A strong internal promoter drives massive expression of YEATS-domain devoid MLLT3 transcripts in HSC and most lethal AML
Abbreviations
-
- AF9
-
- Protein AF9
-
- AFF1/4
-
- ALF Transcription Elongation Factor 1 or 4
-
- AHD
-
- ANC1 (nuclear anchorage protein 1) homology domain
-
- AML
-
- Acute myeloid leukemia
-
- BCOR
-
- BCL6 (B Cell Lymphoma 6) corepressor
-
- CBX8
-
- Chromobox 8
-
- CD14
-
- Monocyte differentiation antigen CD14
-
- CD34
-
- Hematopoietic progenitor cell antigen CD34
-
- DOT1L
-
- Disruptor of telomeric silencing 1 like (a histone lysine methyltransferase)
-
- ELN2017
-
- European LeukemiaNet 2017
-
- GATA2
-
- GATA Binding Protein 2
-
- HSC
-
- Hematopoietic stem cell
-
- IUCT
-
- Institut Universitaire du cancer de Toulouse
-
- KMT2A
-
- Lysine (K) methyl-transferase 2A
-
- MECOM
-
- MDS1 (Myelodysplasia syndrome 1) and EVI1 (Ecotropic virus integration site 1) complex locus
-
- MLLT1/3
-
- Mixed-lineage leukemia translocated to, 1 or 3
-
- PAF1
-
- RNA polymerase II-associated factor 1 homolog
-
- Poly-Ser
-
- Poly-serine
-
- P-TEFb
-
- Positive transcription elongation factor b
-
- RNApol II
-
- RNA polymerase II
-
- RUNX1
-
- Runt-related transcription factor 1
-
- SEC
-
- Super elongation complex
-
- TFIID
-
- Transcription factor II D
-
- TP53
-
- Tumor protein p53
-
- YEATS
-
- Yaf9, ENL, AF9, Taf14, and Sas5 (the five first proteins shown as containing such domain)
The AF9 (protein AF9) transcription factor, encoded by MLLT3 (mixed-lineage leukemia translocated to 3) on chromosome 9, functions as a chromatin reader. Through its N-terminal YEATS (Yaf9, ENL, AF9, Taf14, and Sas5) protein domain, it interacts with acetylated [1] or crotonylated [2] histone H3, as well as with the PAF1 (RNA polymerase II-associated factor 1 homolog) and P-TEFb (positive transcription elongation factor b) components of the super elongation complex (SEC). AF9 also interacts through its poly-serine domain (Poly-Ser) with the TFIID (Transcription factor II D) subunit of the RNA polymerase II (RNApol II) complex. In addition, its C-terminal transactivation domain, AHD (nuclear anchorage protein1 homology domain), binds other SEC components, such as AFF1 and AFF4 (ALF transcription elongation factor 1 or 4), as well as transcription regulators CBX8 (chromobox 8), DOT1L (disruptor of telomeric silencing 1 like), and BCOR (B cell lymphoma 6 corepressor), as reviewed by Kabra & Bushweller [3] (Figure 1A). Thus, MLLT3 is an integral part of the SEC, which is essential for optimizing the catalytic activity of RNApol II transcription at specific genome loci.
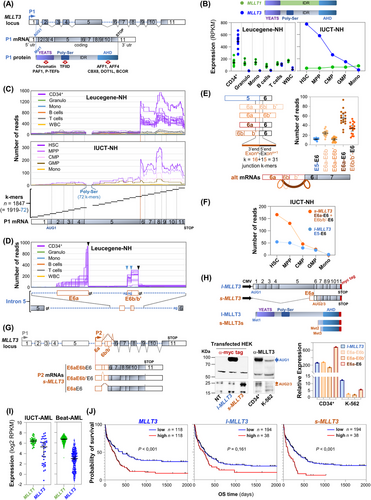
Several studies have indicated that MLLT3 is highly and specifically expressed in hematopoietic stem cells (HSCs), but it is rapidly and significantly downregulated during normal differentiation or immediately after HSCs are placed in ex vivo culture. In both scenarios, this shutdown parallels the rapid loss of stemness. Consistently, ectopic expression of MLLT3 significantly prolongs self-renewal capacity of HSCs, suggesting that MLLT3 is a crucial factor for HSC maintenance [4].
Based on standard quantification of RNA-sequencing reads mapping to the MLLT3 locus, we first confirmed that, compared to the MLLT1 paralogue used as an internal control, MLLT3 expression was significantly higher in CD34+ cells than in mature lymphocytes, granulocytes, or monocytes from healthy samples of the Leucegene dataset (Leucegene-NH, detailed in Supplementary Information) (Figure 1B, left panel). To refine this observation, made in CD34+ cells containing a mixture of progenitors but only a few HSCs, we repeated the analysis in HSCs and various stages of progenitor cells sorted form healthy donors (IUCT-NH, detailed in Supplementary Information). The data clearly confirmed that MLLT3 is highly expressed in HSCs but rapidly declines as differentiation proceeds (Figure 1B, right panel).
However, closer examination using a k-mer approach (described in Materials and Methods in Supplementary Information), which visualized RNA-sequencing read alignment along the 11 exons (E1-E11) of the reference MLLT3 transcript, revealed an unexpected profile. Strikingly, the substantial MLLT3 expression detected in HSCs was driven by a sharp and pronounced increase in reads starting precisely at the first nucleotide of exon E6 (Figure 1C). This unexpected profile was absent when examining MLLT1 expression in the hematopoietic lineage (Supplementary Figure S1). These findings suggest the expression of one or more 5’ end shortened MLLT3 transcripts arising from a hematologic lineage-specific internal promoter located in MLLT3 intron 5.
A new set of specific and successive k-mers covering the entire MLLT3 intron 5 revealed the presence of two novel segments retained in poly(A)+ RNAs. Apart from the 5’ end of the first segment, which lacked a clearly defined starting point consistent with a probable transcription start site, consensus donor and acceptor splice sites flanked these two segments. These unexpected exons were designated as exon E6a and exon E6b/b’ (Figure 1D). Additional k-mer analyses confirmed that these exons were spliced to exon E6, resulting in three possible splice variants: E6a-E6b-E6, E6a-E6b'-E6, and E6a-E6, with the E6a-E6 variant being the most predominant (Figure 1E and Supplementary Figure S2). This assortment of novel CD34+-specific alternative exons was further validated using standard Sanger sequencing following RT-PCR amplification with specific primers (Supplementary Figure S3).
Next, exploration of public CHIP-Seq (chromatin immunoprecipitation followed by sequencing) and CAGE (mRNA 5’ cap analysis of gene expression) datasets revealed the existence of an alternative P2 promoter in addition to the canonical P1 promoter. Associated with an active promoter H3K4me3 mark and a CAGE peak, this P2 promoter, found exclusively in immature CD34+ cells but absent in CD14+ monocytes, was predicted to drive the expression of transcripts beginning with exon E6a (Supplementary Figure S4). Transcript-specific k-mer quantifications confirmed that the exceptionally high overall level of MLLT3 observed in HSCs was primarily due to these shorter transcripts starting with exon E6a. We have collectively named these shorter transcripts s-MLLT3, in contrast to the reference full-length MLLT3 transcript, referred to as l-MLLT3 (Figure 1F).
These findings revealed the existence of an HSC-specific internal promoter (P2) that drives the expression of shorter MLLT3 transcripts (s-MLLT3 mRNAs, Figure 1G). To evaluate the translational potential of these s-MLLT3 transcripts, we cloned a C-terminal myc-tagged version of the predominant E6a-E6 variant into an expression vector (Figure 1H, top), and assessed its protein expression capacity in the HEK (human embryonic kidney) cell line by western blotting. Compared with a similar vector encoding l-MLLT3, the E6a-E6 s-MLLT3 transcript produced shorter s-MLLT3 proteins, (Figure 1H, bottom left). A western blotting analysis of endogenous proteins confirmed the existence of these shorter forms in CD34+ cells but not in the K-562 leukemic cell line, which served as a low-expressing control (Figure 1H, bottom middle). Isoform-specific RT-qPCR quantification further corroborated that, compared to CD34+ cells and full-length l-MLLT3, expression of the three shorter forms (E6a-E6b, E6a-E6b’, and E6a-E6) was very low in K-562 cells (Figure 1H, bottom right). These western blot results identified at least two distinct s-MLLT3 proteins, likely arising from alternative translation initiation codons (AUG2 and AUG3) located in exon 7, producing proteins that retain the C-terminal AHD transactivation domain but lack the YEATS and Poly-Ser domains (Supplementary Figure S5).
Interestingly, the shorter MLLT3 alternative transcripts initiate within intron 5, which is also a frequent site of chromosomal translocation in acute myeloid leukemia (AML). Specifically, MLLT3 intron 5 is the primary site of the t(9;11) chromosomal translocation, leading to fusion with KMT2A (lysine (K) methyl transferase 2A) [5]. KMT2A is a chromatin writer that deposits epigenetic marks indicating active transcription at specific loci, particularly the HOX genes required for hematopoiesis [6]. The resulting KMT2A-MLLT3 fusion transcript encodes a chimeric protein composing the N-terminal third of KMT2A fused to the C-terminal portion of MLLT3, which lacks the YEATS domain but retains the AHD transactivation domain (Supplementary Figure S6). To investigate global MLLT3 expression in AML, we analyzed two RNA-Seq datasets: IUCT-AML [7] and Beat-AML [8] (detailed in Supplementary Information). Compared with MLLT1, the expression range of MLLT3 was broader, showing a > 120-fold amplitude across samples (Figure 1I). Isoform- specific k-mers targeting alternative (E6a-E6 + E6b/b'-E6 for s-MLLT3) and canonical (E5-E6 for l-MLLT3) exon-exon junctions revealed no correlation between s-MLLT3 and l-MLLT3 expressions. Approximately 20% of samples exhibited high s-MLLT3 levels, whereas in other samples, s-MLLT3 was undetectable despite l-MLLT3 expression (Supplementary Figure S7). This finding suggests differential regulation of the two promoters and/or the resulting transcripts in AML.
We next investigated whether the ∼20% of AML samples with high s-MLLT3 expression represented a distinct clinical entity. Samples which were either MECOM+ (myelodysplasia syndrome 1 and EVI1 complex locus) or GATA2-MECOM (Supplementary Figure S8), and those with mutant RUNX1 (runt-related transcription factor 1) and/or TP53 (tumor protein p53) showed higher levels of l-MLLT3, with even greater levels of s-MLLT3 (Supplementary Figure S9, left and middle). No correlation with the KMT2A-MLLT3 translocation was observed. Conversely, NPM1 (nucleophosmin 1)-mutated samples exhibited very low levels of both transcripts (Supplementary Figure S9, left and middle). Notably, elevated s-MLLT3 or l-MLLT3 levels were associated with an adverse ELN2017 (European LeukemiaNet 2017) score (Supplementary Figure S9, right). Median expression-based group separation revealed that patients with the highest overall MLLT3 expression had worse survival outcomes (Figure 1J, left). However, given the lack of correlation between s-MLLT3 and l-MLLT3 expression in AML samples, we assessed their independent impacts on survival. Isoform-specific k-mers showed that s-MLLT3 (but not l-MLLT3) expression significantly influenced poor patient survival (Figure 1J, middle and right panels).
In conclusion, these findings demonstrate the existence of an internal promoter within the MLLT3 locus, driving expression of 5’-end-shortened transcripts encoding an AF9 protein lacking the YEAST chromatin reader domain. These alternative transcripts are highly expressed in HSCs and in ∼20% of AML patients with the worst survival outcomes. These results suggest that the role of the MLLT3 locus in HSCs and in AML should be re-evaluated, considering the expression of this YEATS-domain-devoid AF9 transcription factor.
AUTHOR CONTRIBUTIONS
Stéphane Pyronnet wrote the manuscript. Chloé Bessière, Ahmed Zamani, Sandra Dailhau, Christian Récher, Marina Bousquet, and Stéphane Pyronnet contributed to the study design, conception, and data analysis. Ahmed Zamani, Romain Pfeifer, and Marina Bousquet designed and performed biological experiments. Chloé Bessière, Sandra Dailhau, Camille Marchet, Benoit Guibert, Anthony Boureux, Raïssa Silva Da Silva, Nicolas Gilbert, and Thérèse Commes developed the k-mer-based bioinformatics tools. Fabienne Meggetto, Christian Touriol, and Marina Bousquet provided comments on and contributed to editing the manuscript.
ACKNOWLEDGEMENTS
Some of the results presented in this publication are based on data generated by the Leucegene group, primarily based at IRIC in Montreal, Canada, and supported by Genome Canada and Genome Québec. This data was made possible through human AML specimens provided by the BCLQ in Montreal, Canada.
CONFLICT OF INTEREST STATEMENT
Christian Récher declares a consulting or advisory role with Abbvie, Amgen, Astellas, BMS, Boehringer, Jazz Pharmaceuticals, and Servier, and has received research funding from Abbvie, Amgen, Astellas, BMS, Iqvia, and Jazz Pharmaceuticals. All other authors declare no conflict of interest.
FUNDING INFORMATION
This work was funded by INSERM, Institut Universitaire du Cancer-Toulouse (IUCT), Labex Toucan, Fondation Leucémie Espoir, Ligue Régionale Contre le Cancer, Fondation ARC, Association Laurette Fugain, Agence Nationale de la Recherche (ANR-18-CE45-0020 Transipedia) and (ANR-22-CE45-0007 full-RNA). Chloé Bessière was supported by Fondation de France, Ahmed Zamani and Raïssa Silva Da Silva by Ligue Nationale Contre le Cancer, Romain Pfeifer by Ministère de l'Enseignement Supérieur et de la Recherche and Fondation ARC, Sandra Dailhau by Ministère de la Santé and Institut National du Cancer (INCA, PRT-K-2022-184CircOma).
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
In accordance with French law, each anonymous volunteer donor or patient was informed, and the HIMIP collection has been declared to the Ministère de l'Enseignement Supérieur et de la Recherche (DC 2008-307). A transfer agreement was obtained (AC 2008-129) after approval by the local ethical committee, Comité de Protection des Personnes Sud-Ouest et Outremer II, and the local Research Ethics Committee of the Etablissement Français du Sang (Toulouse, France, agreement #21PLER2021-007). Clinical and biological annotations have also been declared to the Comité National Informatique et Libertés (CNIL). This study was conducted in accordance with the Declaration of Helsinki.
Open Research
DATA AVAILABILITY STATEMENT
The raw and processed RNA-sequencing data generated in this study have been deposited at the National Center for Biotechnology Information Gene Expression Omnibus (repository number GSE62852).




