
RNA Interference
Silencer Select predesigned small interfering RNA (siRNA) for Kif5B (5′-GACAUGUCGCAGUUACAATT-3′), dynein (Dynch1; 5′-GAAAGAUCCGCAACACGAATT-3′), Milton (TRAK1; 5′-GCAACGUGGUCCUCGAUAATT-3′) and scrambled controls (scRNA) were used in the knockdown experiments (Ambion). To produce knockdowns, the media was switched to pre-transfection media (DMEM and 10% FBS). The following day, the media was changed to DMEM and myoblasts were incubated with mito-DsRED2, and either 50 nM of siRNA specific for Kif5B or dynein, or 100 nM of siRNA for TRAK1, along with 10 μl of Lipofectamine 2000 for 6 hrs. Concomitantly, scrambled non-targeting siRNA underwent the same conditions. Forty-eight hours post-transfection, cells were used for image analysis or were collected for protein extracts.
Immunoblotting
Protein extracts from myoblasts were separated on 6–15% SDS-PAGE and subsequently transferred onto nitrocellulose membranes. Membranes were then blocked (1 hr or 1.5 hr) with a 5% skim milk in 1× TBS-Tween20 solution (Tris-buffered saline–Tween-20: 25 mM Tris–HCl, pH 7.5, 1 mM NaCl and 0.1% Tween-20) at room temperature, followed by incubation in blocking solution with antibodies directed towards Kif5B (1:500; Santa Cruz), Dynein heavy chain (1:1000; Santa Cruz), PGC-1α (1:500; Millipore), COX IV (1:250; Abcam), COX I (1:500; MitoSciences), or α-tubulin (1:2000; Calbiochem) overnight at 4°C. Subsequently, membranes were washed 3× 5 min with TBS-Tween20, followed by incubation at room temperature (1 hr) with the appropriate secondary antibody conjugated to horseradish peroxidase, and washed again 3× for 5 min each with TBS-Tween20. Membranes were developed using Western Blot Luminol Reagent (Santa Cruz). Films were subsequently scanned and quantified via densitometric analysis for the intensity of signals with SigmaScanPro software (version 5, Jandel Scientific).
Real-Time PCR
Total RNA was extracted from C2C12 cells 48 hrs after transfection, as previously described (Ostojic et al., 2013). The concentration and purity of the RNA was assessed using absorbance readings at 260 and 280 nm (Ultrospec 2100 Pro). In accordance with the manufacturer's recommendations, SuperScript® III reverse transcriptase (Invitrogen) was used to reverse transcribe the RNA into cDNA. Using sequences from GenBank, primers were designed with Primer 3 v. 0.4.0 software (Massachusetts Institute of Technology, Cambridge, MA) for Milton as the gene of interest, with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β2-microglobulin (β2M) serving as the endogenous controls. The primers used were as follows: Milton forward: 5′-GGGAACGAGGACCACAATAA-3′, Milton reverse: 5′-CCTCCGCTCAGACAGGTAGT-3′, GAPDH forward: 5′-AACACTGAGCATCTCCCTCA-3′, GAPDH reverse: 5′-GTGGGTGCAGCGAACTTTAT-3′, β2M forward: 5′- GGTCTTTCTGGTGCTTGTCT-3’, β2M reverse: 5′- TATGTTCGGCTTCCCATTCT-3’. Quantification of the gene of interest was corrected using the endogenous controls and analyzed as previously described (Ostojic et al., 2013).
Oxygen Consumption
Oxygen consumption was determined based on fluorescence quenching by oxygen using OxoPlates (Innovative Instruments). Cells were added to each well at a density of 8000 cells/well. The following day, cells were pre-incubated (1 hr) with the microtubules and/or actin disrupting agents. Optical oxygen sensors at the bottom of microplates contained an indicator and a reference dye. Wells containing air-saturated (100%) and oxygen-free (0%) solutions were used for calibration. Fluorescence was assessed for 3.5 hrs in 3 min intervals in dual kinetic mode (Synergy HT microplate reader), using filter 1 (530/645 nm) and filter 2 (530/590) settings in order to detect fluorescence of the indicator and reference dyes, respectively. From these values, ratios were calculated and compared to the standard calibrations, thus allowing quantification of oxygen consumption.
Calcium Imaging
Cells were washed with GM without phenol red and incubated in the dark for 40 mins in a solution containing the calcium-sensitive dye fura-2-acetoxymethyl ester (Fura-2 AM, 5 µM) and 0.02% pluronic F-127. Fura-2 AM-loaded cells were washed twice and the media was replaced with GM without phenol red for an additional 20 mins to allow the de-esterification of fura-2 AM to occur. Prior to imaging, the media was replaced. Ratiometric imaging at absorption wavelengths of 340 and 380 nm (emission 510 nm) was taken at 30 sec intervals. Changes in the ratio of fluorescence intensity obtained at 340 nm (fura-2-Ca2+ bound) to that of 380 nm (Ca2+ free fura-2) were used to assess Ca2+ levels following the addition of either ionomycin or thapsigargin to the media, to stimulate the release of calcium, and following the chelation of calcium by EGTA or BAPTA-AM.
Statistical Analyses
Data were analyzed with GraphPad 4.0 software and expressed as means ± SE. T-tests and one-way ANOVAs were performed where appropriate, and differences were considered statistically significant if P < 0.05.