The interplay between natural killer cells and pancreatic stellate cells in pancreatic ductal adenocarcinoma
Abbreviations
-
- AM
-
- acetyoxymethyl
-
- aPSC
-
- activated pancreatic stellate cell
-
- ATRA
-
- all-trans retinoic acid
-
- CAF
-
- cancer associated fibroblast
-
- CXCL
-
- C-X-C chemokine ligand
-
- ECM
-
- extracellular matrix
-
- hTERT
-
- telomerase reverse transcriptase
-
- MFI
-
- mean fluorescence intensity
-
- NK
-
- natural killer
-
- PDAC
-
- pancreatic ductal adenocarcinoma
-
- PSC
-
- pancreatic stellate cell
-
- qPSC
-
- quiescent pancreatic stellate cell
-
- RAR
-
- retinoic acid receptor
-
- SASP
-
- senescence-associated secretory phenotype
-
- STARPAC
-
- stromal targeting for pancreatic cancer
-
- TME
-
- tumour microenvironment
Pancreatic ductal adenocarcinoma (PDAC) remains one of medicine's most urgent areas of unmet need. With 5-year survival rates of ∼11%, PDAC is set to become the second leading cause of cancer related deaths by 2040 [1]. The complex tumour microenvironment (TME) in PDAC, responsible for poor prognosis, is comprised of extracellular matrix (ECM) proteins and multiple cell types; with pancreatic stellate cells (PSCs), which become activated cancer associated fibroblasts (CAFs), being regarded as key orchestrators of the TME. We have demonstrated that treatment with all-trans retinoic acid (ATRA) can render activated PSCs (aPSC) to a quiescent (qPSC) phenotype (shift to G1 phase of cell cycle and other features [2]), resulting in stromal remodelling and thus, influencing cancer cell co-targeting with chemotherapy in patients [3]. This has resulted in the use of ATRA along with standard-of-care chemotherapy in the Stromal TARgeting for PAncreatic Cancer (STARPAC) clinical trial, with promising results [4]. These clinically relevant [5], exciting potential therapeutic benefits of stromal co-targeting through rendering PSCs quiescent [6], along with predictive inflammation-related biomarkers [7], and increased focus on cellular therapeutics such as NK cells, led us to postulate potential targetable PSC-immune cell interactions which may uncover a comprehensive therapeutic strategy for treating hitherto, incurable PDAC.
We identified differential NK-92 (a cell line representing NK cells) cytotoxicity against qPSCs (telomerase reverse transcriptase (hTERT) immortalised PS1 cell line rendered quiescent by administering ATRA for seven days at 1 µmol/L daily [2]) and aPSC phenotypes as assessed by surface expression of CD107a/b, and Calcein Acetyoxymethyl (AM) cytotoxicity assays (Supplementary Figure S1A-B). Furthermore, qPSC or aPSC education of NK-92 cells resulted in altered and distinct cytotoxicity towards pancreatic cancer cells (BxPC3, Capan2, MiaPaca2) as indicated by surface CD107a/b expression (Figure 1A) and complemented by Calcein AM cytotoxicity assays (data not shown).
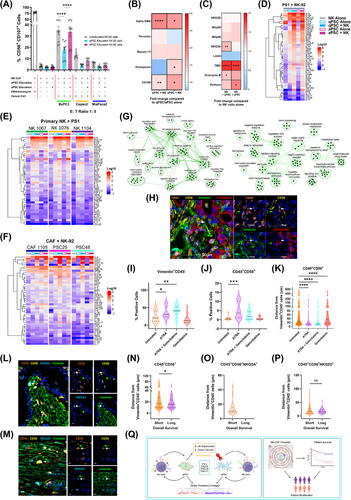
Surface and intracellular markers for CAF subtypes (pCAFassigner subtypes A-D [3]; CD105+/− CAFs [8]) and surface activating/inhibitory receptors and intracellular functional markers for NK cells, as assessed by spectral flow cytometry and Luminex ELISA secretome analysis, demonstrated that this interaction is, indeed, bidirectional. We demonstrated stellate cell polarisation to a myofibroblastic activation state [2] in response to direct contact with NK cells as assessed by alpha-SMA abundance (geometric mean fluorescence intensity (MFI)), as well as upregulation of CD105 expression, a CAF marker of anti-tumour immunity [8], irrespective of previous PSC activation status (Figure 1B), a fact not observed in Transwell™ separated co-culture (Supplementary Figure S1C) or conditioned media from NK cells (Supplementary Figure S1D), underlining the importance of direct cell-to cell interaction.
Concomitantly, we observed an upregulation of TIM3 on NK cells in response to direct co-culture with both qPSC and aPSCs, but a differential increase in NKG2A expression in response to qPSC but not aPSC. Functional markers, such as Granzyme B and Perforin, were also found to be differentially expressed (Figure 1C) in direct co-culture but not in Transwell™ separated conditions (Supplementary Figure S1E). Secretome analysis of direct co-culture demonstrated clear upregulation of interferon-γ, C-X-C chemokine ligand (CXCL) 9, CXCL10, CXCL11, and other cytokines and chemokines, not observed in indirect co-culture conditions, explaining the obligatory NK cell-PSC contact requirement to reciprocally modulate cellular phenotype and function as well as NK cell-mediated cancer cell cytotoxicity (Figure 1D, Supplementary Figure S1F).This preliminary cell line work could be confirmed in multiple, independent, primary, PDAC-patient-derived NK cells and CAFs grown from tissues of patients with PDAC (Figure 1E-F). In fact, these functional and phenotypic changes were manifest due to involvement of multiple intra-cellular pathways such as protein folding, interferon and cytokine signalling, chromatin remodelling and nucleotide biosynthesis, as demonstrated by proteomic analysis from cell lysates after direct co-culture (Figure 1G, Supplementary Figure S1G-H). Thus, we demonstrated, a convincing in vitro bidirectional, global PSC-NK interaction affecting cancer cell cytotoxicity using cell lines and primary patient derived material; where ATRA was used to modulate PSC, but not NK or cancer cells.
We confirmed these bidirectional interactions in an in vivo murine autochthonous, transgenic model of PDAC (Figure 1H) [9], which had been treated with cancer targeting chemotherapy (Gemcitabine) and/or stromal modulating agent (ATRA), compared to vehicle treated animals [9]. We demonstrated stromal modulation with ATRA which resulted in increased Vimentin+CD45− cellular density (Figure 1I). Surprisingly, even with just one week's treatment, we saw an increase in overall NK cell infiltrate (CD45+CD56+) upon stromal modulation with ATRA (Figure 1J). Further, cellular proximity analysis determined that upon stromal modulation (ATRA), CD45+CD56+ NK cells were significantly closer to Vimentin+CD45− CAFs when compared to those animals treated with vehicle control (Figure 1K), confirming that ATRA mediated PSC modulation changes NK cell proximity and infiltrate. Recent independent work complemented our findings by demonstrating that the activation of the retinoic acid receptor (RAR) induced a tumour-suppressive senescence-associated secretory phenotype (SASP) in cancer cells, which can lead to a robust anti-tumour immune response. Thus, combination of RAR activation and chemotherapy enhanced NK-cell-mediated tumour clearance as well as NK cell recruitment to the tumour [10].
Whilst cognate cell surface NK cell receptors are difficult to explore in murine tissues, we demonstrated the importance of cellular proximity in NK: PSC (CAF) interaction in human samples, to further validate this finding. We utilised multiplex immunohistochemistry to explore total NK populations as well as subsets expressing specific activating/inhibitory receptors and their relationship to CAFs (pan-CAF marker vimentin [8]) in human PDAC samples. To investigate the prognostic importance of NK cell infiltrate in PDAC, we dichotomised patients into short (< 30 months, n = 48) or long (> 30 months, n = 16) survivors, with both groups expressing similar tumour variables known to have a prognostic implication (Supplementary Figure S2). We confirmed the well documented intra- and inter-tumoral heterogeneity among CAF subtype distribution (Supplementary Figure S3A-B) as well as identified wide NK cell phenotypic heterogeneity (Figure 1L-M, Supplementary Figure S3C- D). Interestingly, no difference was identified in total NK cell infiltrate between short and long survivors (Supplementary Figure S3E). However, CD45+CD56+NKG2A+ cells (but not other subsets) were found to be absent in long survivors (Supplementary Figure S3F-I).
Next, we assessed the role of NK-CAF proximity in PDAC. Using HALO's Spatial Analysis module (Indica Labs) we identified CD45+CD56+ (NKG2D+/NKG2A+/NKp46+/LAG3+) cells within PDAC tissues and assessed their absolute distance to Vimentin+CD45− CAFs (Figure 1N-P, Supplementary Figure S3J-K). Long survivors demonstrated shorter spatial distance between CD45+CD56+ NK cells and CAFs than did short survivors, suggesting that it is the NK cell proximity to CAFs, and not the global NK infiltrate, which impacts patient prognosis (Figure 1N). Notably, activation/quiescence of CAFs in human samples could not be evaluated due to lack of reliable markers to assess these phenotypic states; however, this would be valuable area for further investigation. Taken together, these results suggested that NK interaction with PSC/CAFs in PDAC may play a prognostic role and consequently may be used for patient stratification.
In conclusion, this study suggested a bidirectional relationship between NK cells and PSCs in PDAC. We offer novel observational insights into the cell-cell interactions at the phenotypic, functional, and proteomic levels. Additionally, we demonstrated prognostic implications of NK-CAF proximity in PDAC and suggested its potential use as a marker for patient stratification (Figure 1Q). Further work is needed to fully evaluate the mechanistic underpinnings of this dynamic relationship, as well as the up/down regulation of specific NK cell and CAF markers. This may perhaps be achieved through genetically engineered mouse models with modified NK or CAF populations as well as spatial transcriptomics, facilitating a deeper understanding of the long-term sequalae of this dynamic interaction; thus, uncovering potential targets to exploit for the treatment of PDAC.
AUTHOR CONTRIBUTIONS
Design of work: Rachel Elizabeth Ann Fincham, Parthiban Periasamy, Joe Poh Sheng Yeong, and Hemant Mahendrakumar Kocher. Acquisition and analysis: Rachel Elizabeth Ann Fincham, Parthiban Periasamy, Craig Ryan Joseph, Jia Meng, Jeffrey Chun Tatt Lim, Felicia Wee, Jiangfeng Ye, Li Yen Chong, Joe Poh Sheng Yeong, and Hemant Mahendrakumar Kocher. Methodological troubleshooting: Rachel Elizabeth Ann Fincham, Parthiban Periasamy, Craig Ryan Joseph, Jia Meng, Jeffrey Chun Tatt Lim, Felicia Wee, Konstantinos Stasinos, Michelle Rodrigues Goulart, Joe Poh Sheng Yeong, and Hemant Mahendrakumar Kocher. Interpretation: Rachel Elizabeth Ann Fincham, Parthiban Periasamy, Joe Poh Sheng Yeong, and Hemant Mahendrakumar Kocher. Writing: Rachel Elizabeth Ann Fincham, Denise Goh, Parthiban Periasamy, Joe Poh Sheng Yeong, Hemant Mahendrakumar Kocher, and Bijin Veonice Au. All authors read and approved the final manuscript.
ACKNOWLEDGMENTS
We thank members of the Kocher (Centre for Tumour Biology, Barts Cancer Institute-a Cancer Research United Kingdom [CRUK] Centre of Excellence, Queen Mary University of London, London, UK) and Yeong (Institute of Molecular and Cell Biology [IMCB], Agency of Science, Technology and Research [A*STAR], Singapore) laboratories for critical appraisal and suggestions throughout this project. We thank Radoslow Sobota (Functional Proteomics Laboratory, Institute of Molecular and Cell Biology [IMCB], Agency of Science, Technology and Research [A*STAR], Singapore) for his support with mass spectrometry sample acquisition.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
FUNDING INFORMATION
This research is partly supported by Barts Charity and A*STAR Research Attachment Programme (ARAP) PhD studentship. Pancreatic Cancer Research Fund Tissue Bank is funded by Pancreatic Cancer Research Fund. Hemant Kocher is supported by NIHR Barts Biomedical Research Centre. Barts Cancer Institute is supported by Cancer Research UK.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Ethics approval for the use of human pancreatic cancer tissue was obtained from City and East London Research Ethics Committee (REC0029 07/0705/87). Each participant signed an informed consent before participating to this study. Primary cells were provided by Pancreatic Cancer Research Fund Tissue Bank.
Open Research
DATA AVAILABILITY STATEMENT
Data is available from the corresponding author on reasonable request.




