Blocking ITGA5 potentiates the efficacy of anti-PD-1 therapy on glioblastoma by remodeling tumor-associated macrophages
Rongrong Zhao and Ziwen Pan contributed equally to this work.
Abstract
Background
Glioblastoma (GBM) is largely refractory to antibodies against programmed cell death 1 (anti-PD-1) therapy. Fully understanding the cellular heterogeneity and immune adaptations in response to anti-PD-1 therapy is necessary to design more effective immunotherapies for GBM. This study aimed to dissect the molecular mechanisms of specific immunosuppressive subpopulations to drive anti-PD-1 resistance in GBM.
Methods
We systematically analysed single-cell RNA sequencing and spatial transcriptomics data from GBM tissues receiving anti-PD-1 therapy to characterize the microenvironment alterations. The biological functions of a novel circular RNA (circRNA) were validated both in vitro and in vivo. Mechanically, co-immunoprecipitation, RNA immunoprecipitation and pull-down assays were conducted.
Results
Mesenchymal GBM (MES-GBM) cells, which were associated with a poor prognosis, and secreted phosphoprotein 1 (SPP1)+ myeloid-derived macrophages (SPP1+ MDMs), a unique subpopulation of MDMs with complex functions, preferentially accumulated in non-responders to anti-PD-1 therapy, indicating that MES-GBM cells and SPP1+ MDMs were the main anti-PD-1-resistant cell subpopulations. Functionally, we determined that circular RNA succinate dehydrogenase complex assembly factor 2 (circSDHAF2), which was positively associated with the abundance of these two anti-PD-1-resistant cell subpopulations, facilitated the formation of a regional MES-GBM and SPP1+ MDM cell interaction loop, resulting in a spatially specific adaptive immunosuppressive microenvironment. Mechanically, we found that circSDHAF2 promoted MES-GBM cell formation by stabilizing the integrin alpha 5 (ITGA5) protein through N-glycosylation. Meanwhile, the N-glycosylation of the ITGA5 protein facilitated its translocation into exosomes and subsequent delivery to MDMs to induce the formation of SPP1+ MDMs, which in turn maintained the MES-GBM cell status and induced T-cell dysfunction via the SPP1-ITGA5 pathway, ultimately promoting GBM immune escape. Importantly, our findings demonstrated that antibody-mediated ITGA5 blockade enhanced anti-PD-1-mediated antitumor immunity.
Conclusions
This work elucidated the potential tissue adaptation mechanism of intratumoral dynamic interactions between MES-GBM cells, MDMs and T cells in anti-PD-1 non-responders and identified the therapeutic potential of targeting ITGA5 to reduce anti-PD-1 resistance in GBM.
Abbreviations
-
- GBM
-
- glioblastoma
-
- MES
-
- mesenchymal
-
- TME
-
- tumor microenvironment
-
- anti-PD-1/PD-L1
-
- antibodies against programmed cell death (/ligand) 1
-
- TAM
-
- tumor-associated macrophages
-
- MDM
-
- myeloid-derived macrophages
-
- MG
-
- microglia
-
- scRNA-seq
-
- single-cell RNA sequencing
-
- ST
-
- spatial transcriptomics
-
- CircRNA
-
- circular RNAs
-
- UMAP
-
- uniform manifold approximation and projection
-
- DEGs
-
- differentially expressed genes
-
- GSEA
-
- Gene Set Enrichment Analysis
-
- TF
-
- transcription factor
-
- EMT
-
- epithelial-mesenchymal transition
-
- GSCs
-
- Glioma stem cells
-
- ELDA
-
- extreme limiting dilution assay
-
- siRNA
-
- small interfering RNA
-
- qPCR
-
- quantitative Real-time PCR
-
- RNase R
-
- Ribonuclease R
-
- FISH
-
- RNA fluorescence in situ hybridization
-
- Co-IP
-
- co-immunoprecipitation
-
- RIP
-
- RNA immunoprecipitation
-
- ChIP
-
- chromatin immunoprecipitation
-
- DAPI
-
- 2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride
-
- NKT
-
- natural killer T cell
-
- NK
-
- natural killer cell
-
- Treg
-
- regulatory T cells
-
- circRNA
-
- circular RNA
-
- GAPDHglyceraldehyde-3-phosphate dehydrogenase;18S
-
- glyceraldehyde-3-phosphate dehydrogenase;18S18S ribosomal RNA
-
- U6
-
- U6 small nuclear RNA
-
- SDHAF2
-
- succinate dehydrogenase complex assembly factor 2
-
- CCL2
-
- chemokine (C-C motif) ligand 2
-
- Bp
-
- Base Pair
-
- SPP1
-
- secreted phosphoprotein 1
-
- ITGA5
-
- integrin alpha 5
-
- RUNX1
-
- runt-related transcription factor 1
-
- FAK
-
- protein tyrosine kinase 2
-
- P-FAK
-
- phosphorylated protein tyrosine kinase 2
-
- STAT3
-
- signal transducer and activator of transcription 3
-
- P-STAT3
-
- phosphorylated signal transducer and activator of transcription 3
-
- CHI3L1
-
- chitinase 3-like 1
-
- B4GALT1
-
- beta-1,4-galactosyltransferase 1
-
- β-acitin
-
- beta-actin
-
- GZMB
-
- Granzyme B
1 BACKGROUND
Glioblastoma (GBM), the most prevalent primary brain tumor, is characterized by a grim prognosis and is surrounded by a highly immunosuppressive tumor microenvironment (TME) [1]. Among immune checkpoint blockade agents, antibodies against programmed cell death (/ligand) 1 (anti-PD-1/PD-L1) have shown favorable efficacy in multiple solid cancers [2]. However, the highly suppressive TME severely limits its efficacy [2], and the great diversity of tumor cell states, cellular compositions and phenotypic characteristics make it difficult to classify GBM accurately and find effective therapeutic approaches [3, 4]. Considering that immune suppression and drug resistance in the TME lead to unsatisfactory anti-PD-1 therapy efficacy, it is important to explore the connections between the TME and patient outcomes in patients receiving anti-PD-1 therapy and to identify accurate biomarkers for predicting treatment response.
Tumor-associated macrophages (TAMs), encompassing both microglias (MGs) and myeloid-derived macrophages (MDMs), are central to the immunosuppressive nature of the GBM tumor microenvironment (TME). Their interactions with diverse immune cells and cytokines within the TME are critically important in driving tumor immune evasion [5, 6]. These cells and cytokines may be promising therapeutic targets [7]. Recent advancements in single-cell omics technologies have greatly expanded our knowledge of the molecular heterogeneity within tumor-associated macrophages (TAMs) [8, 9]. What molecular characteristics and regulatory pathways distinguish TAMs in their role in fostering the malignant transformation of GBM cells and in conferring resistance to anti-PD-1 therapy? Addressing this question may help us identify attractive therapeutic targets to overcome immunotherapy resistance in patients with end-stage cancer in the clinic. Therefore, comprehensive and high-resolution cellular and molecular analyses of tumor cells and immune cells with single-cell RNA sequencing (scRNA-seq) and spatial transcriptomics (ST) techniques are urgently needed to elucidate the formation of the immunosuppressive TME and help develop more precise treatment strategies for GBM.
Circular RNAs (circRNAs) are a type of non-coding RNAs that form a single-stranded, covalently closed loop. They are known for their exceptional stability, prevalence, and evolutionary conservation. Recently, we reported that circRNA dysregulation was frequently observed in cancers, revealing that the aberrant expression of circRNAs can contribute to tumorigenesis and tumor progression [10]. However, very little is known about whether these circRNAs participate in immune escape, especially anti-PD-1 resistance, in GBM.
In this study, we systematically revealed key TME alterations induced by anti-PD-1 therapy in non-responders using scRNA-seq and ST data, and performed a systematic screen for circRNAs involved in this tumor tissue adaptation. Additionally, we performed in vitro and in vivo assays to explore the involvement of circRNAs in the adaptive immunosuppressive TME, and developed a combinational immunotherapeutic strategy against GBM. This study explored dynamic intercellular regulatory crosstalk among GBM cells, MDMs, and T cells and developed a combinational therapeutic strategy to enhance the efficacy of anti-PD-1 therapy in GBM.
2 MATERIALS AND METHODS
2.1 scRNA-seq data processing
The R toolkit Seurat V4.0 pipeline (http://satijalab.org/seurat/) for single-cell transcriptomics [11] was employed for marker gene and cell type identification following initial data filtration and normalization. The processed scRNA-seq data and associated metadata, derived from 11 GBM patients undergoing neoadjuvant immunotherapy, were analyzed. These patients were categorized into 5 non-responders and 6 responders, with the data accessible via Figshare (https://doi.org/10.6084/m9.figshare.22434341) [12]. Since the scRNA-seq data were already pre-normalized, they were directly imported into Seurat without further scaling or centering. Next, 3,000 highly variable genes, were identified using the “FindVariableFeatures” function in Seurat and served as inputs for dimensionality reduction through principal component analysis (PCA). Principal components were assessed alongside correlated genes to determine the optimal number for downstream analyses. Uniform manifold approximation and projection (UMAP) was applied to the top 20 principal components to create a two-dimensional representation of the cells. To identify specific differentially expressed genes in each cluster, the “FindAllMarkers” function in Seurat was utilized, selecting genes expressed in at least 10% of cells, with a log-fold change exceeding 0.25, and an adjusted P value below 0.05 according to the Wilcoxon test. Finally, marker genes were used to annotate cell clusters, enabling the identification of biological cell types.
2.2 Functional analysis of differentially expressed genes (DEGs)
DEG analysis across all cell types was carried out using the Wilcoxon test integrated into the Seurat package. To estimate pathway activity at the single-cell level, the Gene set variation analysis (GSVA) package (version 1.22.4) [13] was employed with default settings. Gene Set Enrichment Analysis (GSEA) was conducted using version 4.0.3 of the GSEA software, which incorporates predefined gene sets from the Molecular Signatures Database (MSigDB v7.1, https://www.gsea-msigdb.org/gsea/msigdb).
2.3 Public GBM data collection and related analysis
The Cancer Genome Atlas (TCGA)-GBM RNA sequencing (RNA-seq) transcriptome data and corresponding clinicopathological parameters were obtained from the TCGA database (http://cancergenome.nih.gov/). The Gravendeel-GBM microarray dataset and clinicopathological parameters were extracted from the GlioVis database (http://gliovis.bioinfo.cnio.es/).
Survival analysis was performed via the R package “survival”. Kaplan-Meier survival curves were generated via the “survfit” function. The endpoint was overall survival (OS), defined as the period from the date of diagnosis until the date of death or last follow-up. The “maxstat.test” function of the R package maxstat was used to iteratively test all potential cut-off points to find the maximum rank statistic for dichotomizing subpopulation enrichment or gene expression, and then patients were divided into two groups based on the selected maximum log statistic. To evaluate the gene signatures associated with hypoxic MDM and MES-GBM cell infiltration, we first divided the TCGA-GBM and Gravendeel-GBM cohorts into four groups (MES-GBMhigh + hypoxic MDMhigh; MES-GBMhigh + hypoxic MDMlow; MES-GBMlow + hypoxic MDMhigh; and MES-GBMlow + hypoxic MDMlow) based on the dichotomization described above.
We used the Estimation of Stromal and Immune cells in MAlignant Tumor tissues using Expression data (ESTIMATE) algorithm to calculate the immune scores of the GBM TME for each sample.
2.4 ST data processing
The ST data for GBM were obtained from Figshare [12] and processed following the recommended guidelines (https://satijalab.org/seurat/articles/spatial_vignette.html). Briefly, data normalization was performed using the SCTransform function, and dimensionality reduction was achieved through PCA and UMAP. Clustering was conducted based on the resolution of the first 20 principal components. Gene expression patterns were visualized using the SpatialFeaturePlot function, and signature enrichment score for MES-GBM cells, SPP1+ MDM, epithelial-mesenchymal transition (EMT) and hypoxia was performed with the multimodal intersection analysis (MIA) function [14].
2.5 Cell culture
Glioma stem cells (GSCs), including GSC20, GSC267, GSC28, GSC11 and GSC8-11, were generously provided by Dr. Frederick F Lang and Dr. Krishna P.L. Bhat (M.D. Anderson Cancer Center, University of Texas, Houston, TX, USA). Culture media, including DMEM/F12 (#10565018; Gibco, Grand Island, NY, USA) and B-27 without serum supplement (#17504044; Gibco), were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Recombinant human epidermal growth factor (EGF, #236-EG) and Recombinant human basic fibroblast growth factor (bFGF, #233-FB) protein were purchased from R&D Systems, Inc. (Minneapolis, MN, USA). GSCs were maintained in DMEM/F12 supplemented with 2% B-27 without serum supplement, 20 ng/mL recombinant human bFGF and 20 ng/mL recombinant human EGF.
The human THP1 cells and mouse glioblastoma CT2A cell line was sourced from the Chinese Academy of Sciences Cell Bank (Shanghai, China). THP1 cells were cultured in RPMI1640 (#A4192301, Gibco) with 100 U/mL streptomycin-, 100 U/mL penicillin (#C0222, Beyotime, Shanghai, China), and 10% fetal bovine serum (FBS, #SA201, Cellmax, Beijing, China). CT2A cells were cultured in DMEM/F12 (#CGM104.05, Cellmax) with 100 U/mL streptomycin-penicillin (#C0222, Beyotime), and 10% fetal bovine serum (FBS, #SA201, Cellmax). All cell lines were maintained in a humidified incubator at 37°C with 5% CO2 and were validated using analysis of short tandem repeat.
2.6 Quantitative real-time PCR (qPCR)
For qPCR, total RNA was extracted from GSCs, THP1-differentiated macrophages and CD3 T cells (extracted from the serum of GBM patients) using TRIzol™ Reagent (#15596026CN, Thermo Fisher Scientific). RNA from exosomes was extracted with the SeraMir™ Exosome RNA Extraction Kit (#RA806A-1, System Biosciences, Palo Alto, CA, USA). Reverse transcription was performed using the ReverTra Ace qPCR RT Kit (#FSQ-101, TOYOBO, Shanghai, China), and quantitative PCR was carried out with TB Green™ Premix Ex Taq™ (#RR820A, Takara, Tokyo, Japan). β-actin was employed as internal control for mRNA and U6 for miRNA, respectively. Reverse transcription was carried out using the ReverTra Ace qPCR RT Kit (#FSQ-101, TOYOBO). The quantity and concentration of RNA were measured using a spectrophotometer (#DS-11, Denovix, Guangzhou, China). Quantitative PCR was performed with TB Green™ Premix Ex Taq™. Each 10 µL reaction mixture included 1 µL of DNA extract, 1 µL of each forward and reverse gene-specific primers, 5 µL of TB Green, and 3 µL ddH2O. Thermal cycling conditions consisted of an initial denaturation step at 95°C for 30 s, followed by 40 cycles of denaturation at 95°C for 5 s, annealing at 60°C for 34 s. Each experiment was conducted a minimum of three times for replication. The expression levels of β-actin and U6 served as internal controls, and the 2-ΔΔCT method was employed to process the data. Details of the primers used are provided in Supplementary Table S1.
2.7 Circular RNA succinate dehydrogenase complex assembly factor 2 (circSDHAF2) structure validation
CircBase database (http://www.circbase.org/) was consulted to confirm that circSDHAF2 originates from the back-to-back splicing of exons 2 and 3 from the SDHAF2 gene. We designed primers targeting the back splice sites and performed qPCR followed by Sanger sequencing to validate the circular structure of circSDHAF2.
2.8 Actinomycin D assays
In 6-well plates, GSC20 and GSC267 cells were seeded at a density of 2 × 10⁵ cells per well. Cells were treated with 2 µg/mL actinomycin D (#SBR00013, Sigma-Aldrich, St. Louis, MO, USA) and harvested at specified time intervals. The stability of circSDHAF2 and SDHAF2 transcripts was analyzed by qPCR.
2.9 Ribonuclease R (RNase R) assays
Two micrograms of total RNA were treated with RNase R (#RNR07250, Epicentre Technologies, Madison, WI, USA) at a concentration of 5 U/µg at 37°C for 15 min, followed by analysis using qPCR.
2.10 Transfection of small interfering RNA (siRNA) and plasmid
siRNA, plasmid, and corresponding negative control (NC), as well as qPCR primers were obtained from Boshang Biotechnology Co., Ltd. (Shanghai, China). Human sh-circSDHAF2 and circSDHAF2 sequences were inserted into the pLVX-IRES-Puro vector (Boshang Biotechnology Co., Ltd.) to achieve stable knockdown and overexpression, respectively, with the empty vector serving as a control. PCR amplified specific ITGA5 fragments were cloned into the pLVX-IRES-Puro vector with a Flag tag. All cloned fragments were validated through DNA sequencing. Mutations, including MUT84, MUT182, MUT297, MUT307, and 2NQ, were introduced by mutagenic primers, using QuikChange Multi Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA, USA). The generation mutants were identified by sequence analysis. siRNA and plasmid transfections were performed using the Lipofectamine 3000 kit (#L3000015, Invitrogen, Carlsbad, CA, USA). Sequences are provided in Supplementary Table S2. Cells transfected with circSDHAF2 and ITGA5 were subjected to puromycin selection to establish stable cell lines.
2.11 Nuclear-cytoplasmic fractionation
RNA from both the nucleus and cytoplasm was isolated using the Cytoplasmic & Nuclear RNA Purification Kit (#78833, Thermo Fisher Scientific), following the provided guidelines. Subsequently, the expression levels of the circSDHAF2 within these cellular compartments were quantified using qPCR.
2.12 RNA fluorescence in situ hybridization (FISH)
GSC20 and GSC267 cells were first fixed in 4% paraformaldehyde for 15 min, then rinsed with 1× PBS. Afterward, they were treated with 1% pepsin (#9001-75-6, Sigma-Aldrich) prepared in 10 mmol/L HCl. For denaturation, a buffer containing a 20 nmol/L FISH probe in hybridization was incorporated in cells, followed by an incubation at 73°C. After 5 min, hybridization was then conducted at 37°C for 24 h. Following this, the slides were washed with 1× PBS, dehydrated, and stained with 4',6-diamidino-2-phenylindole (DAPI). The RNA FISH probes were designed and synthesized by GenePharma (Shanghai, China).
2.13 Neurosphere formation assay
GSC20, GSC267 and GSC28 cells were dissociated using an Accutase solution (#A6964, Sigma-Aldrich) and seeded in 6-well plates (2,000 cells/well). Neurospheres were visualized, and their diameters were measured using a microscope (DM2500, Leica, Wetzlar, Germany) for subsequent quantification analysis.
2.14 Extreme limiting dilution assay (ELDA)
In 96-well plates, GSC20, GSC267 and GSC28 cells were plated at densities of 0, 2, 4, 8, 16, 32, 64, and 128 cells/well, with 12 replicates for each condition. After 14 days of incubation, the number of wells containing colonies was recorded. Data were analyzed using the ELDA software (http://bioinf.wehi.edu.au/software/elda/).
2.15 Western blotting
For Western blotting, proteins were extracted from GSCs (or their exosomes) using RIPA buffer (#P0013B, Beyotime). Antibodies utilized for Western blotting analysis are listed in Supplementary Table S3. The specific experimental methods: firstly, separating proteins by gel electrophoresis via PAGE Gel Quick Preparation Kit (#20327ES, Yeasen, shanghai, China), transferring them to a PVDF membrane (#36126ES, Yeasen), blocking non-specific binding sites, probing with a primary antibody diluted in a primary antibody diluent (#36206ES, Yeasen) to the target protein, washing via TBST (#60145ES, Yeasen), applying a secondary antibody that binds to the primary and is linked to a detection system, and finally visualizing the protein bands through a fluorescent signal.
2.16 Animal study
Male C57BL/6 or BALB/c nude mice (aged 4-6 weeks), were obtained from GemPharmatech Co., Ltd. (Nanjing, Jiangsu, China) and housed at the Neurosurgery Laboratory of Qilu Hospital of Shandong University (Jinan, Shandong, China).
To investigate the GBM-promoting effects circSDHAF2 in vivo, THP1 cells were treated with 100 ng/mL phorbol 12-myristate 13-acetate (PMA, #HY-18739, MedChemExpress, Shanghai, China) for 8 h to induce them into macrophages, and then GSC267 cells expressing luciferase (5 × 10⁵ per mouse), either alone or combined with THP1-differentiated macrophages (1 × 105 per mouse), were implanted intracranially into BALB/c nude mice (10 mice per group). Tumor progression was tracked using the IVIS Spectrum in vivo imaging system (IVIS; PerkinElmer Inc., Waltham, MA, USA) on days 4, 11, 14 and 18 post-implantation. Survival analysis was conducted, with survival time defined as the period from tumor implantation to death.
For combination therapy experiments, luciferase-labeled CT2A cells (1 × 10⁶ per mouse) were implanted into the brains of C57BL/6 mice. Treatments included intraperitoneal injections of anti-PD-1 reagent (#BP0273, clone 29 F.1A12TM, BioXcell, West Lebanon, NH, USA), anti-ITGA5 antibody (#MAB1984-I, clone BMA5, Sigma-Aldrich), or isotype control antibody (#BP0089, BioXcell) at a dosage of 250 µg per mouse per injection on days 7, 10, 14, 18, and 22 after tumor implantation. Bioluminescence imaging was used to assess the tumor volume, using the IVIS Spectrum in vivo imaging system (IVIS; PerkinElmer Inc.).
All the mice were housed in a specific pathogen free breeding barrier with individually ventilated cages. Isoflurane (#R510-22-10, RWD Life Science, Shenzhen, Guangdong, China) was used to anaesthetize mice. Anesthesia induction was carried out in the gas mixture composed of 2% to 2.5% isoflurane and oxygen, and anaesthesia maintenance was performed in a gas mixture composed of 1.5% to 1.8% isoflurane and oxygen. We used two criteria to identify the moribund animals based on our animal use protocol: 1) mouse has difficulty breathing, eating, or drinking; 2) mouse loses ≥ 15% body weight in 4 days. The animals were exposed to high-concentration CO₂ (30%) for a minimum of 3 min to alleviate suffering until complete cessation of breathing was observed, followed by cervical dislocation to ensure death. The euthanasia procedures were performed in accordance with Institutional Animal Care and Use Committee (IACUC) guidelines and were approved by the IACUC of the Qilu Hospital of Shandong University (Approval No. DWLL-2021-122).
2.17 Weighted gene co-expression network analysis (WGCNA)
The expression profiles of 282 potential proteins, binding to circSDHAF2, were obtained from the TCGA-GBM dataset, and then be used to construct a co-expression network using the WGCNA package [15] in R. The modules were generated using the blockwise modules function with a power of 6, the minimum module size of 30, and a merged cut-off height of 0.3536. The correlation coefficient (R) represents the correlation between the expression value of each gene and the phenotypic trait: 0 means that the gene is not correlated with the trait, and 1 means that it is highly correlated. If a gene in a module has a higher R value for this trait, it means that the gene is more highly correlated with this module.
2.18 Pseudotime analysis
Single-cell trajectory analysis was conducted using a cell-by-gene expression matrix, processed with Monocle2 [16]. Cell type-specific differentially expressed genes with a log-fold change exceeding 1 and an adjusted P value below 0.05, which were identified via the “FindAllMarkers” function in Seurat, were utilized to construct dynamic trajectories, arranging cells along a pseudotime continuum. Initially, transcript expression levels for each gene were calculated. Genes were then ranked based on their coefficient of variation relative to the mean, and the top 20 genes for each cell type were chosen as key features. Finally, velocity estimates were mapped onto the UMAP embedding generated in Seurat, facilitating visualization of the inferred cellular dynamics.
2.19 Prediction of RNA secondary structure and protein-protein interaction
For RNA secondary structure prediction, we utilized the RNAfold WebServer (http://rna.tbi.univie.ac.at/) to predict the secondary structure of circSDHAF2. According to the secondary structure, circSDHAF2 was divided into four segments. To predict the binding sequences of ITGA5 and B4GALT1, we used AlphaFold-Multimer (https://github.com/google-deepmind/alphafold3), a deep learning-based tool for predicting protein-protein complex structures. The amino acid sequences of ITGA5 and B4GALT1 were entered into the software, and the top-ranked prediction models were selected based on the basis of confidence scores (pLDDT and interface quality indicators). The predicted complex structures were analyzed by PyMOL (https://github.com/schrodinger/pymol-open-source), and the key amino acid residues on the binding interface were identified. Finally, experimental verification was combined to confirm the accuracy of the prediction results.
2.20 RNA pull-down assays
RNA pull-down experiments were conducted using the Pierce™ Magnetic RNA-Protein Deposition Kit (#20164, Thermo Fisher Scientific). Biotin-labeled circSDHAF2 and its antisense sequence were synthesized by RiboBio (GenePharma). GSC20 and GSC267 cell lysates were incubated with the biotinylated circSDHAF2 probe, followed by streptavidin-conjugated agarose magnetic beads for 30 min at room temperature. Proteins bound to circSDHAF2 were identified using Western blotting, silver staining and mass spectrometry (MS).
2.21 Silver staining
The gel containing proteins was placed in the fixing solution provided by the kit (#G7210, Solarbio Science&Technology, Beijing, China) for 30 min. After fixation, the gel was transferred to the sensitizing solution and incubated for 60 s. The gel was rinsed with distilled water 3-5 times, with each rinse lasting 5-10 min. The gel was soaked in silver staining solution for 20 min. The gel was quickly rinsed with distilled water for 5-10 s to wash away the unbound silver ions. The gel was placed into color development solution, and the protein bands were allowed to develop color. When the color development reached a satisfactory level, the gel was put into the terminating solution and soaked for 10-15 min to terminate the color development reaction and preserve the gel.
2.22 RNA immunoprecipitation (RIP) assays
Magna RIP RNA-Binding Protein Immunoprecipitation Kit (#17-700, Millipore, Billerica, MA, USA) was used for RIP assays. GSC20 and GSC267 cells (1 × 107) were lysed in 1 mL of RIP lysis buffer supplemented with RNase inhibitors. The lysates were rotated at 4°C overnight with beads coated with either IgG or anti-ITGA5 antibodies (#ZRB1122, Sigma-Aldrich). RNA was then extracted using the RNeasy MinElute Cleanup Kit (#74204, Qiagen, Duesseldorf, Germany) and subjected to qPCR analysis.
2.23 Co-immunoprecipitation (Co-IP) experiments
Thermo Scientific Pierce Co-IP Kit (#88804, Thermo Fisher Scientific) was used for Co-IP. GSC20 and GSC267 cells were lysed in 500 µL radio immunoprecipitation assay lysis buffer containing phenylmethanesulfonyl fluoride (#ST506, Beyotime, Shanghai, China), followed by centrifuging at 12,000 × g for 15 min at 4°C. After obtaining the supernatants, magnetic beads and antibodies were added, and the combination was incubated at 4°C for 12 h. The beads were then washed four times with Co-IP buffer, and the bound proteins were eluted for analysis by Western blotting.
2.24 Enzymatic deglycosylation
N-linked glycosylation (N-glycosylation) sites of ITGA5 were predicted using the UniProt database (https://www.uniprot.org/). The deglycosylation experiments were carried out on the protein extracts of the specified cells using Peptide N-Glycosidase F (PNGase F) (#P2318L, Beyotime) according to the manufacturer's protocols. Briefly, denaturing reaction conditions were employed for the treatment of PNGase F, as described below. The cell lysates were denatured in PNGase F denaturing buffer at 100°C for 10 min. The denatured lysates were subsequently cooled and mixed with concentrated PNGase F reaction buffer, 0.5 mol/L sodium phosphate (pH 7.5), or 10% NP-40 at a volume ratio of 1/10. The samples were subsequently digested with PNGase F at 37°C for 1 h. The reaction was terminated by adding sample buffer containing 2-mercaptoethanol. Before Western blotting, the mixture was incubated at 100°C for 5 min.
2.25 Tunicamycin (TM) and cycloheximide (CHX) treatment
GSC20 and GSC267 cells were treated with PBS or 2, 4, or 8 µg/mL TM (#S7894, Selleck Chemicals, Houston, TX, USA) or 8 h. Subsequently, Western blotting was used to detect changes in the expression level and N-glycosylation level of ITGA5. GSC20 and GSC267 cells were treated with 8 µg/mL TM for 0, 2, 4, or 6 h. Subsequently, Western blotting was used to detect changes in the expression level and molecular weight of ITGA5. GSC20 and GSC267 cells were treated with 8 µg/mL TM and 8 µg/mL CHX (#HY-12320, MedChemExpress) for 0, 2, 4, or 6 h. Subsequently, Western blotting was used to detect changes in the expression level, half-life and molecular weight of ITGA5.
2.26 Exosome isolation
Culture media from GSC20 and GSC267 cells was collected and subjected to sequential centrifugation steps at 4°C at 2,000 × g for 30 min and 12,000 × g for 45 min to remove cell precipitate. The resulting supernatant was then ultracentrifuged at 100,000 × g for 70 min at 4°C to pellet exosomes. The exosome pellets were resuspended in PBS and stored at -80°C, ensuring repeated freeze-thaw cycles were avoided.
2.27 Transcription factor (TF) regulon analysis
Regulon analysis was conducted using pySCENIC [17]. Regulon activity, quantified as Area Under the Curve (AUC), was assessed using the AUCell module in pySCENIC based on the default AUCell threshold. Differentially expressed regulons were determined via the Seurat package's “FindAllMarkers” function, employing the Wilcoxon rank-sum test with the following parameters: min.pct = 0.1, logfc.threshold = 0.25, pseudocount.use = FALSE, and only.pos = TRUE. Scaled regulon activity expression values were utilized to create a heatmap, while a rank diagram was used to highlight transcription factors specifically enriched in distinct cell subtypes.
2.28 Analysis of TF binding to targeted gene promoter regions
Initially, we utilized the hTFtarget database (http://bioinfo.life.hust.edu.cn/hTFtarget) to identify Runt-related transcription factor 1 (RUNX1) as a high-confidence TF candidate based on conserved binding motifs in the SPP1 promoter region. Then, the NCBI database was accessed to retrieve the promoter sequence located 1-2,000 kb upstream of SPP1. The JASPAR database (https://jaspar.elixir.no/) was subsequently used to identify the motif and potential binding sites of RUNX1.
2.29 Chromatin immunoprecipitation (ChIP) assay
DNA bound to the anti-RUNX1 antibody was precipitated using the ChIP assay kit (#P2078, Beyotime), and the purified DNA was processed using the DNA Cleanup Kit (#D0041M, Beyotime).
2.30 Luciferase reporter assays
The reporter plasmids pGL3-SPP1-WT, pGL3-SPP1-mut1 and pGL3-SPP1-mut2 were cotransfected with siNC, siRUNX1#1 or siRUNX1#2 into GSC267 cells. Two days later, a dual luciferase reporter assay kit (#16186, Thermo Fisher Scientific) was used to measure reporter activity according to the manufacturer's instructions. All reporter plasmids were synthesized by Boshang Biotechnology Co., Ltd.
2.31 Cell-cell communication analysis
The CellChat package (version 1.6.1) was used to assess cell–cell communication via interaction network analysis [18]. A Seurat object was used as the input for CellChat following the standard protocol described at https://github.com/sqjin/CellChat.
2.32 Flow cytometry
Cells were stained with Fluorescein Isothiocyanate (FITC)-conjugated anti-CD11b antibodies (#101205, Biolegend, San Diego, CA, USA) to detect CD11b+ macrophages, with isotype controls (#400605, Biolegend) included as references. Flow cytometry analysis was conducted using a NovoCyte Quanteon flow cytometer (Agilent Technologies).
2.33 Enzyme-linked immunosorbent assay (ELISA)
SPP1 in THP1-differentiated macrophages treated with supernatant collected from GSC20/267 in the NC and ITGA5-overexpression groups, or co-cultured with anti-ITGA5 (#HY-P99333, MedChemexpress) and blocking SPP1 antibodies (anti-SPP1, #AF1433, R&D systems, Minneapolis, MN, USA) as indicated, was detected through ELISA kits (#DOST00, R&D systems).
2.34 Immunofluorescence (IF) assays
For IF, GSCs (2,000 per well) were seeded in μ-Slide 8-well plates (#PEZGS0896, Millipore) overnight. The cells were then fixed with 4% paraformaldehyde for 20 min and treated with 0.1% Triton X-100 in PBS for 10 min. The cells were blocked with 5% BSA for 30 min at 37 °C, incubated with primary antibody overnight at 4 °C, and washed with PBS three times. Then, the cells were incubated with DAPI for 30 min. All the images were viewed under a confocal microscope (Leica TCS SP8).
2.35 Statistical analysis
Statistical analyses were performed with GraphPad Software 8 (GraphPad, CA, USA) using R Studio (version 4.3.1). Kaplan-Meier curves were analyzed using log-rank tests, and Cox regression models (α = 0.05) were applied via the coxph function in the R survival package, using the Breslow method for tied events. Correlations were assessed with Pearson tests. Data are shown as mean ± SD. Student's t-test, Wilcoxon test, or one-way ANOVA were used for group comparisons as appropriate. P < 0.05 was considered significant.
3 RESULTS
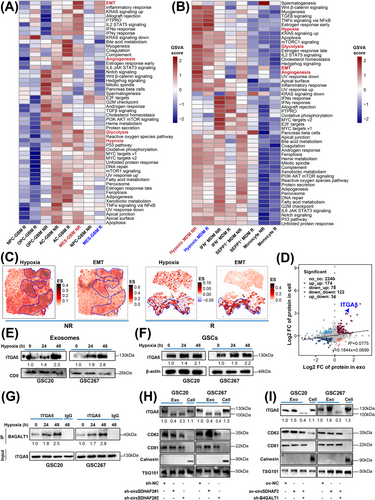
3.1 MES-GBM cells and hypoxic MDMs persisted in GBM receiving anti-PD-1 treatment
To understand the immunological landscape disparities in the TME between non-responders and responders receiving anti-PD-1 treatment, we obtained scRNA-seq data from Figshare [12]. Clustering analysis revealed that the cells were clustered into ten major cell compartments, including malignant cells, TAMs (consisting of MDMs and MGs), stromal cells (fibroblasts and endothelial cells), conventional dendritic cells, lymphocytes, oligodendrocytes and proliferating cells (Figure 1A, Supplementary Figure S1, Supplementary Table S4). We found that the proportion of lymphocytes in responders tended to increase, whereas the proportion of MDMs in non-responders was markedly increased in the GBM microenvironment (Supplementary Figure S2A-B). Furthermore, GSVA revealed that, compared with MGs, MDMs were enriched mainly in classical procarcinogenic signaling pathways related to hypoxia, EMT and angiogenesis (Supplementary Figure S2C). We further subdivided the tumor cells, TAMs and lymphocytes into different subtypes, which included 4 GBM cell subclusters, 5 MG subclusters, 4 MDM subclusters and 5 lymphocyte subclusters (Supplementary Figure S2D, Supplementary Table S5). Further analysis revealed that the population of MDMs with a hypoxic signature (hypoxic MDMs) was greater in non-responders than in responders, and MES-GBM cells, which are strongly associated with therapy resistance and MDMs infiltration [19-22], also showed an up-regulation trend in non-responders (P < 0.1) (Figure 1B), whereas we observed no significant changes in other cell types (Supplementary Figure S2E), indicating that the crosstalk between hypoxic MDM and MES-GBM subpopulations might play key roles in the therapeutic resistance of anti-PD1. Further Cox survival analyses revealed consistent associations between MES-GBM cell and hypoxic MDM signature upregulation and reduced survival in both TCGA-GBM (Figure 1C) and Gravendeel-GBM cohorts (Supplementary Figure S2F). Patients with high proportions of both MES-GBM cell and hypoxic MDM had shorter OS than did those with lower proportions of both cell subpopulations (Figure 1D, Supplementary Figure S2G). Further correlation analyses revealed consistent positive associations between the enrichment of MES-GBM and MDM gene sets in both TCGA-GBM and Gravendeel-GBM datasets (Supplementary Figure S2H). ST data analysis further demonstrated that the expression of conventional MES-GBM cell markers CD44 and Chitinase 3 Like 1/2 (CHI3L1/2) was spatially co-localized with that of hypoxic MDM markers CD14, versican (VCAN) and SPP1, and this expression was greater in non-responders (Supplementary Figure S3). More importantly, the ST data analysis also revealed that patients with greater proportions of MES-GBM cell and hypoxic MDM signatures had less benefit from anti-PD-1 therapy, and MIA revealed that MES-GBM cells and hypoxic MDMs highlighted co-localization in the same region (Figure 1E), suggesting that these two cell subpopulations could collectively promote GBM progression and anti-PD-1 therapy failure.
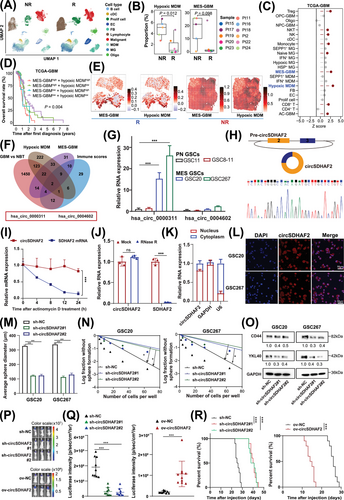
3.2 circSDHAF2 promoted MES-GBM cell formation and tumorigenesis
As MES-GBM is the major cell subtype associated with poor anti-PD-1 therapeutic efficacy, we next narrowed our list of candidates by selecting circRNAs that were highly associated with MES GBM cells, hypoxic MDMs and the immune scores derived from the TCGA-GBM dataset, revealing that two circRNAs were associated with these signatures, which were also highly expressed in GBM versus normal brain tissue (GBM vs. NBT) (Figure 1F). hsa_circ_0000311 was the most highly upregulated circRNA in MES glioma stem cells (GSCs) compared with pro-neural GSCs (Figure 1G) and attracted our attention. Analysis of the circBase database revealed that hsa_circ_0000311 was formed by head-to-tail splicing of exons 2 and 3 of SDHAF2, which is subsequently termed as circSDHAF2 in the remainder of the article (Figure 1H). The back-splice junction site of circSDHAF2 was validated via Sanger sequencing (Figure 1H). We further determined that circSDHAF2 was more stable than its linear form by treatment with actinomycin D and RNase R (Figure 1I-J). Moreover, nuclear-cytoplasmic fractionation experiments and FISH assays revealed that circSDHAF2 was mainly located in the cell cytoplasm (Figure 1K-L).
To determine the functional importance of circSDHAF2 in MES GSCs, we constructed GSC20, GSC267 and GSC28 cells in which circSDHAF2 was knocked down or overexpressed. The expression efficiency was confirmed via qPCR (Supplementary Figure S4A-B). We then performed sphere formation assays and ELDA on MES GSCs (GSC20, GSC267 and GSC28 cells), which revealed that circSDHAF2 significantly promoted neurosphere expansion and the sphere-forming ability of GSCs (Figure 1M-N, Supplementary Figure S4C-F). Moreover, circSDHAF2 upregulated MES-GBM cell markers CD44 and YKL40 (also known as CHI3L1) (Figure 1O, Supplementary Figure S4G). Additionally, in vivo data also showed that knockdown of circSDHAF2 dramatically restrained tumor progression and improved the prognosis of mice, whereas overexpression of circSDHAF2 showed the reverse results (Figure 1P-R). Moreover, IF analysis showed that knockdown of circSDHAF2 downregulated MES-GBM cell markers (Supplementary Figure S4H). Collectively, our findings suggested that circSDHAF2 had an important function in regulating MES-GBM cell formation, suggesting a possible relationship with acquired anti-PD-1 resistance.
3.3 circSDHAF2 stabilized the ITGA5 protein to promote MES-GBM cell formation
A multitude of research has demonstrated that circRNAs retained in the cytoplasm are capable of binding proteins to regulate cellular processes [23]. We then conducted RNA pull-down and MS sequencing to look for the potential proteins that combine with circSDHAF2, and 282 proteins were detected (Supplementary Table S6). Furthermore, we selected the survival time, MES-GBM, hypoxic MDM and immune scores of each sample from the TCGA-GBM cohort as the traits, constructed a scale-free co-expression network for these 282 genes via the R package “WGCNA”, and generated four modules. The results showed that the brown module was strongly correlated with these traits (Figure 2A, Supplementary Figure S5A-B). We found that ITGA5 was most strongly correlated with the brown module (module membership R = 0.82, Supplementary Table S7), thus we speculated that it was a potential interacting protein for circSDHAF2. We further verified that circSDHAF2 could interact with ITGA5 using RNA pull-down and RIP-qPCR assays (Figure 2B-C, Supplementary Figure S5C). Subsequent FISH-IF assays confirmed the cytoplasmic colocalization of endogenous circSDHAF2 and ITGA5 in MES GSCs (Supplementary Figure S5D). We then applied the RNAfold WebServer database to predict the secondary structure of circSDHAF2, which was divided into four major substructures (S1, S2, S3 and S4) (Figure 2D). RNA pull-down assays revealed that circSDHAF2 S2 bound to ITGA5 as efficiently as did the full-length circSDHAF2, while the other substructures lost their binding ability (Figure 2E). These findings indicated that circSDHAF2 physically interacted with ITGA5 in the cytoplasm. We further showed that circSDHAF2 knockdown decreased the protein level of ITGA5 (Figure 2F), and decreased the half-life of the ITGA5 protein via CHX-chase assays (Figure 2G), but did not affect the RNA expression of ITGA5 (Supplementary Figure S5E). Moreover, ITGA5 ubiquitination was elevated in circSDHAF2-knockdown GSC20 and GSC267 cells compared with that in NC groups (Figure 2H), indicating that circSDHAF2 regulated the ITGA5 protein expression through ubiquitin-proteasome activity.
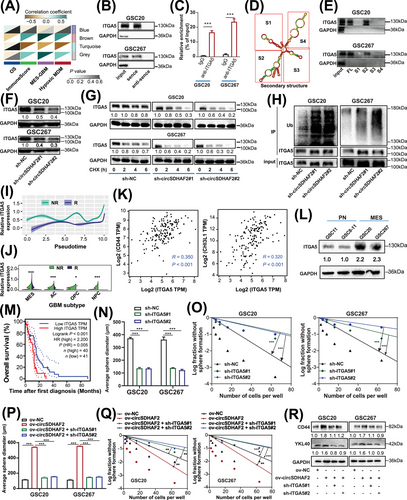
A better understanding of the end-stage differentiation status of GBM cell subsets may provide attractive therapeutic targets for treating GBM in the clinic. We then conducted Monocle2 pseudotime analysis, and unveiled a progressive rise in ITGA5 expression levels alongside the progression of GBM, with notably higher expression in non-responders compared with responders (Figure 2I). Furthermore, a violin plot revealed that ITGA5 was highly expressed in MES-GBM cells and was more highly expressed in non-responders than in responders (Figure 2J). Further analysis revealed that ITGA5 was positively correlated with MES-GBM markers CD44 and CHI3L1 (Figure 2K). Furthermore, ITGA5 expression was upregulated in MES-GSCs (Figure 2L). Additionally, the survival analysis revealed that GBM patients with high ITGA5 expression presented a worse prognosis, compared with those with low ITGA5 expression in the TCGA-GBM dataset (Figure 2M). IF of GBM patients further demonstrated that ITGA5 expression was higher in tumor tissues than in peripheral normal tissues (Supplementary Figure S5F). Furthermore, our functional experiments revealed that ITGA5 significantly upregulated the expression of MES-GBM markers (Supplementary Figure S5G) and promoted tumorsphere expansion and sphere formation in GSCs (Figure 2N-O, Supplementary Figure S5H-J). Further we found that circSDHAF2 overexpression increased sphere-forming ability and upregulated the expression of MES markers, which could be reversed by knocking down ITGA5 (Figure 2P-R, Supplementary Figure S6). To sum up, our findings demonstrated that circSDHAF2 physically interacted with ITGA5 in the cytoplasm to increase its stability, which in turn promotes MES-GBM cell formation to acquire anti-PD-1 resistance.
3.4 circSDHAF2 stabilized the ITGA5 protein by facilitating B4GALT1-mediated N-glycosylation
The second most highly correlated protein with the WGCNA brown module was B4GALT1 (Supplementary Table S7), an N-glycosyltransferase that can catalyze protein N-glycosylation. N-glycosylation is a prevalent posttranslational modification that can stabilize proteins, help proteins migrate to the right location, or guide proper folding of molecular chaperones [24, 25]. We found that ITGA5 has abundant N-glycosylation sites, as predicted using the UniProt database). However, whether N-glycosylation affects the protein stability of ITGA5 has not been fully investigated. We subsequently explored the glycosylation pattern of ITGA5 in MES-GSCs. We observed that the N-glycosylation (130 kDa) and protein expression (100 kDa) of ITGA5 were downregulated after treatment with the N-glycosylation inhibitor TM at different doses (Figure 3A), whereas the RNA level of ITGA5 was not changed (Supplementary Figure S7A). Further analysis revealed that the ITGA5 protein degraded more rapidly in GSC20 and GSC267 cells in the TM-treated group, compared with the vehicle group (Figure 3B), suggesting that N-glycosylation stabilized the ITGA5 protein. Further investigation revealed that circSDHAF2 overexpression increased the protein level of ITGA5, which was abrogated by TM treatment (Figure 3C). Further analysis revealed that circSDHAF2 overexpression increased the sphere-forming ability, which could be reversed by TM (Supplementary Figure S7B-D). To determine whether circSDHAF2 antagonized ITGA5 glycosylation by promoting its ubiquitination to stabilize protein expression, we treated cells with TM and found that the ubiquitination level of ITGA5 was increased (Figure 3D). Further rescue experiments confirmed that the circSDHAF2-induced decrease in the ubiquitination level of the ITGA5 protein could be abrogated by TM inhibitors (Figure 3E). In summary, our findings indicated that circSDHAF2 enhanced the N-glycosylation of ITGA5, thereby inhibiting its ubiquitination and degradation.
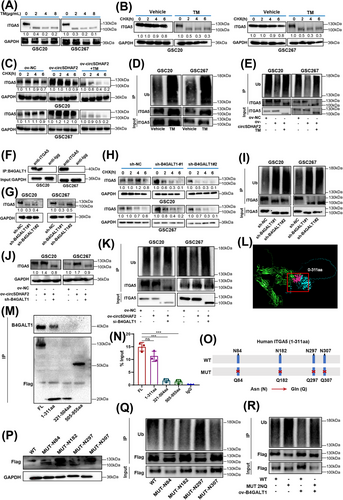
We next asked whether B4GALT1 regulates the aberrant glycosylation of ITGA5 in MES-GSCs. Firstly, we found that knockdown of circSDHAF2 did not affect protein expression of B4GALT1 (Supplementary Figure S7E). Co-IP assays demonstrated that B4GALT1 interacted with ITGA5 in GSC20 and GSC267 cells (Figure 3F). We then constructed GSC20 and GSC267 cell lines in which B4GALT1 was overexpressed or knocked down. The expression efficiency was verified by Western blotting assays (Supplementary Figure S7F-G). Compared with the NC group, B4GALT1 knockdown decreased the ITGA5 protein level (Figure 3G), while the RNA level of ITGA5 did not change (Supplementary Figure S7H). Then GSCs were treated with PNGase F, revealing that PNGase F blocked B4GALT1-mediated glycosylation of ITGA5 (Supplementary Figure S7I). We further observed that the B4GALT1-knockdown group presented a shorter half-life via the CHX-chase assays (Figure 3H), and increased the ubiquitination level of ITGA5, compared with the NC group (Figure 3I). The increase in the expression level and decrease in the ubiquitination level of ITGA5 induced by circSDHAF2 overexpression could be reversed by inhibiting B4GALT1 (Figure 3J-K).
To identify the N-glycosylation sites of ITGA5, we first predicted the protein interaction region by using the alphafold2 sequence and found that the 0-311 aa region of ITGA5 could be a potential binding site for B4GALT1 (Figure 3L). A subsequent Co-IP assay confirmed this result (Figure 3M). Further RIP-qPCR assays demonstrated that circSDHAF2 also interacted with this structural domain of ITGA5 (Figure 3N). Next, we mutated the potential glycosylation site, which predicted via UniProt database, from asparagine (Asn, N) to glutamine (Gln, Q) to suppress N-glycosylation (Figure 3O). Further studies revealed that mutations at sites N84 and N297 greatly reduced glycosylation and increased the ubiquitination of ITGA5, compared to mutations at other sites (Figure 3P-Q). Furthermore, the reduction in the ubiquitination level of the ITGA5 protein caused by B4GALT1 overexpression disappeared after mutation of the identified sites (Figure 3R). Collectively, these results indicated that circSDHAF2 stabilized ITGA5 by facilitating B4GALT1-mediated N-glycosylation, thereby preventing its ubiquitin-mediated degradation.
3.5 circSDHAF2 promoted ITGA5 translocation into exosomes by facilitating B4GALT1-mediated N-glycosylation
A hypoxic microenvironment profoundly affects the biological behavior and malignant phenotype of cancer cells and generates local TME niches that induce immune tolerance through complex mechanisms [26-28]. The GSVA of scRNA-seq data revealed that both MES-GBM cells and hypoxic MDMs were typically enriched in pro-oncogenic pathways, such as hypoxia, glycolysis, EMT and angiogenesis, and all enrichment scores of these traits were greater in non-responders, compared with responders (Figure 4A-B). Further ST data analysis revealed that compared with responders, MES-GBM cell and hypoxic MDM subsets were highly enriched in extensive hypoxic regions, and had higher hypoxia and EMT scores in non-responders (Figure 4C). We then isolated the exosomes of hypoxia-stimulated GSCs, which exhibited similar typical cup-shaped morphologies, sizes, and concentrations (Supplementary Figure S8A-B), and performed proteomic sequencing to analyze the protein compositions of both GSCs and exosomes under hypoxic conditions. We found that the integrin membrane protein ITGA5 was significantly upregulated in both the cells and the exosomes (Figure 4D), which was further validated by Western blotting (Figure 4E-F). Glycosylation has been reported to facilitate protein targeting to the plasma membrane [29], a step that is a prerequisite for the sorting of protein cargoes into exosomes. Therefore, we hypothesized that the B4GALT1-regulated N-glycosylation of ITGA5 mediated its translocation to exosomes. We found that hypoxia increased the binding intensity of B4GALT1 to ITGA5 (Figure 4G). Further experiments demonstrated that circSDHAF2 knockdown downregulated ITGA5 expression in exosomes (Figure 4H). Moreover, circSDHAF2 overexpression upregulated the expression of ITGA5 in exosomes, but this effect was abrogated by B4GALT1 knockdown (Figure 4I). These results indicated that circSDHAF2 promoted ITGA5 translocation to exosomes by facilitating the B4GALT1-mediated N-glycosylation of ITGA5.
3.6 Exosomal ITGA5 protein derived from MES-GBM promoted hypoxic MDM polarization
Recent studies have revealed that the integrin family is strongly enriched in tumor cell exosomes and plays an important role in the formation of an immunosuppressive TME [30-33]. Previous studies have shown that the MES-GBM TME is characterized by increased MDM infiltration via C-C motif chemokine ligand 2 (CCL2) [34-36]. We found that knockdown of ITGA5 suppressed signal transducer and activator of transcription 3 (STAT3) activity (Supplementary Figure S5G), a core pathway of MES-GBM formation, which could induce the secretion of CCL2, a classical macrophage chemotactic factor [37]. Our Western blotting also showed that knockdown of ITGA5 inhibited the protein level of CCL2 (Supplementary Figure S5G). Thus, we hypothesized that ITGA5high MES-GBM cells could promote MDM infiltration through CCL2. Our GSEA analysis showed that compared with responders, the macrophage chemotaxis and migration signatures were highly enriched in non-responders (Figure 5A). The expression of CCL2 was higher in MES-GBM cells of non-responders compared with those in responders (Figure 5B). Additionally, pseudotime analysis using Monocle2 indicated a gradual increase in CCL2 expression levels in correlation with tumor progression in non-responders (Figure 5C). Our qPCR results also confirmed that circSDHAF2 upregulated the expression of CCL2, which could be abrogated by ITGA5 knockdown (Figure 5D). We then co-cultured GSCs with THP1-differentiated macrophages (Supplementary Figure S8C), to explore the role of circSDHAF2 in promoting macrophage chemotaxis. Our findings indicated that the conditioned medium derived from GSCs overexpressing circSDHAF2 markedly enhanced the migratory capacity of THP1-differentiated macrophages, an effect that was reversed by the knockdown of ITGA5 (Figure 5E). Consequently, we hypothesized that the hypoxic microenvironment might form a positive feedback loop that promotes the formation of MES-GBM and hypoxic MDM subsets, which were highly enriched in extensive hypoxic regions (Figure 1, 4), leading to the generation of spatially specific adaptive transcriptional programs and ultimately promoting the immune escape of GBM. Subsequent analyses revealed that, compared with monocytes, the hypoxic MDM subset highly expressed SPP1 (Figure 5F), an immunosuppressive molecule with multiple complex functions [38-40]. Thus, we also termed hypoxic MDMs as SPP1+ MDMs. Further TF regulon analysis revealed that RUNX1 was a specific transcription factor for the SPP1+ MDM subpopulation (Figure 5G). We found that SPP1 was a potential transcriptional target of RUNX1 (Supplementary Table S8), as predicted by the hTFtarget database [41]. We also confirmed that RUNX1 knockdown successfully inhibited SPP1 expression (Figure 5H). We further predicted two binding sites of RUNX1 on the SPP1 promoter (Figure 5I) via the JASPAR database. ChIP-qPCR verified that RUNX1 could bind to the promoter of SPP1 (Figure 5J), and subsequent luciferase reporter assays confirmed that site 1 of the SPP1 promoter region was recognized by RUNX1 (Figure 5K). Expression analysis revealed that both SPP1 and RUNX1 were highly expressed in the SPP1+ MDM subpopulation and were more highly expressed in non-responders than in responders (Figure 5L). Exosomes have been shown to deliver integrins to receptor cells and activate focal adhesion kinase (FAK) signalling [42], which has been shown to regulate RUNX1 activity [43, 44]. Therefore, we proposed that exosomes could transmit the ITGA5 protein to MDMs and activate FAK/RUNX1 signaling. Our subsequent Western blotting analysis revealed that exosomes collected from ITGA5-knockdown MES-GSCs inhibited FAK/RUNX1 pathway activity and downregulated SPP1 expression in THP-1 differentiated macrophages (Figure 5M). However, ITGA5-overexpressing MES-GSC-derived exosomes activated the FAK/RUNX1 pathway and upregulated SPP1 expression, which could be abrogated by inhibiting exosomal ITGA5 expression with a neutralizing anti-ITGA5 antibody (Figure 5N), suggesting that the MES-GSC exosome-derived ITGA5 membrane protein mediated the formation of immunosuppressive SPP1+ MDMs.
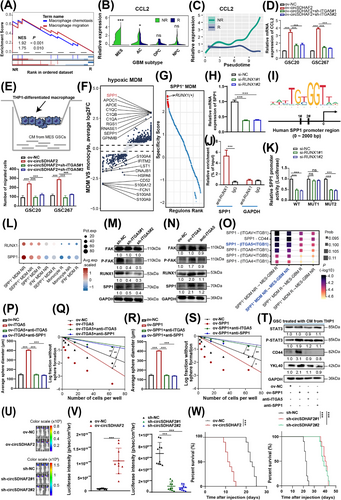
Previous studies have shown that infiltrating MDMs in the TME secreted SPP1 to bind to cell surface integrins to promote tumor progression [45, 46]. Our further scRNA CellChat analyses revealed that SPP1+ MDMs could interact with MES-GBM through the SPP1-ITGA5 pathway and that the interaction intensity of this pathway was the strongest among hypoxic MDM non-responders and MES-GBM non-responders (Figure 5O). We detected the expression of SPP1 in each group of macrophages and confirmed that GSCs overexpressing ITGA5 could significantly promote the expression of SPP1 in macrophages, and this phenomenon could be rescued by anti-ITGA5 or anti-SPP1 (Supplementary Figure S9A). Subsequent co-culture experiments indicated that ITGA5 overexpression increased the sphere-forming ability of GSCs, which could be attenuated by a neutralizing anti-SPP1 and anti-ITGA5 (Figure 5P-Q, Supplementary Figure S9B). Moreover, the overexpression of SPP1 in macrophages increased the sphericality of GSCs and upregulated the expression of MES markers and activated STAT3 signaling, which could be attenuated by blocking the crosstalk pathway with anti-ITGA5 or anti-SPP1 (Figure 5R-T, Supplementary Figure S9C). Our findings indicated that the SPP1-ITGA5 pathway played a significant role in the interactions between MES-GBM cells and SPP1+ MDMs. Moreover, to further investigate the immunosuppressive polarization effect of circSDHAF2 on macrophages in vivo, MES-GSCs with circSDHAF2 overexpression or knockdown and the corresponding NC vector were co-implanted with macrophages into the nude mouse brains. Compared with the NC group, circSDHAF2-overexpressing group dramatically promoted tumor progression and shortened the survival time of mice, whereas circSDHAF2 knockdown had the opposite effect (Figure 5U-W). Further IF experiments revealed that SPP1 expression in macrophages (CD68+) was upregulated in the circSDHAF2 overexpression group (Supplementary Figure S9D). Therefore, these results demonstrated that the circSDHAF2-mediated ITGA5-SPP1 pathway participated in a self-amplifying mechanism involving crosstalk between MES-GBM cells and SPP1+ MDMs, promoting the formation of localized immunosuppressive TME niches.
3.7 SPP1+ MDMs promoted GBM immune escape through SPP1-induced T-cell dysfunction
Dysfunctional T-cell status in the TME is a major contributor to immunotherapy resistance [47, 48]. Recently, a study reported that macrophages can induce T-cell exhaustion by acting on the ITGB1 receptor on T cells via the secreted SPP1 protein [49]. CellChat communication analyses of SPP1+ MDMs and T cells revealed that SPP1+ MDMs could interact with multiple T-cell subpopulations via SPP1-integrin α5β1 (ITGA5+ITGB1) signaling, and the interaction intensity were stronger between SPP1+ MDM (hypoxic MDM) non-responders and CD4/8+ T cell non-responders, compared with the interactions between other groups (Figure 6A). Furthermore, we overexpressed SPP1 in macrophages and established a co-culture system with primary human T cells. We found that the proliferative capacity and IFN-γ expression levels of T cells in the SPP1-overexpressing group were significantly reduced, whereas the expression of the exhaustion marker thymocyte selection associated high mobility group box (TOX) was significantly increased, which could be abrogated by anti-ITGA5 (Figure 6B-C). Our survival analyses of patients in the TCGA-GBM cohort revealed that patients with simultaneous high expression of SPP1 and ITGA5 presented the worst prognosis (Figure 6D). Further spatial transcriptome analysis revealed that patients with high ITGA5 and SPP1 expression had less therapeutic benefit (Figure 6E). We also found that ITGB1 was highly enriched in SPP1+ MDM enriched area (Supplementary Figure S10A). Patients with high expression of Signatures (SPP1, ITGA5 and ITGB1) exhibited the shortest survival time in the TCGA-GBM cohort (Supplementary Figure S10B). Therefore, we proposed that the formation of SPP1+ MDMs induced by MES-GBM cells via exosomal ITGA5 could increase T-cell dysfunction through SPP1-ITGA5 signaling and ultimately promoted anti-PD-1 resistance in GBM. Blocking SPP1+ MDM-mediated immunosuppressive TME formation with anti-ITGA5 may be an effective way to improve the therapeutic efficacy of anti-PD-1 in GBM. To validate our conclusion, we found that ITGA5 and SPP1 were highly enriched in mouse CT2A cells (Supplementary Figure S11A). Then we confirmed that B4GALT1 mediated the N-glycosylation of ITGA5 and stabilized its protein expression (Supplementary Figure S11B-H). Further in vivo experiments showed that anti-ITGA5 could effectively improve the therapeutic efficacy of anti-PD-1 in mice (Figure 6F-G). Moreover, IF staining showed that the combination therapy could significantly promote the killing function of T cells (Supplementary Figure S12A-B) and inhibited the infiltration of SPP1+ MDMs, while suppressing the expression of CD44 in GBM cells (Supplementary Figure S12C-D). Sum up, these results indicated that targeting the SPP1-ITGA5 pathway is a viable strategy for improving the efficacy of anti-PD-1 immunotherapy.
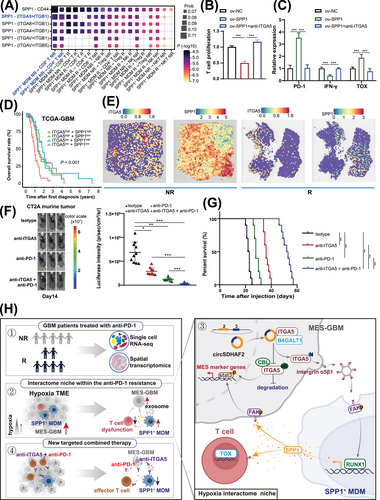
4 DISCUSSION
Neoadjuvant immune checkpoint blockade therapy is effective in only a small percentage of GBM patients, and the mechanism of non-responsiveness have not been fully revealed. In this study, we used scRNA-seq and ST data from published datasets to identify the factors affecting the response to anti-PD-1 therapy, revealing that MES-GBM cells (with the worst prognosis) and SPP1+ MDMs (with complex functions), which form niche-specific regions of space enrichment, preferentially accumulated in non-responders. We then determined that circSDHAF2 was positively associated with these two anti-PD-1-resistant cell subpopulations. We functionally confirmed that circSDHAF2 facilitated the formation of localized MES-GBM and SPP1+ MDM subpopulations, leading to a spatially specific adaptive immunosuppressive microenvironment. Mechanistically, we found that circSDHAF2 promoted MES-GBM formation by stabilizing the ITGA5 protein through B4GALT1-mediated N-glycosylation. Moreover, the N-glycosylation of the ITGA5 protein facilitated its translocation to exosomes and subsequent transmission to MDMs to activate the FAK/RUNX1 pathway to induce the formation of immunosuppressive SPP1+ MDM subpopulations, which in turn maintained the MES-GBM status and activated the SPP1-ITGA5 pathway to induce T-cell dysfunction, ultimately promoting immune escape in GBM. Our findings demonstrated that the therapeutic efficacy of anti-PD-1 can be enhanced by inhibiting the interactions between MES-GBM cells, SPP1+ MDMs and T cells through the use of an ITGA5-blocking antibody (Figure 6H).
Recent single-cell multiomics techniques for clustering subpopulations in the GBM tissues in an unbiased manner have greatly contributed to our understanding of the molecular diversity of GBM [50, 51], especially for TAMs, and thus targeting TAM therapies are also emerging as important precision medicine strategies [8, 52, 53]. Our study demonstrated the heterogeneity of MDMs in the GBM tumor milieu and revealed that MDMs may have different impacts depending on their subpopulations, casting doubt on the traditional M1/M2 dichotomous polarization system. MDM phenotypes are likely to be complex and should be further explored for their potential use as potential biomarkers or therapeutic targets. In this work, we identified a new subset of MDMs with high expression of SPP1 that are activated by the transcription factor RUNX1 (Figure 5); these cells could form localized interaction loops with MES-GBM cells, leading to the generation of spatially specific adaptive transcriptional programs that ultimately promote anti-PD-1 resistance in GBM. Additionally, we found that the MES-GBMhigh combined with hypoxic MDMhigh group seemed to have very low survival (Figure 1D). Recently, several studies identified hypoxic TAMs as distinct MDM clusters enriched in the hypoxic ecological niche of GBMs and formed by hypoxic tumor cues to stimulate angiogenesis [54] and sequestered cytotoxic T lymphocytes (CTLs), where they were reprogrammed toward a dysfunctional state [55]. Thus, hypoxic MDMs promoted multiple aspects of tumor progression, and their abundance is strongly associated with poor prognosis in multiple cancer types [56].
Osteopontin (OPN), officially designated as SPP1, is an intriguing and multifunctional protein that is expressed in various cell types. It is involved in intercellular communication as well as in the composition and function of the extracellular matrix [46, 57]. Nevertheless, there is limited understanding of SPP1 regulation within the central nervous system (CNS). Integrins, which form heterodimeric complexes, possess the distinctive capability of transducing signals from the cell surface to the nucleus and vice versa. Their signaling specificity is further enhanced by their varied expression patterns that are tailored to specific cell and tissue types [58, 59]. Key insights into SPP1's role in disease progression hinge on identifying the cells that express OPN/SPP1 and the receptors activated within the local microenvironment. Our research has shown that the integrin family member ITGA5 is highly expressed in MES-GBM cells. Furthermore, it can be transported to exosomes through glycosylation, thereby inducing the formation of SPP1-positive MDMs (Figures 3-5).
This study has some limitations. To explore the interaction between GSCs and MDMs, we selected THP1-differentiated macrophages as a model to validate our findings. Monocytes derived from human peripheral blood mononuclear cell (PBMC) as a research model is undoubtedly optimal, while the high variability from individual donors has become a prevalent problem in the application of monocytes derived from PBMC, resulting in less reproducible results [60]. Thus, we made a compromise choice based on the previous study, which showed that co-culture experiments of THP-1 cells with other cell types could serve as an alternate for studying cell-to-cell interactions in vitro and could better simulate in vivo conditions by comparing them with primary monocytes [60, 61]. Going forward, we hope to utilize human GBM-specific mutation-driven mouse models as preclinical platforms to validate newly identified targets, which in turn will enable the development of better therapeutic strategies to provide meaningful clinical outcomes.
5 CONCLUSIONS
Our study reported that MES-GBM cells and SPP1+ MDMs were the main anti-PD-1-resistant cell subpopulations. Furthermore, we revealed that circSDHAF2 promoted MES-GBM cell formation by stabilizing the ITGA5 protein through B4GALT1-mediated N-glycosylation. Meanwhile, the N-glycosylation of the ITGA5 protein facilitated its translocation into exosomes and subsequent delivery to MDMs to induce the formation of immunosuppressive SPP1+ MDM subpopulations, which in turn maintained the MES-GBM cell status and induced T-cell dysfunction via the SPP1-ITGA5 pathway, ultimately promoting GBM immune escape. More importantly, we demonstrated that anti-ITGA5 enhanced anti-PD-1-mediated antitumor immunity. This study laid a theoretical foundation for in-depth analyses of the mechanism of specific immune tolerance in MES GBMs to develop more precise treatment strategies for GBM.
AUTHOR CONTRIBUTIONS
Rongrong Zhao and Ziwen Pan designed the study and performed all the experiments. Yanhua Qi, Wei Qiu and Jiawei Qiu were responsible for clinical sample collection and subsequent sample delivery. Boyan Li, Zijie Gao, Weijie Tang and Xiaofan Guo helped to revise the manuscript. Gang Li, Hao Xue and Lin Deng supervised the study. All authors have read and approved the article.
ACKNOWLEDGMENTS
We thank Dr. Frederick F. Lang and Dr. Krishna P.L. Bhat for providing the GSCs. We appreciate the help from members in the Laboratory of Brain Function Remodeling, Neurosurgery, and patients who participated in these studies. Thanks to KS Official Customer Team (Chongqing, China) team for their help of data analysis tutorial and visual code. This work was supported by grants from the National Natural Science Foundation of China (No. 82473403, 82273286, 82273195), the Fundamental Research Funds for the Central Universities (2022JC019), Jinan Science and Technology Bureau of Shandong Province (2021GXRC029), Natural Science Foundation of Shandong Province of China (ZR2021LSW025, ZR2023LZL004), Taishan Pandeng Scholar Program of Shandong Province (tspd20210322), Youth Taishan Scholar Program of Shandong Province (tsqn202211316).
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The human primary GBM samples in this study were obtained from Qilu Hospital of Shandong University, according to the protocol approved by the Ethics Committee of Qilu Hospital of Shandong University. All participants provided written informed consent, and the study was approved by the Ethics Committee on Scientific Research of Shandong University Qilu Hospital (approval number: KYLL-2022(ZM)-439).
All the experimental procedures for animals were approved by the Animal Care and Use Committee of the Qilu Hospital of Shandong University (Approval No. DWLL-2021-122).
Open Research
DATA AVAILABILITY STATEMENT
The circRNA sequencing data of our local samples have been deposited in the Genome Sequence Archive (CRA002339). The RNA pull down and MS data used in this study have been deposited to the ProteomeXchange Consortium via the iProX partner repository (IPX0008241000). The remaining data are available within the article, supplementary information or available from the authors upon request.