Grayscale image statistics of COVID-19 patient CT scans characterize lung condition with machine and deep learning
Edited by Yi Cui
Abstract
Background
Grayscale image attributes of computed tomography (CT) of pulmonary scans contain valuable information relating to patients with respiratory ailments. These attributes are used to evaluate the severity of lung conditions of patients confirmed to be with and without COVID-19.
Method
Five hundred thirteen CT images relating to 57 patients (49 with COVID-19; 8 free of COVID-19) were collected at Namazi Medical Centre (Shiraz, Iran) in 2020 and 2021. Five visual scores (VS: 0, 1, 2, 3, or 4) are clinically assigned to these images with the score increasing with the severity of COVID-19-related lung conditions. Eleven deep learning and machine learning techniques (DL/ML) are used to distinguish the VS class based on 12 grayscale image attributes.
Results
The convolutional neural network achieves 96.49% VS accuracy (18 errors from 513 images) successfully distinguishing VS Classes 0 and 1, outperforming clinicians’ visual inspections. An algorithmic score (AS), involving just five grayscale image attributes, is developed independently of clinicians’ assessments (99.81% AS accuracy; 1 error from 513 images).
Conclusion
Grayscale CT image attributes can be successfully used to distinguish the severity of COVID-19 lung damage. The AS technique developed provides a suitable basis for an automated system using ML/DL methods and 12 image attributes.
Highlights
-
Grayscale image statistics of CT scans can effectively classify lung abnormalities
-
Graphical trends of grayscale statistics distinguish visual assessments COVID-19 classes
-
Machine/deep learning algorithms predict severity from image grayscale attributes
-
Algorithmic class systems can be established using just five grayscale attributes
-
Confusion matrices provide detailed insight to algorithm prediction capabilities
1 INTRODUCTION
The COVID-19 virus continues to have devastating impacts worldwide involving millions of lives lost, repeated nationwide lockdowns, and substantial damage to the global economy. Methods that can quickly and accurately classify the severity of patient lung damage caused by COVID-19 offer the potential to assist in establishing and rapidly implementing appropriate treatment plans. Most pulmonary examinations now include computed tomography (CT) scanning as they can be executed rapidly and painlessly. Analysis of CT is crucial in establishing the degree to which lung functionality is impacted by various diseases, including cancers, various forms of pneumonia and, since early 2020, COVID-19.1, 2 Radiological image analysis, both X-ray and CT, help to diagnose COVID-19 and provide more details of patient conditions combined with virus' nucleic acid by real-time reverse transcription-polymerase chain reaction (rRT-PCR) tests.3 Moreover, CT scan analysis can achieve up to 98% sensitivity in diagnosing COVID-19.4 Lung CT-scan-slice analysis also provide useful insight by identifying and/or confirming the nature of pulmonary impacts caused by COVID-19.5 However, it takes time for radiological/medical assessments of CT scans to be conducted, making automated classification techniques desirable.6
Thoracic CT images taken from COVID-19 patients are typically used to support the diagnosis of the condition, establishing prognoses and treatments for patients.7 They are also used to monitor symptom progression and patient responses during treatment.8 The CT images tend not to be routinely used to quantify the severity of lung impacts caused by COVID-19, although some studies have focused on that objective.9 Attempts to achieve this aided by deep- and/or machine-learning (DL/ML) have to overcome a number of challenges.10, 11
Pulmonary-CT-scan images display a range of distinguishing features in some COVID-19 sufferers, that vary according to the conditions severity. Granularly opaque image areas, chaotic arrangements of lineations, agglomerated masses, opaque alveoli, inverse halo and/or atoll features, thick-polygonal forms with variable and indistinct linear opacities are some of the common features observed.12 Lung features observed in radiological images are likely to be affected by some pre-existing abnormalities in some patients acting to confuse COVID-19 CT-scan interpretations.13 Nevertheless, linking characteristic features observed in CT scan images has the potential to be exploited for systematically classifying the severity of lung conditions in COVID-19 sufferers.14 In particular, DL/ML techniques can be exploited to identify the common lung features associated with COVID-19 with some degree of accuracy.15-17 Some DL methods have applied convolutional neural networks (CNN) combined with automated feature-extraction routines applied directly to the CT images to achieve this.18, 19 Such approaches require effective image-segmentation processing.16, 20, 21
Customizing effective and rapid CT-scan-image analysis offers the potential to accelerate COVID-19 patient diagnosis, and treatment plan implementation and improve patients’ long-term prognoses. CNN coupled with effective feature-extraction algorithms offers one route for such analysis.10 However, such approaches often ignore the underlying relationships between the various grayscale-image attributes associated with the distinctive CT image features typically observed in COVID-19 patients.
Many DL studies using X-ray and CT-scan images are focused on the early diagnosis of COVID-19.22, 23 This involves a binary assessment assigning a negative or positive result.24, 25 The novelty of this study is that it considers in detail the grayscale image attributes of lung CT images from patients with and without COVID-19. Moreover, it links the combinations of those grayscale statistical attributes to the severity of the pulmonary effects impacting those patients with COVID-19.
The research objective of this study was to automate the interpretation of thoracic CT-scan images using grayscale image statistical attributes to provide a reliable classification of the severity of lung conditions afflicting COVID-19 sufferers. It does this applying CNN and several ML algorithms and comparing their performance. The analysis initially focuses on predicting a clinicians’ visual assessments of the CT scan images. It does this in terms of a binary distinction between patients with and without COVID-19 and classifying the degree of severity of the lung impacts. The study additionally develops an effective algorithmic classification of the severity of lung impacts of COVID-19 based on CT-scan image grayscale statistics. It assesses how that novel classification system could be adapted to provide an effective and rapid automated expert system for classifying the severity of lung impacts from pulmonary CT images. DL and ML models are applied to predict that algorithmic classification. The study evaluates the prediction accuracy of a combination of the proposed algorithmic classification of lung conditions with DL/ML prediction models. It also compares the results of the visual and algorithmic classification analysis of the CT images.
2 METHOD
2.1 Overview of proposed CT-image analysis
Grayscale statistics extracted from CT pulmonary-parenchyma images can be exploited to provide valuable insight into the lung's alveolar tissue employed in respiratory processes, and this study focuses on that information. CT-image slices are rapidly analyzed to yield values for a range of grayscale statistical attributes. Interpretations considering combinations of these statistics with graphic representations identify distributions related to the severity of lung impacts related to COVID-19 that, to some extent, overlap with each other. However, DL/ML methods can be applied to combinations of these overlapping, grayscale-attribute distributions to accurately the severity of lung abnormalities of each CT-image slice assessed.
2.2 Ethical approval
The CT analyses of human patients used in this study were performed in accordance with the relevant guidelines and regulations of Namazi Medical Centre and the Medical School of Shiraz University of Medical Sciences (Shiraz, Iran). In particular, the consent for analysis was obtained from each patient and their anonymity was guaranteed. Consequently, the confidentiality of the biographical information pertaining to each patient has been maintained at all times. All CT analytical protocols applied were approved by the Medical School of Shiraz University of Medical Sciences.
2.3 CT-scan slice procurement
The suites of lung CT images were obtained from a representative set of patients. Most of them were shown by laboratory testing to be afflicted with COVID-19. However, images from several patients with negative COVID-19 tests were included in the suite of images compiled. These patients were all under treatment at the Namazi Medical Centre, Shiraz, Iran. The Philips Ingenuity CT-scanning equipment (Philips) located at that medical center provided suites of CT-scan-slice images (0.625 mm thickness) for all patients assessed. Such multislice-CT-scan equipment is now widely used to rapidly deliver high-resolution images.26
CT images considered are all noncontrasted, which are typical of those widely used to assess various lung conditions in addition to those caused by coronaviruses. The CT-scan-slice images utilized are derived from 49 COVID-positive patients displaying a substantial range of visible features known to be commonly associated with COVID-19. Additionally, multiple CT-scan-slice images associated with eight COVID-negative patients are also assessed for comparative purposes. The COVID-negative patients consisted of four women with ages ranging from 18 to 68 years, together with four men with ages varying from 24 to 71 years. These images were compared with those from 23 female COVID-positive patients with ages ranging from 22 to 74 years, together with 26, COVID-positive, male patients with ages ranging from 32 to 86 years. These patients’ images were selected to provide well-balanced distributions of patients’ genders and ages.
2.4 CT-scan-slice image assessment
2.4.1 Classifications involving visual assessment by clinicians
The suites of CT-slice images from each patient were assessed by a clinician (visual inspection). That assessment resulted in each image being assigned a number from 0 to 4 (five classes in total) to distinguish the degree of severity of visible, abnormal lung features they exhibited. Those five classes are defined as follows: Class 0, COVID-19-negative test lacking visual traces of abnormal lung features; Classes 1–4, apply to all patients with COVID-19-positive tests; Class 1, none, or trace, indications visible of abnormal lung features; Class 2, minimal but visible indications of abnormal lung features; Class 3, sporadic but pervasive visible signs of abnormal lung features; Class 4, extensive visible signs of severe abnormal lung features.
This visual-scoring (VS) classification scheme is considered as the initial objective function for predictive DL/ML algorithms assessed using grayscale-image attribute statistics.
Example CT-scan-slice images with associated rectilinear extract images for VS Classes 0 and 1 (minimal or no visual sign of lung abnormalities) are displayed in Supporting Information: Figure S1. Rectangular-extract segments from the images displayed in Supporting Information: Figure S1 are representative of those used for grayscale—statistical analysis are shown in Supporting Information: Figure S2.
2.4.2 Rectangular-extract-image segment analysis
A total of 392 rectilinear image extracts were compiled from the 49 COVID-19-positive individuals (approximately eight extracts/person). Additionally, 121 rectilinear image extracts were compiled from the eight COVID-19-negative individuals (approximately 15 extracts/person).
The dimensions of the 513 rectilinear images range from 2000 to 80,000 pixels, with average dimensions of approximately 25,000 pixels. This range occurs because the CT-image extracts are positioned to sample just the parenchyma portion of each lung. The rectilinear extracts are also positioned to avoid the pleura, diaphragm, or mediastinum portions of the lung. Consequently, areal extent in each CT image extract varies.
Every pixel in a monochromatic CT image possesses one grayscale value varying from 0 (black) to 255 (white). Values on that scale from 1 to 254 represent progressively lighter shades of gray. The pixel values are expressed in an 8-bit-integer digital format. This simple grayscale-image intensity scale means that the collective sets of pixels making up an image form grayscale distributions. Those distributions can be assessed with statistical confidence as thousands of pixels are involved in each image extract. OpenCV functions coded in Python are employed in this study to conduct a comprehensive grayscale statistical analysis of each image extract.27 These are (1) the number of pixels present in an image; (2) average pixel grayscale value (a number between 0 and 255 on the grayscale); (3) the number of pixels in an image possessing its average pixel grayscale value; (4) percent of pixels in an image possessing its average pixel grayscale value; (5) variance of all pixel grayscale values in an image (a number between 0 and 255 on the grayscale); (6) ratio of pixel variance to pixel average pixel in an image; (7) standard deviation of all pixel grayscale values in an image (a number between 0 and 255 on the grayscale); (8) standard error of the mean of all pixel grayscale values in an image; (9) minimum grayscale value of all pixels in an image; (10) tenth percentile (P10) of the grayscale values of all pixels in an image; (11) fiftieth percentile (P50) of the grayscale values of all pixels in an image; (12) ninetieth percentile (P90) of the grayscale values of all pixels in an image; (13) maximum grayscale value of all pixels in an image.
Clearly, the number of pixels in an image extract that possesses the average pixel grayscale value (attribute#3), and the standard error of the mean (attribute#8), are statistics that are dependent on the image extract size. Attribute#8 is calculated as the grayscale standard deviation divided by the square root of the number of pixels in the image extract. Apart from the two mentioned, all other grayscale attribute statistics listed are not dependent of image size.
The standard error of the mean values are an indicator of the degree of uncertainty associated with the average (mean) grayscale value of each image. The standard error of the mean values is all <0.7 for the images analyzed. Compared to the grayscale range of 0–255, a standard error of the mean of <0.7 confirms that there is a very limited uncertainty range associated with the average (mean) grayscale value of all the image extracts.
2.4.3 Deep/machine learning (DL/ML) models configured to exploit grayscale statistics
Twelve of the grayscale attribute statistics determined for each image extract are used as input variables for the DL/ML analysis to predict the five VS Classes (0–4) as a dependent variable. The number of pixels in each image is not used as an input variable as it is dependent on image size. Each DL/ML classifier is configured and executed in Python. The objective function minimized by the DL/ML algorithms is the mean squared error (MSE) for between the actual (clinician determined) VS class (VSact) and the predicted VS class (VSpred) taking into account the full suite of 500 and 13 image extracts.
The 10 ML methods and 1 DL method employed are described in Table 1.
| Algorithm name | Abbreviation | Description |
|---|---|---|
| Adaptive Boosting “Adaboost” | ADA | Boosted decision tree28, 29 |
| Convolutional Neural Network | CNN | DL neural network18, 19 |
| Decision Tree | DT | A single tree configuration30-32 |
| Extreme Learning Machine | ELM | Single-layer feed-forward neural network33-35 |
| Gaussian Process Classification | GPC | Gaussian probability applying Laplace approximation36, 37 |
| K-nearest Neighbor | KNN | Data matching algorithm15, 38 |
| Multilayer perceptron | MLP | Classic neural network39 |
| Naïve Bayes Classifier | NBC | Simple Bayes’ theorem classifier15, 40 |
| Random Forest | RF | Tree-based ensemble method using bagging41, 42 |
| Quadratic Discriminant Analysis | QDA | Nonlinear classifier43 |
| Support Vector Classifier | SVC | Non-probabilistic classifier44, 45 |
These algorithms are well known and extensively applied for many industrial and academic purposes, including image analysis and classification. Published examples of their specific applications to classify image datasets are cited in Table 1. Distinct mathematical, statistical, and logical concepts are associated with these algorithms. Most of these algorithms derive and build their predictions upon hidden regression relationships, driven by correlations between the input variables and dependent variables. The K-nearest Neighbor (KNN) algorithm is an exception in that it uses data-record matching to establish its predictions rather than correlations.
Each of the DL/ML algorithms requires its hyperparameters to be optimized. These algorithm control values were established using trial and error and grid search techniques (SciKit-Learn).46 The configurations of each algorithms, expressed in terms of their optimized hyperparameters, are listed in Supporting Information: Table S1.
Trial-and-error and K-fold cross-validation analysis was used to determine the optimum allocation of data records between the training subset and testing subset evaluated by each of the DL/ML methods. That analysis indicated that a random 80% allocation of data records to the training subset with the remaining 20% of data records assigned to the testing subset worked effectively with the CT-scan-extract image data set.
2.4.4 Statistical metrics used to assess the classification accuracy of DL/ML models
The following statistical measures of classification accuracy are used in this study to determine the VS classification performance achieved by each DL/ML method applied.
Mean square error (MSE)
Root mean squared error (RMSE)
Percent deviation (PDi)
Average percent deviation (APD)
Absolute average percent deviation (AAPD)
Standard deviation (SD)
Correlation coefficient (R)
R between variables Xi and Yi is expressed on a scale between −1 and +1.
Coefficient of determination = R2, which is expressed on scale between 0 and 1.
2.4.5 Workflow of the methodology applied to assess CT images
Figure 1 summarizes the workflow applied in this study to assess and classify thoracic CT images in terms of their COVID-19 impacts.
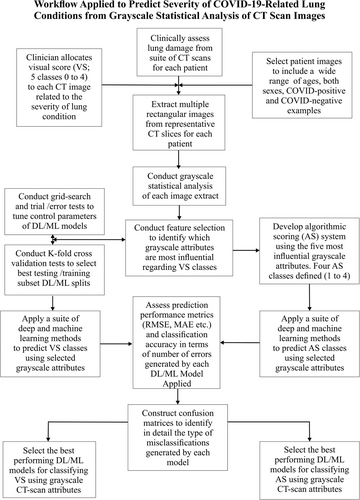
3 RESULTS
3.1 Grayscale image statistics
The grayscale statistical-attribute distributions of the CT-extract images evaluated are described in Supporting Information: Table S2. Figure 2 displays and compares box and whisker plots for selected CT-extract-image grayscale attribute distributions relating to the 121 COVID-19-negative individuals and the 392 COVID-19-positive individuals.
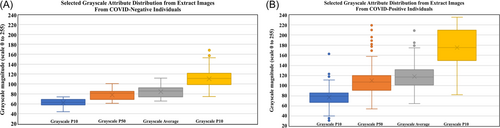
Pixel P50, pixel average, and pixel P90 grayscale statistics display some the most obvious distinctions between distributions for COVID-19-negative and COVID-19-positive individuals (Figure 2). It is these more obvious differences, plus the more subtle differences associated with other grayscale attribute statistics that the DL/ML models use in their VS class predictions.
Some of the grayscale statistics relating to the CT-extract images also display strong correlations with their VS classification (Table 2). The Pearson's correlation coefficient (R) values are highly positive for VS versus grayscale, P50, P90, average, variance, and standard deviation. R values for VS versus P10, standard error, and variance/average ratio are moderately positive. On the other hand, the R value for VS versus the percentage of pixels at the average grayscale value is highly negative. The dispersion of the grayscale-attribute distributions, and their correlations with VS, are indicative of systematic variations occurring in these attributes that are spread across the VS classes throughout the CT-extract-image data set.
| Image grayscale statistical attributes | Attribute identifier | Pearson correlation coefficient (R) with VS | Spearman correlation coefficient (p) with VS |
|---|---|---|---|
| Number of Pixels in Extract | 1 | −0.0882 | −0.1191 |
| Grayscale Average | 2 | 0.7834 | 0.8059 |
| Number of Pixels at Grayscale Average | 3 | −0.5140 | −0.5552 |
| Percentage of Pixels at Grayscale Average | 4 | −0.7456 | −0.7978 |
| Grayscale Variance | 5 | 0.6749 | 0.7184 |
| Grayscale Variance/Average | 6 | 0.3740 | 0.4117 |
| Grayscale Standard Deviation | 7 | 0.6911 | 0.7184 |
| Grayscale Standard Error | 8 | 0.4148 | 0.4539 |
| Grayscale Minimum | 9 | 0.1837 | 0.2373 |
| Grayscale P10 | 10 | 0.5511 | 0.5640 |
| Grayscale P50 | 11 | 0.7242 | 0.7534 |
| Grayscale P90 | 12 | 0.8667 | 0.8776 |
| Grayscale Maximum | 13 | 0.3162 | 0.2236 |
- Note: The Spearman correlation coefficient uses the same formula as the Pearson correlation coefficient (Equation 9) but uses rank positions in the distributions rather than absolute values.
- Abbreviations: CT, computed tomography; VS, visual scores.
Spearman's rank correlation coefficient (p) is also calculated for VS versus the grayscale statistical attributes and displayed in Table 2. A feature of the R statistic is that an assumption is made that the two-variable distributions compared both conform to normal distributions, that is, they display parametric behavior. The nonparametric p statistic avoids that assumption as it is calculated based on the ranked position of each data record in the respective distributions instead of their absolute values. For VS versus the grayscale attributes the p and R values are in relatively close agreement (Supporting Information: Figure S3). This implies that the grayscale attribute distributions are relatively symmetrical and not highly skewed.
3.2 CT-extract image grayscale attribute dispersions across VS classes
Figure 3 displays a ternary plot for three grayscale attributes: average; variance; and, pixel% at the average value. The scaling factors applied to the grayscale attributes serve to centralize the spread of data points on this ternary plot. In such plots, the actual values of the three variables are normalized to sum to a constant value (the value of 100 is used for Figure 3).
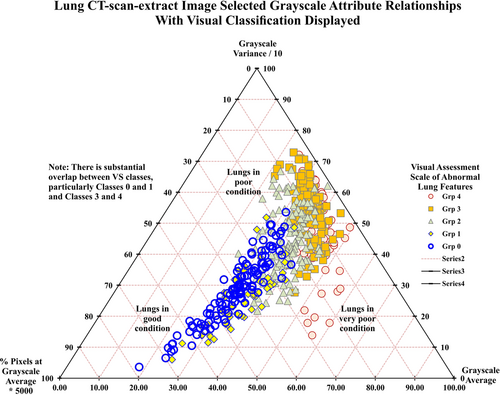
These normalized values cannot then vary in the diagram independently of each other. Thus, if the normalized values of two of the variables plotted are known, the value of the third can be determined. This configuration creates only two degrees of freedom in the normalized system, allowing the three variables to be displayed in the two-dimensional ternary plots (Figure 3).
The three grayscale attributes (average; variance; pixel% at the average) perform relatively good discrimination between the VS classes (Figure 3). There is an almost continuous progression from VS = 0 (lower-left in ternary plot) to VS = 4 (middle-right in ternary plot). VS Classes 0 and 3 appear in distinct regions on the ternary plot. Nevertheless, overlaps are apparent among VS Classes 1, 2, and 4 and between those classes and VS Classes 0 and 3. Lungs with little or no evidence of abnormal features tend to be associated with relatively low average and variance grayscale values. As abnormal features materialize in the lung-image extracts, grayscale average and variance values progressively increase. At the stage where abnormal features become extensive in the lung-image extracts, the percentage of image pixels at the average grayscale for the image tends to decrease. This tendency forms a distinctive characteristic in the VS Class 4 images.
The images displaying the severest abnormal lung features plot towards the lower-right portion of the ternary plot (Figure 3). Such images show a prevalence of lighter-gray pixels. This causes the grayscale variance value to decline and the grayscale average value to increase. From VS = 0 to VS = 3 the image positions tend to move from south-west to north-east position in Figure 3. Whereas, from VS = 3 to VS = 4 the image positions tend to move in a southerly direction in Figure 3.
Three-variable plots (Figures S4 and S5) reveal other progressive and distinctive trends that exist involving grayscale percentage statistics. The involvement of grayscale P90 in Supporting Information: Figure S4 displays a continuous trend from the base-right portion (VS = 0) to top-left left portion (VS = 3 or 4) with substantial overlap between VS Classes 3 and 4. By involving both grayscale P10 and P90 with grayscale average (Figure S5) the overlap between classes VS = 3 and VS = 4 is somewhat reduced.
The distributions of grayscale statistics from CT-extract images (Supporting Information: Table S2, Figures S4 and S5, and Figure 3) include substantial dispersion and inter-relationships. These can be exploited graphically (and by DL/ML methods) to classify the degree of abnormalities in lungs affected by COVID-19. The three-statistical-attribute combinations displayed in Figures 3, Supporting Information: S4 and S5 discriminate quite well between some of the VS classes. However, overlapping positions, particularly between VS Classes 0 and 1 and VS Classes 3 and 4 make a graphical approach unsuited to definitive VS classification. Involving DL/ML methods to consider multiple grayscale attributes as input variables offers the potential to improve upon the graphical approach.
3.3 Feature selection for DL/ML analysis
Thirteen grayscale-attribute statistics were extracted and computed for each of the (513) CT-extract images (Supporting Information: Table S2). All of these attributes could be employed as independent (input) variables with VS class as the single dependent variable for DL/ML model evaluation. However, as some of these attributes show relatively low correlations with VS class (Table 2 and Supporting Information: Figure S3) trial and error sensitivity analysis was conducted to determine their impact on the DL/ML prediction performance. The following sensitivity tests were conducted:
Test #1: 9 input variables (pixel#, standard error, minimum, variance/average ratio excluded)
Test #2: 10 input variables (pixel#, standard error, variance/average ratio excluded)
Test #3: 11 input variables (pixel# and variance/average ratio excluded)
Test #4: 12 input variables (pixel# excluded)
Test #5: 13 input variables
The results showed that Test #4 with 12-input variables included achieved the highest VS prediction accuracy. This suggests that all 12 of those attributes can provide useful input to the DL/ML models. Those 12 input features were, therefore, selected for detailed DL/ML model analysis.
3.4 K-fold cross-validation analysis
K-fold cross-validation is applied to evaluate, select and justify the percentage splits of the data records between training and testing subsets to be applied in ML/DL analysis. Four specific folds or cases are evaluated for five algorithms. Those cases are: fourfold (75% training: 25% testing), fivefold (80% training: 20% testing), 10-fold (90% training: 10% testing), and 15-fold (93% training: 7% testing). Three runs are executed for each k-fold analysis case, thereby generating meaningful statistics for each case. Twelve distinct splits are evaluated for fourfold case (3 times 4). The number of splits evaluated increases to 45 for the 15-fold analysis (3 times 15). The mean MAE and the standard deviation MAE are recorded for all the splits evaluated for each fold (Table 3).
| K-fold cross-validation results | Visual scoring | Algorithmic scoring | ||||||
|---|---|---|---|---|---|---|---|---|
| Model | K-fold | Training: testing percentage splits | Splits evaluated in each run | Distinct cases for three repeated runs | MAE mean | MAE standard deviation | MAE mean | MAE standard deviation |
| ADA | 4-fold | 75:25 | 4 | 12 | 0.3841 | 0.0597 | 0.0286 | 0.0151 |
| 5-fold | 80:20 | 5 | 15 | 0.3248 | 0.0709 | 0.0256 | 0.0281 | |
| 10-fold | 90:10 | 10 | 30 | 0.3588 | 0.0845 | 0.0287 | 0.0230 | |
| 15-fold | 93:7 | 15 | 45 | 0.3607 | 0.0946 | 0.0338 | 0.0339 | |
| DT | 4-fold | 75:25 | 4 | 12 | 0.3613 | 0.0459 | 0.0176 | 0.0132 |
| 5-fold | 80:20 | 5 | 15 | 0.3346 | 0.0475 | 0.0135 | 0.0172 | |
| 10-fold | 90:10 | 10 | 30 | 0.3527 | 0.0898 | 0.0150 | 0.0192 | |
| 15-fold | 93:7 | 15 | 45 | 0.3470 | 0.0872 | 0.0150 | 0.0267 | |
| KNN | 4-fold | 75:25 | 4 | 12 | 0.3191 | 0.0372 | 0.0721 | 0.0197 |
| 5-fold | 80:20 | 5 | 15 | 0.2985 | 0.0512 | 0.0608 | 0.0216 | |
| 10-fold | 90:10 | 10 | 30 | 0.3112 | 0.0819 | 0.0663 | 0.0235 | |
| 15-fold | 93:7 | 15 | 45 | 0.3193 | 0.0988 | 0.0677 | 0.0377 | |
| RF | 4-fold | 75:25 | 4 | 12 | 0.2743 | 0.0322 | 0.0240 | 0.0098 |
| 5-fold | 80:20 | 5 | 15 | 0.2582 | 0.0479 | 0.0222 | 0.0173 | |
| 10-fold | 90:10 | 10 | 30 | 0.2652 | 0.0846 | 0.0231 | 0.0200 | |
| 15-fold | 93:7 | 15 | 45 | 0.2614 | 0.0893 | 0.0234 | 0.0267 | |
| SVM | 4-fold | 75:25 | 4 | 12 | 0.5563 | 0.0954 | 0.1170 | 0.0257 |
| 5-fold | 80:20 | 5 | 15 | 0.4532 | 0.0709 | 0.1063 | 0.0276 | |
| 10-fold | 90:10 | 10 | 30 | 0.5392 | 0.1038 | 0.1092 | 0.0482 | |
| 15-fold | 93:7 | 15 | 45 | 0.5361 | 0.1153 | 0.1040 | 0.0535 | |
- Note: Model abbreviations are listed in Table 1.
- Abbreviations: ADA, Adaptive Boosting “Adaboost”; AS, algorithmic scale; DT,Decision Tree; KNN, K-nearest Neighbor; MAE, mean absolute error; RF, Random Forest; SVM, support vector machine.
All four K-folds evaluated generate credible prediction results for the VS and algorithmic scale (AS) classifications (low MAE means and low MAE standard deviations). However, the fivefold (80%:20%) split is selected as this generates the lowest mean MAE and a lower standard deviation than the 10- and 15-fold analysis. Also, the 80%:20% split involves a substantial number (103) of randomly selected images in each testing subset.
3.5 DL/ML model VS class prediction performance
Table 4 shows the VS class prediction accuracy (VSpred vs. VSact) for the 11 DL/ML models developed. The results refer to the trained and validated models applied to the entire data set. The number and percentages of misclassified data records is also listed in Table 4. The algorithms are ranked, with the best performing shown as rank 1, and the worst-performing shown as rank 11. The rankings are made based on RMSE and classification error numbers (Table 4). The CNN model is ranked 1 in the VS classification task incurring only 18 misclassifications from the 513 images (96.49% accuracy). Moreover, the CNN model achieved RMSE = 0.19 (with reference to the VS scale of 0–4) and R2 = 0.98. The RF model was ranked second in its VS classifications (26 misclassifications; RMSE = 0.26; R2 = 0.96; 94.93% accuracy). Also, the ADA, DT, ELM, and KNN models performed well in classifying VS. On the other hand, the MLP, NBC, and QDA did not perform well in classifying VS. The percentage accuracies achieved for this multiclass VS scale prediction analysis compare favorably with the percentage accuracies achieved by recent deep learning studies published for the binary (COVID negative vs. COVID positive) predictions from CT and X-ray scans. For instance, Arora et al.6 analyzed 812 CT images and achieved a binary prediction accuracy of 98%; Polsinelli et al.24 analyzed 100 images using CNN and achieved a binary prediction accuracy of 85%; Dansana et al.23 analyzed 360 CT images using CNN and achieved a binary prediction accuracy of 91%; and Bharati et al.22 analyzed 3411 CT images using DL models and achieved a binary prediction accuracy of 82.42%. On the other hand, Bharati et al.25 analyzed 5606 X-ray images and achieved a binary prediction accuracy of 73%.
| Prediction accuracy visual score (VS: 0–4) using 12 grayscale input variables | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ML/DL Model | RMSE (VS) | MAE (VS) | APD (%) | AAPD (%) | Sdev (VS) | R2 | Number of errors | % correct | Rank by RMSE | Rank by errors |
| ADA | 0.3175 | 0.0698 | −0.6686 | 3.0459 | 0.3186 | 0.9445 | 28 | 94.54 | 5 | 4 |
| CNN | 0.1868 | 0.0349 | 0.3165 | 1.2209 | 0.1869 | 0.9805 | 18 | 96.49 | 1 | 1 |
| DT | 0.3294 | 0.0775 | −0.3424 | 2.9199 | 0.3305 | 0.9404 | 32 | 93.76 | 6 | 5 |
| ELM | 0.3144 | 0.0872 | −1.3372 | 3.7984 | 0.3156 | 0.9453 | 42 | 91.81 | 4 | 6 |
| GPC | 0.4176 | 0.1589 | 0.2778 | 4.8643 | 0.4175 | 0.9041 | 78 | 84.80 | 7 | 8 |
| KNN | 0.2887 | 0.0640 | 0.3230 | 2.1641 | 0.2890 | 0.9542 | 28 | 94.35 | 3 | 4 |
| MLP | 0.9139 | 0.5291 | 12.9167 | 18.7629 | 0.8225 | 0.6949 | 196 | 61.79 | 11 | 11 |
| NBC | 0.7587 | 0.4205 | −9.3346 | 21.8992 | 0.7609 | 0.7024 | 177 | 65.50 | 10 | 10 |
| QDA | 0.5171 | 0.2054 | −5.1583 | 9.8934 | 0.5184 | 0.8524 | 90 | 82.46 | 9 | 9 |
| RF | 0.2604 | 0.0562 | −0.6944 | 2.3094 | 0.2614 | 0.9620 | 26 | 94.93 | 2 | 2 |
| SVM | 0.4313 | 0.1202 | 0.7526 | 4.6932 | 0.4285 | 0.9005 | 46 | 91.03 | 8 | 7 |
- Note: VS system involves classes 0 to 4 assigned by a clinician.
- Abbreviations: AAPD, absolute average percentage deviation; ADA, Adaptive Boosting “Adaboost”; APD, average percentage deviation; CNN, Convolutional Neural Network; DT, Decision Tree; ELM, Extreme Learning Machine; KNN, K-nearest Neighbor; MAE, mean absolute error; RMSE, root mean squared error; Sdev, standard deviation; SVM, support vector machine. Model abbreviations are listed in Table 1.
Table 4 reveals that certain ML models achieve different rankings based on RMSE compared to rankings based on error numbers. For example, the ELM model has an RMSE-rank of 4 but error rank of 6, whereas ADA has RMSE-rank of 5 but error rank of 4 (Table 4). This is a consequence of the nature of the misclassification errors involved. Misclassifications involving adjacent VS classes have a smaller impact on RMSE than misclassifications that are more than one class away from the correct class.
3.6 Confusion matrix assessment of VS classification
Classification errors made by the DL/ML models may increase, while for the same analysis, RMSE values might decrease (Table 4). Confusion matrices deliver a detailed assessment of multiclass misclassifications on a VS class by class basis. Confusion matrices reveal VS classes predicted with most and least accuracy. The three models considered (CNN; KNN; ADA; Figure S6) perform quite distinctly. CNN (Figure S6A) performs best for VS Class 0 (just 1 misclassification) and, in percentage error terms worst for VS Class 1, although it misclassifies the most data records (6 out of 129) associated with VS Class 3, CNN predicts VS Classes 0, 1, and 2 more convincingly than VS classes 3 and 4. CNN misclassifications only involve erroneous predictions involving adjacent VS classes. Confusion matrices for KNN (Figure S6B) and ADA (Figure S6C) reveal that both models result in 28 misclassifications associated with the 513 CT-extract images evaluated. Nevertheless, the distribution of these misclassifications differs considerably for those two models. KNN has five misclassifications that are beyond the adjacent VS classes, whereas ADA results in eight such extreme misclassifications. Consequently, KNN (RMSE = 0.2887) outperforms ADA (RMSE = 0.3175) overall in its VS class prediction. KNN and ADA struggle most to correctly classify VS Class 2 (11 misclassifications; about 40% of total misclassifications). KNN performs best in classifying VS Classes 0 and 1 (better than CNN). ADA does not deal as well with VS Class 0 as the CNN and KNN models but performs almost as well as the CNN model in its classification of VS Classes 3 and 4.
Analysis of confusion matrices provides a complementary perspective on the RMSE/error count analysis of each model in assessing their overall multiclass classification prediction strengths and weaknesses.
4 DISCUSSION
4.1 Formulaic/algorithmic scoring approach to classifying CT-extract images
The relatively good discriminatory performances among the VS classes of the graphical relationships (Figure 3, Supporting Information: Figures S4 and S5), involving just grayscale attributes, makes them exploitable for automated formulaic classification. Such an algorithmic/formulaic scoring system (AS) has the potential to provisionally classify the extent of abnormal lung features in CT-extract images before inspection by a clinician and/or DL/ML analysis. AS scores can complement the VS score assigned by a clinician (i.e., VS based on clinician's expert evaluation; AS based on a defined algorithm). Automating CT-extract-image assessments has the attraction of speed and objectivity. This can provide provisional guidance for a subsequent more detailed expert visual assessment.
An example AS approach involves just five grayscale attributes (P10, average, P90, variance and pixel% at the average value; attributes 10, 2, 12, 5, and 4 in Table 2 and Supporting Information: Figure S3, respectively) corresponding to those used in Figure 3, Supporting Information: Figure S4 and S5 for VS analysis.
- 1.
The AS formulas involve just four groups of rules: AS = 4 (extensive presence of abnormal lung features) is established for those images that satisfy three grayscale attribute levels: P10 grayscale ≥ 100; average grayscale ≥ 150; P90 grayscale ≥ 200.
- 2.
AS = 1 (few if any abnormal lung features) is then established (after AS = 4 images have been allocated and removed from the selection) for those remaining images that satisfy four grayscale attribute levels: P10 grayscale < 80; P90 grayscale <125; variance <1000; Pixel% at the average value > 1.5%.
- 3.
AS = 2 (minor abnormal lung features) is then established (after AS = 4 and AS = 1 images have been allocated and removed from the selection) for those remaining images that satisfy two grayscale attribute levels: Average grayscale < 125; P90 grayscale < 150.
- 4.
AS = 3 (substantial abnormal lung features) is then established (after AS = 4, AS = 1, and AS = 2 images have been allocated and removed from the selection) with all remaining CT-extract images not assigned to other groups.
Table 5 and Supporting Information: Figure S7 displays the Pearson and Spearman correlation coefficients between the grayscale attribute variables and the AS classes for all CT-extract images. As should be expected, the R and p values are slightly higher than those for correlations between the same variables and the VS classes (Table 2). This is particularly the case for those grayscale variables involved in the formulaic definition of the AS classes.
| Image grayscale statistical attributes | Attribute identifier | Pearson correlation coefficient (R) with algorithmic score (AS) | Spearman correlation coefficient (p) with AS |
|---|---|---|---|
| Number of Pixels in Extract | 1 | −0.0980 | −0.1344 |
| Grayscale Average | 2 | 0.8202 | 0.8531 |
| Number of Pixels at Grayscale Average | 3 | −0.5560 | −0.5682 |
| Percentage of Pixels at Grayscale Average | 4 | −0.7786 | −0.7876 |
| Grayscale Variance | 5 | 0.6559 | 0.7340 |
| Grayscale Variance/Average | 6 | 0.3661 | 0.4144 |
| Grayscale Standard Deviation | 7 | 0.6988 | 0.7340 |
| Grayscale Standard Error | 8 | 0.4364 | 0.4871 |
| Grayscale Minimum | 9 | 0.2656 | 0.3304 |
| Grayscale P10 | 10 | 0.6338 | 0.6516 |
| Grayscale P50 | 11 | 0.7459 | 0.7897 |
| Grayscale P90 | 12 | 0.9015 | 0.9174 |
| Grayscale Maximum | 13 | 0.3821 | 0.2892 |
The 513 CT-extract images assigned to AS classes are shown in Figure 4. An increased degree of segregation of the AS classes is discernible in Figure 4 compared to that achieved for the VS classes in Figure 3. This outcome confirms that the value of AS is a preliminary, automated, objective CT-extract image assessment. The five-attribute AS classification system can be further improved upon by applying DL/ML algorithms to consider all the available attributes.
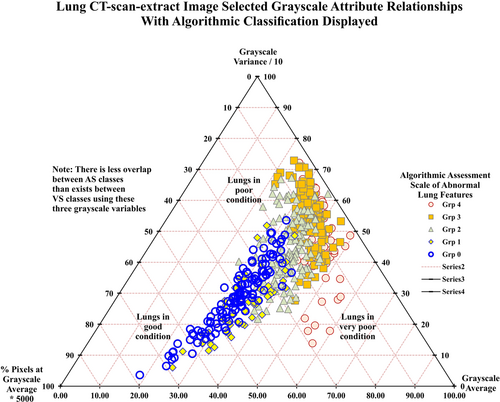
Table 6 summarizes AS classification prediction accuracy determined by DL/ML models using the same 12 input variables (grayscale attributes) recorded for each of 513 CT-extract images. The results confirm that the DL/ML methods find it easy (fewer misclassifications and much lower RMSE/MAE values) to classify the CT-extract images using the AS classes than the VS classes.
| Prediction accuracy algorithmic score (AS: 1–4) using 12 grayscale input variables | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ML/DL model | RMSE (VS) | MAE (VS) | APD (%) | AAPD (%) | Sdev (VS) | R2 | Number of errors | % correct | Rank by RMSE | Rank by errors |
| ADA | 0.0623 | 0.0039 | 0.1292 | 0.1292 | 0.0624 | 0.9955 | 2 | 99.61 | 3 | 3 |
| CNN | 0.1245 | 0.0155 | 0.1906 | 0.3844 | 0.1248 | 0.9821 | 8 | 98.44 | 6 | 6 |
| DT | 0.0440 | 0.0019 | 0.0646 | 0.0646 | 0.0442 | 0.9978 | 1 | 99.81 | 2 | 2 |
| ELM | 0.1460 | 0.0213 | 0.0258 | 0.6072 | 0.1465 | 0.9753 | 11 | 97.86 | 7 | 7 |
| GPC | 0.0984 | 0.0097 | 0.1421 | 0.2390 | 0.0986 | 0.9888 | 5 | 99.03 | 4 | 4 |
| KNN | 0.1165 | 0.0136 | 0.1098 | 0.3682 | 0.1168 | 0.9843 | 7 | 98.64 | 5 | 5 |
| MLP | 0.3294 | 0.1085 | −0.2584 | 3.5853 | 0.3304 | 0.8760 | 56 | 89.08 | 11 | 11 |
| NBC | 0.3294 | 0.1085 | −0.9496 | 3.7016 | 0.3306 | 0.8777 | 56 | 89.08 | 11 | 11 |
| QDA | 0.2567 | 0.0659 | −0.6008 | 2.2416 | 0.2577 | 0.9243 | 34 | 93.37 | 9 | 9 |
| RF | 0.0440 | 0.0019 | 0.0388 | 0.0388 | 0.0442 | 0.9978 | 1 | 99.81 | 2 | 2 |
| SVM | 0.1761 | 0.0310 | 0.0032 | 1.0368 | 0.1766 | 0.9641 | 16 | 96.88 | 8 | 8 |
- Note: AS system involves Classes 1–4 defined by formulaic/logical rules. Model abbreviations are listed in Table 1.
- Abbreviations: AAPD, absolute average percentage deviation; APD, average percentage deviation; MAE, mean absolute error; RMSE, root mean squared error; Sdev, standard deviation; SVM, support vector machine.
Two of the ML algorithms, RF and DT, make only one classification error (out of 513 CT-extract assignments) in the AS classification exercise. Those two models achieved 99.81% accuracy in classifying AS correctly. That level of classification accuracy is better than that achieved for the VS scale (Section 3.5). It also compares very favorably with the accuracy achieved by published DL studies of CT scans focused on the binary (COVID negative vs. COVID positive) classifications,6, 22-25 as already mentioned in Section 3.5.
The CNN (DL model) ranks 6 out of 11 for its AS classification performance. CNN is outperformed by the RF, DT, ADA, GPC, and KNN models for AS classification (Table 6). Nevertheless, the CNN model still achieves and AS classification accuracy of >98%. The confusion matrix analysis (not shown) reveals that DT misclassifies just one CT-extract image of AS = 2 as belonging to Class 1, with RF misclassifying a CT-extract image of AS = 4 as belonging to Class 3.
Alternative combinations of grayscale attributes (distinct from the five attributes used to define AS), and/or alternative classification scales including more or less than four classes, could also be explored. It is possible that such alternative algorithmic systems could also provide useful algorithmic segregation of the CT-extract images in terms of their severity of lung abnormalities. Further research is justified to evaluate such possibilities. Notwithstanding possible alternative algorithmic approaches, the AS system, as defined, is considered to provide a viable algorithmic CT-extract image classification system based on easy-to-measure grayscale attributes.
4.2 Value of DL/ML models in predicting VS and AS classification systems
VS class definitions benefit from the input of a clinician's expertise, potentially taking account of a wider set of information than just image grayscale attributes. However, human judgement, irrespective of the expertise involved, makes VS class assignments by visual inspection somewhat subjective. This means that classifications are likely to vary, in detail, among clinicians. The magnitude of abnormal lung features associated with COVID-19 clearly varies over a broad range, as reflected by the grayscale attribute dispersions recorded (Figures 3, Supporting Information: Figures S4 and S5). Unfortunately, the measured grayscale attributes do not crisply segregate into clusters (e.g., to facilitate simple COVID-19-negative vs. COVID-19-positive classification). This makes the distinction of arbitrarily imposed visual-inspection classes unreliable.
In summary, the potential disadvantages of the VS method are (1) the risk of inconsistency and subjectivity in the VS class assignments of different clinicians; and (2) the slightly overlapping nature of the grayscale attribute clusters relating to the VS groups. Nevertheless, the classification performance of the CNN DL method applied to the prediction of VS classes is extremely good (just 18 misclassifications out of 513 CT-extract images evaluated; 96.49% accuracy). Based on that performance, evaluations with larger datasets may be able to further improve VS classification accuracy. However, it would be difficult to consistently achieve zero VS misclassifications due to the element of subjectivity involved the visual assessment process. By removing that subjectivity and applying a formulaic grayscale assessment, such as the AS system defined and evaluated, more reliable image classification can be reached (Table 6 and Figure 4), achieving up to 99.81% accuracy by DT and RF models. Although the AS method overcomes the disadvantages of the VS method, a remaining disadvantage of the technique is that the extraction of the grayscale image slices from the CT scans currently lacks automation. Future developments planned for the technique are to automate the image slice extraction process.
A comparison of VS Class 0 (COVID-19-negative individuals) and VS Class 1 (COVID-19-positive individuals displaying minimal visual signs in terms of abnormal lung features) classification performance by DL/ML models is worth reflecting upon. CNN and KNN 12-variable models are able to distinguish between these classes with very few errors (Supporting Information: Figure S6). On the other hand, clinicians can only do this, in most cases, with the aid of a rRT-PCR COVID-19 test. This outcome highlights the power of the DL/ML models to use the CT-extract grayscale attributes to achieve valuable distinctions between the most difficult-to-separate VS classes based on visual assessment alone. There is clearly valuable grayscale attribute information available in the CT-scan images. The DL/ML methods offer a rapid and highly effective way of extracting that information in a way that could be used positively to better assess lung damage severity in COVID-19 patients and improve early decision-making on the most suitable care and treatment to administer. Although the use of DL methods for CT-image analysis and pattern recognition is expanding, ML methods should not be discounted as shown by the results of the DT and RF models with the AS data set described.
5 CONCLUSIONS
Based upon grayscale attribute analysis of 513 CT-scan-extract images taken from 57 hospitalized individuals (49 with positive COVID-19 tests; 8 with negative COVID-19 tests) the following conclusions can be drawn: (1) Grayscale attributes can be used to classify, with reasonable accuracy using graphical methods, based a clinician's VS scale of VS = 0 (COVID-19-negative) to VS = 4 (COVID-19-positive with severe lung abnormalities visible) involving five classes. (2) Due to overlap among the VS classes, graphical analysis of grayscale image attributes does not on its own generate highly reliable VS classifications. (3) Deep- and machine-learning (DL/ML) methods can achieve much better VS classifications based on 12 grayscale image attributes than graphical analysis. (4) Of 11 DL/ML models evaluated, the CNN provided the best VS classifications; just 18 misclassifications from the 513-image data set. This represents an accuracy of 96.49%. That multiclass prediction accuracy compares favorably with the range of published prediction accuracies (73%–98%) achieved for DL models configured to address the binary classification issue (COVID negative vs. COVID positive) using CT-scan and/or X-ray images. (5) A simple formula/logic-based AS system, based on just five grayscale-image attributes is able to separate all the CT-extract images evaluated into four classes; AS = 1 (minimal abnormal lung features) to AS = 4 (severe abnormal lung features) involving four classes without considering COVID-19 test information. (6) Applying DL/ML models with 12 grayscale attribute input variables to predict the AS classes, decision tree (DT) and random forest (RF) models outperformed the other models. Those two ML models did so by classifying all but one of the images into the correct AS classes. This represents a multiclass classification accuracy of 99.81%. That multiclass prediction accuracy exceeds the range of published prediction accuracies (73%–98%) achieved for DL models configured to address the binary classification issue (COVID negative vs. COVID positive) using CT-scan and/or X-ray images. (7) The CNN model performed less well classifying using AS scale and grayscale attributes than several of the ML methods evaluated but still achieved 98.44% accuracy. (8) The algorithmic approach, AS coupled with DL/ML models, offers the potential of providing an accurate, automated expert system for classifying into multiple classes the severity of lung abnormalities from CT-scan information based on grayscale image attributes.
Based on these findings, the methods proposed in this study, and grayscale attributes of CT scans in general, are worthy of further evaluation with larger datasets of CT images.
AUTHOR CONTRIBUTIONS
Clinical assessment of CT images and reviewing manuscript: Sara Ghashghaei. Methodology, CT image statistical analysis, machine and deep learning analysis, data interpretation, writing, reviewing, and editing subsequent drafts: David A. Wood. Reviewing and editing manuscript drafts: Erfan Sadatshojaei. Procurement and initial assessment of CT images and reviewing analysis: Mansooreh Jalilpoor.
ACKNOWLEDGMENT
No external funding was received in association with this research.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
ETHICS STATEMENT
The data handling procedures of Namazi Medical Centre (Shiraz, Iran) were observed at all times. The images used for this study with procured with the patients’ permissions with patient confidentiality and anonymity maintained.
Open Research
DATA AVAILABILITY STATEMENT
There is no shared data available with this article.




