Peptide Bond Cleavage by Ni(II) Ions within the Nuclear Localization Signal Sequence
Corresponding Author
Tomasz Frączyk
Department of Immunology, Transplantology and Internal Medicine, Medical University of Warsaw, Nowogrodzka 59, 02-006 Warsaw, Poland
Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Pawińskiego 5a, 02-106 Warsaw, Poland
Search for more papers by this authorArkadiusz Bonna
Department of Biochemistry, University of Cambridge, Tennis Court Road, CB2 1QW Cambridge, United Kingdom
Search for more papers by this authorEwelina Stefaniak
Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Pawińskiego 5a, 02-106 Warsaw, Poland
Search for more papers by this authorNina E. Wezynfeld
Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Pawińskiego 5a, 02-106 Warsaw, Poland
Faculty of Chemistry, Warsaw University of Technology, Noakowskiego 3, 00-664 Warsaw, Poland
Search for more papers by this authorWojciech Bal
Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Pawińskiego 5a, 02-106 Warsaw, Poland
Search for more papers by this authorCorresponding Author
Tomasz Frączyk
Department of Immunology, Transplantology and Internal Medicine, Medical University of Warsaw, Nowogrodzka 59, 02-006 Warsaw, Poland
Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Pawińskiego 5a, 02-106 Warsaw, Poland
Search for more papers by this authorArkadiusz Bonna
Department of Biochemistry, University of Cambridge, Tennis Court Road, CB2 1QW Cambridge, United Kingdom
Search for more papers by this authorEwelina Stefaniak
Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Pawińskiego 5a, 02-106 Warsaw, Poland
Search for more papers by this authorNina E. Wezynfeld
Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Pawińskiego 5a, 02-106 Warsaw, Poland
Faculty of Chemistry, Warsaw University of Technology, Noakowskiego 3, 00-664 Warsaw, Poland
Search for more papers by this authorWojciech Bal
Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Pawińskiego 5a, 02-106 Warsaw, Poland
Search for more papers by this authorAbstract
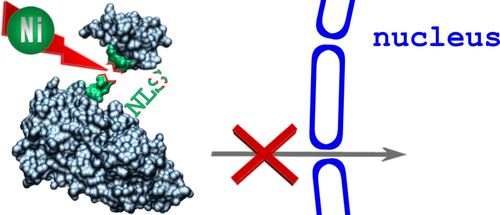
Nickel is harmful to humans, being both carcinogenic and allergenic. However, the mechanisms of this toxicity are still unresolved. We propose that Ni(II) ions disintegrate proteins by hydrolysis of peptide bonds preceding the Ser/Thr-Xaa-His sequences. Such sequences occur in nuclear localization signals (NLSs) of human phospholipid scramblase 1, Sam68-like mammalian protein 2, and CLK3 kinase. We performed spectroscopic experiments showing that model nonapeptides derived from these NLSs bind Ni(II) at physiological pH. We also proved that these sequences are prone to Ni(II) hydrolysis. Thus, the aforementioned NLSs may be targets for nickel toxicity. This implies that Ni(II) ions disrupt the transport of some proteins from cytoplasm to cell nucleus.
Graphical Abstract
References
- 1F. Alinaghi, N. H. Bennike, A. Egeberg, J. P. Thyssen, J. D. Johansen, ‘Prevalence of contact allergy in the general population: A systematic review and meta-analysis’, Cont. Derm. 2019, 80, 77–85.
- 2M. G. Ahlström, J. P. Thyssen, T. Menné, K. Midander, A. Julander, C. Lidén, C. R. Johnsen, J. D. Johansen, ‘Short contact with nickel causes allergic contact dermatitis: an experimental study’, Br. J. Dermatol. 2018, 179, 1127–1134.
- 3C. R. Hamann, D. Hamann, in ‘Metal Allergy’, Eds. J. K. Chen, J. P. Thyssen, Springer, Cham, 2018, pp. 137–162.
- 4J. P. Thyssen, D. J. Gawkrodger, I. R. White, A. Julander, T. Menné, C. Lidén, ‘Coin exposure may cause allergic nickel dermatitis: a review’, Cont. Derm. 2012, 68, 3–14.
- 5M. Aquino, T. Mucci, M. Chong, M. D. Lorton, L. Fonacier, ‘Mobile phones: potential sources of nickel and cobalt exposure for metal allergic patients’, Pediatr. Allergy Immunol. Pulmonol. 2013, 26, 181–186.
- 6K. Midander, J. Kettelarij, A. Julander, C. Lidén, ‘Nickel release from white gold’, Cont. Derm. 2014, 71, 108–128.
- 7K. Midander, A. Hurtig, A. Borg Tornberg, A. Julander, ‘Allergy risks with laptop computers – nickel and cobalt release’, Cont. Derm. 2016, 74, 353–359.
- 8P. Olmedo, W. Goessler, S. Tanda, M. Grau-Perez, S. Jarmul, A. Aherrera, R. Chen, M. Hilpert, J. E. Cohen, A. Navas-Acien, A. M. Rule, ‘Metal concentrations in e-cigarette liquid and aerosol samples: the contribution of metallic coils’, Environ. Health Perspect. 2018, 126, 027010.
- 9R. S. Pappas, M. R. Fresquez, N. Martone, C. H. Watson, ‘Toxic metal concentrations in mainstream smoke from cigarettes available in the USA’, J. Anal. Toxicol. 2014, 38, 204–211.
- 10E. Mann, U. Ranft, G. Eberwein, D. Gladtke, D. Sugiri, H. Behrendt, J. Ring, T. Schäfer, J. Begerow, J. Wittsiepe, U. Krämer, M. Wilhelm, ‘Does airborne nickel exposure induce nickel sensitization?’, Cont. Derm. 2010, 62, 355–362.
- 11IARC, ‘Nickel and nickel compounds’, IARC Monogr. Eval. Carcinog. Risks Hum. 2012, 100c, 169–218.
- 12J. E. Goodman, R. L. Prueitt, S. Thakali, A. R. Oller, ‘The nickel ion bioavailability model of the carcinogenic potential of nickel-containing substances in the lung’, Crit. Rev. Toxicol. 2011, 41, 142–174.
- 13B. Schultze, P. M. Lind, A. Larsson, L. Lind, ‘Whole blood and serum concentrations of metals in a Swedish population-based sample’, Scand. J. Clin. Lab. Invest. 2013, 74, 143–148.
- 14H. J. Raithel, K. H. Schaller, A. Reith, K. B. Svenes, H. Valentin, ‘Investigations on the quantitative determination of nickel and chromium in human lung tissue’, Int. Arch. Occup. Environ. Health 1988, 60, 55–66.
- 15J. M. Benson, I. Y. Chang, Y. S. Cheng, F. F. Hahn, C. H. Kennedy, E. B. Barr, K. R. Maples, M. B. Snipes, ‘Particle clearance and histopathology in lungs of F344/N rats and B6 C3F1 mice inhaling nickel oxide or nickel sulfate’, Fundam. Appl. Toxicol. 1995, 28, 232–244.
- 16T. H. Petersen, M. H. Jee, A. Ø. Gadsbøll, J. D. Schmidt, J. J. Sloth, G. F. Sonnenberg, C. Geisler, J. P. Thyssen, C. M. Bonefeld, ‘Mice with epidermal filaggrin deficiency show increased immune reactivity to nickel’, Cont. Derm. 2019, 80, 139–148.
- 17A. Schunch, J. Schwitulla, ‘Decrease in nickel allergy in women after the second EU nickel directive’, Cont. Derm. 2013, 69, 253–256.
- 18J. C. Wataha, N. L. O′Dell, B. B. Singh, M. Ghazi, G. M. Whitford, P. E. Lockwood, ‘Relating nickel-induced tissue inflammation to nickel release in vivo’, J. Biomed. Mater. Res. 2001, 58, 537–544.
- 19T. Schwerdtle, A. Hartwig, ‘Bioavailability and genotoxicity of soluble and particulate nickel compounds in cultured human lung cells’, Mat.-Wiss. Werkstofftech. 2006, 37, 521–525.
- 20Y. Yao, M. Costa, ‘Toxicogenomic effect of nickel and beyond’, Arch. Toxicol. 2014, 88, 1645–1650.
- 21K. S. Kasprzak, F. W. Sunderman Jr., K. Salnikow, ‘Nickel carcinogenesis’, Mutat. Res. 2003, 533, 67–97.
- 22E. Kopera, A. Krężel, A. M. Protas, A. Belczyk, A. Bonna, A. Wysłouch-Cieszyńska, J. Poznański, W. Bal, ‘Sequence-specific Ni(II)-dependent peptide bond hydrolysis for protein engineering: reaction conditions and molecular mechanism’, Inorg. Chem. 2010, 49, 6636–6645.
- 23A. A. Karaczyn, W. Bal, S. L. North, R. M. Bare, V. M. Hoang, R. J. Fisher, K. S. Kasprzak, ‘The octapeptidic end of the C-terminal tail of histone H2 A is cleaved off in cells exposed to carcinogenic nickel(II)’, Chem. Res. Toxicol. 2003, 16, 1555–1559.
- 24N. E. Wezynfeld, K. Bossak, W. Goch, A. Bonna, W. Bal, T. Frączyk, ‘Human annexins A1, A2, and A8 as potential molecular targets for Ni(II) ions’, Chem. Res. Toxicol. 2014, 27, 1996–2009.
- 25N. E. Wezynfeld, A. Bonna, W. Bal, T. Frączyk, ‘Ni(II) ions cleave and inactivate human alpha-1 antitrypsin hydrolytically, implicating nickel exposure as a contributing factor in pathologies related to antitrypsin deficiency’, Metallomics 2015, 7, 596–604.
- 26E. Kurowska, J. Sasin-Kurowska, A. Bonna, M. Grynberg, J. Poznański, L. Knizewski, K. Ginalski, W. Bal, ‘The C2H2 zinc finger transcription factors are likely targets for Ni(II) toxicity’, Metallomics 2011, 3, 1227–1231.
- 27L. M. McLane, A. H. Corbett, ‘Nuclear localization signals and human disease’, IUBMB Life 2009, 61, 697–706.
- 28N. Arashiki, Y. Takakuwa, ‘Maintenance and regulation of asymmetric phospholipid distribution in human erythrocyte membranes: implications for erythrocyte functions’, Curr. Opin. Hematol. 2017, 24, 167–172.
- 29J. S. Lee, J. G. Burr, ‘Salpα and Salpβ, growth-arresting homologs of Sam68’, Gene 1999, 240, 133–147.
- 30A. Eisenreich, V. Y. Bogdanov, A. Zakrzewicz, A. Pries, S. Antoniak, W. Poller, H. P. Schultheiss, U. Rauch, ‘Cdc2-like kinases and DNA topoisomerase I regulate alternative splicing of tissue factor in human endothelial cells’, Circ. Res. 2009, 104, 589–599.
- 31M. H. Chen, I. Ben-Efraim, G. Mitrousis, N. Walker-Kopp, P. J. Sims, G. Cingolani, ‘Phospholipid scramblase 1 contains a nonclassical nuclear localization signal with unique binding site in importin alpha’, J. Biol. Chem. 2005, 280, 10599–10606.
- 32K. E. Süel, H. Gu, Y. M. Chook, ‘Modular organization and combinatorial energetic of proline-tyrosine nuclear localization signals’, PLoS Biol. 2008, 6, e137.
- 33L. A. Schneider, A. Korber, S. Grabbe, J. Dissemond, ‘Influence of pH on wound-healing: a new perspective for wound-therapy?’, Arch. Dermatol. Res. 2007, 298, 413–420.
- 34S. Schreml, R. J. Meier, K. T. Weiß, J. Cattani, D. Flittner, S. Gehmert, O. S. Wolfbeis, M. Landthaler, P. Babilas, ‘A sprayable luminescent pH sensor and its use for wound imaging in vivo’, Exp. Dermatol. 2012, 21, 948–976.
- 35M. Naghavi, R. John, S. Naguib, M. S. Siadaty, R. Grasu, K. C. Kurian, W. B. van Winkle, B. Soller, S. Litovsky, M. Madjid, J. T. Willerson, W. Casscells, ‘pH Heterogeneity of human and rabbit atherosclerotic plaques; a new insight into detection of vulnerable plaque’, Atherosclerosis 2002, 164, 27–35.
- 36W. Bal, H. Kozlowski, R. Robbins, L. D. Pettit, ‘Competition between the terminal amino and imidazole nitrogen donors for coordination to Ni(II) ions in oligopeptides’, Inorg. Chim. Acta 1995, 231, 7–12.
- 37M. N. Chahine, G. N. Pierce, ‘Therapeutic targeting of nuclear protein import in pathological cell conditions’, Pharmacol. Rev. 2009, 61, 358–372.
- 38T. Stelma, A. Chi, P. J. van der Watt, A. Verrico, P. Lavia, V. D. Leaner, ‘Targeting nuclear transporters in cancer: diagnostic, prognostic and therapeutic potential’, IUBMB Life 2016, 68, 268–280.
- 39N. Arashiki, M. Saito, I. Koshino, K. Kamata, J. Hale, N. Mohandas, S. Manno, Y. Takakuwa, ‘An unrecognized function of cholesterol: regulating the mechanism controlling membrane phospholipid asymmetry’, Biochemistry 2016, 55, 3504–3513.
- 40N. Andraka, L. Sánchez-Magraner, M. García-Pacios, F. M. Goñi, J. L. Arrondo, ‘The conformation of human phospholipid scramblase 1, as studied by infrared spectroscopy. Effects of calcium and detergent’, Biochim. Biophys. Acta 2017, 1859, 1019–1028.
- 41I. Ben-Efraim, Q. Zhou, T. Wiedmer, L. Gerace, P. J. Sims, ‘Phospholipid scramblase 1 is imported into the nucleus by a receptor-mediated pathway and interacts with DNA’, Biochemistry 2004, 43, 3518–3526.
- 42W. Humphrey, A. Dalke, K. Schulten, ‘VMD: visual molecular dynamics’, J. Mol. Graph. 1996, 14, 33–38.
- 43T. Wiedmer, Q. Zhou, D. Y. Kwoh, P. J. Sims, ‘Identification of three new members of the phospholipid scramblase gene family’, Biochim. Biophys. Acta 2000, 1467, 244–253.
- 44Q. Pan, O. Shai, L. J. Lee, B. J. Frey, B. J. Blencowe, ‘Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing’, Nat. Genet. 2018, 40, 1413–1415.
- 45J. Chen, W. A. Weiss, ‘Alternative splicing in cancer: implications for biology and therapy’, Oncogene 2015, 34, 1–14.
- 46H. K. Kim, M. H. C. Pham, K. S. Ko, B. D. Rhee, J. Han, ‘Alternative splicing isoforms in health and disease’, Pflugers Arch. 2018, 470, 995–1016.
- 47https://www.proteinatlas.org/ENSG00000131773-KHDRBS3/tissue; Accessed October 7, 2019.
- 48Y. Matsumoto, J. Itou, F. Sato, M. Toi, ‘SALL4 - KHDRBS3 network enhances stemness by modulating CD44 splicing in basal-like breast cancer’, Cancer Med. 2018, 7, 454–462.
- 49K. F. Lei, B. Y. Liu, Y. F. Wang, X. H. Chen, B. Q. Yu, Y. Guo, Z. G. Zhu, ‘SerpinB5 interacts with KHDRBS3 and FBXO32 in gastric cancer cells’, Oncol. Rep. 2011, 26, 1115–1120.
- 50https://www.proteinatlas.org/ENSG00000179335-CLK3/tissue; Accessed October 7, 2019.
- 51S. Negi, S. Pandey, S. M. Srinivasan, A. Mohammed, C. Guda, ‘LocSigDB: a database of protein localization signals’, Database 2015, bav003.
- 52W. C. Chan, P. D. White, ‘Fmoc solid phase peptide synthesis, a practical approach’, Oxford University Press, New York, 2000.