Targeting the LMP1-ALIX axis in EBV+ nasopharyngeal carcinoma inhibits immunosuppressive small extracellular vesicle secretion and boosts anti-tumor immunity
Fajian He, Yan Gong and Yajuan Zhou contributed equally.
Abstract
Background
Immunotherapy has revolutionized the therapeutical regimen for nasopharyngeal carcinoma (NPC), yet its response rate remains insufficient. Programmed death-ligand 1 (PD-L1) on small extracellular vesicles (sEVs) mediates local and peripheral immunosuppression in tumors, and the mechanism of PD-L1 loading into these vesicles is garnering increasing attention. Latent membrane protein 1 (LMP1), a key viral oncoprotein expressed in Epstein-Barr virus (EBV)-positive NPC, contributes to remodeling the tumor microenvironment. However, the precise mechanisms by which LMP1 modulates tumor immunity in NPC remain unclear. Here, we aimed to investigate the roles and regulatory mechanisms of LMP1 and sEV PD-L1 in NPC immune evasion.
Methods
We analyzed the impact of LMP1 on tumor-infiltrating lymphocyte abundance in NPC tissues and humanized tumor-bearing mouse models using multiplex immunofluorescence (mIF) and flow cytometry, respectively. Transmission electron microscopy and nanoparticle tracking analysis were employed to characterize sEVs. Immunoprecipitation-mass spectrometry was utilized to identify proteins interacting with LMP1. The regulatory effects of sEVs on tumor microenvironment were assessed by monitoring CD8+ T cell proliferation and interferon-γ (IFN-γ) expression via flow cytometry. Furthermore, the expression patterns of LMP1 and downstream regulators in NPC were analyzed using mIF and survival analysis.
Results
High LMP1 expression in NPC patient specimens and mouse models was associated with restricted infiltration of CD8+ T cells. Additionally, LMP1 promoted sEV PD-L1 secretion, leading to inhibition of CD8+ T cell viability and IFN-γ expression in vitro. Mechanistically, LMP1 recruited apoptosis-linked gene 2-interacting protein X (ALIX) through its intracellular domain and bound PD-L1 through its transmembrane domain, thereby facilitating the loading of PD-L1 into ALIX-dependent sEVs. Disruption of ALIX diminished LMP1-induced sEV PD-L1 secretion and enhanced the anti-tumor immunity of CD8+ T cells both in vitro and in vivo. Moreover, increased expression levels of LMP1 and ALIX were positively correlated with enhanced immunosuppressive features and worse prognostic outcomes in NPC patients.
Conclusion
Our findings uncovered the mechanism by which LMP1 interacts with ALIX and PD-L1 to form a trimolecular complex, facilitating PD-L1 loading into ALIX-dependent sEV secretion pathway, ultimately inhibiting the anti-tumor immune response in NPC. This highlights a novel target and prognostic marker for NPC immunotherapy.
Abbreviations
-
- ALIX
-
- apoptosis-linked gene 2-interacting protein X
-
- BCA
-
- bicinchoninic acid
-
- CFSE
-
- carboxyfluorescein succinimidyl ester
-
- CHX
-
- cycloheximide
-
- co-IP
-
- co-immunoprecipitation
-
- CTAR
-
- c-terminal activating regions
-
- DAPI
-
- 4′,6-diamidino-2-phenylindole
-
- EBV
-
- Epstein-barr virus
-
- ELISA
-
- enzyme-linked immunosorbent assay
-
- ESCRT
-
- endosomal sorting complexes required for transport
-
- FBS
-
- fetal bovine serum
-
- GEO
-
- gene expression omnibus
-
- GO
-
- gene ontology
-
- IFN-γ
-
- interferon-γ
-
- IHC
-
- immunohistochemistry
-
- ILVs
-
- intralumenal vesicles
-
- KEGG
-
- kyoto encyclopedia of genes and genomes
-
- LMP1
-
- latent membrane protein 1
-
- mIF
-
- multiplex immunofluorescence
-
- MS
-
- mass spectrometry
-
- MVB
-
- multivesicular body
-
- NF-κB
-
- nuclear factor kappa-B
-
- NPC
-
- nasopharyngeal carcinoma
-
- NTA
-
- nanoparticle tracking analysis
-
- PBMC
-
- peripheral blood mononuclear cell
-
- PD-1
-
- programmed death-1
-
- PD-L1
-
- programmed cell death 1 ligand 1
-
- PRR
-
- proline-rich region
-
- RNA-seq
-
- RNA sequencing
-
- SDS
-
- sodium dodecyl sulfate
-
- SDS-PAGE
-
- SDS-polyacrylamide gel electrophoresis
-
- sEVs
-
- small extracellular vesicles
-
- TEM
-
- transmission electron microscopy
-
- TSA
-
- tyramide signal amplification
-
- TSG101
-
- tumor susceptibility 101.
1 BACKGROUND
The Epstein-Barr virus (EBV) is a pivotal etiological factor in various malignancies, including lymphoma [1], nasopharyngeal carcinoma (NPC) [2], and EBV-associated gastric cancer [3]. Over 90% NPC cases are linked to EBV infection, and many studies highlight the virus's potent capability to drive malignant transformation during NPC progression [4-6]. Recently, immunotherapy emerged as an effective therapeutical strategy for NPC, yet it is challenged by a notable deficiency in response rates [7, 8]. Parallel investigations revealed a substantial link between the status of EBV infection and the outcomes of anti-programmed death-1 (PD-1) immunotherapy [6, 9]. Therefore, it is crucial to investigate the underlying mechanisms through which EBV facilitates immune evasion to enhance the efficacy of immunotherapy for NPC.
The EBV-encoding oncogenic latent membrane protein 1 (LMP1) is acknowledged for its multifunctional role in providing growth signals, impeding apoptosis, and influencing immune regulatory mechanisms in latently infected cells [10, 11]. This pleiotropic oncoprotein is characterized by an N-terminal cytoplasmic domain, 6 transmembrane domains, and a C-terminal cytoplasmic domain that harbors the C-terminal activating regions (CTAR) 1 and 2 [12, 13]. It is well established that LMP1 functions as a constitutively active tumor necrosis factor receptor that triggers the activation of multiple signaling pathways, notably those of the nuclear factor kappa-B (NF-κB) family [10, 14, 15]. Regarding EBV-associated NPC, activation of these pathways by LMP1 bolsters the malignant transformation potential of tumors [16, 17]. The role of LMP1 in tumor immunity has lately gained increased focus. It was reported that LMP1 could upregulate the transcription of programmed cell death 1 ligand 1 (PD-L1) by activating the NF-κB pathway through its CTAR1 and CTAR2 domains [18, 19]. Previous research also indicated that LMP1 upregulated various cellular antigens within B cells, subsequently triggering potent cytotoxic reactions by CD4+ and CD8+ T cells [20]. However, the exact roles of LMP1 in remodeling the tumor immune microenvironment in NPC remain unclear.
Small extracellular vesicles (sEVs) are membrane-bounded particles with a diameter less than 200 nm, carrying bioactive molecules that influence cell-cell communication [21, 22]. Tumor-derived sEVs play essential roles in shaping an immunosuppressive microenvironment that favors tumor growth [23]. Unraveling the intricacies of how cargo is directed into sEVs is vital for comprehending the diversity of the immune microenvironment and bolstering anti-tumor responses [24]. The endosomal sorting complexes required for transport (ESCRT) machinery are essential for sEV biogenesis and cargo sorting [25]. Apoptosis-linked gene 2-interacting protein X (ALIX), an ESCRT-associated protein, was initially thought to primarily involve cytokinesis and virus budding [26]. However, its contribution to intralumenal vesicles (ILVs) formation and sEV cargo sorting has increasingly become a focal point of study [27, 28]. Exporting PD-L1 via sEVs is widely acknowledged as a strategic maneuver by tumor cells to dampen anti-tumor immunity [29-31]. Nevertheless, the precise mechanisms underlying PD-L1 sorting into sEVs have yet to be fully elucidated.
How EBV viral gene products hijack host intracellular signaling pathways to mediate immune evasion is a promising area of research. Previous studies have reported that the assembly and release of viral particles exploit the host cell's multivesicular body (MVB) formation pathway. EBV-encoded LMP1 is believed to alter the quantity and nature of sEVs, thereby promoting tumor malignancy, though the specific mechanisms remain unclear. In this study, we investigated the association between LMP1 and the immunosuppressive microenvironment of NPC, hypothesizing that LMP1 upregulates sEV PD-L1. Using co-immunoprecipitation (co-IP) combined with mass spectrometry (MS), we identified ALIX as a key molecule in LMP1-mediated upregulation of sEV PD-L1 and immune suppression. We further explored the role of the LMP1-ALIX-sEV PD-L1 signaling axis in EBV-mediated immune evasion. Additionally, we also investigated the effects of ALIX inhibition on the effectiveness of immunotherapy in NPC using a peripheral blood mononuclear cell (PBMC)-humanized mouse model.
2 MATERIALS AND METHODS
2.1 Clinical sample collection
NPC samples for plasma analysis and immunohistochemistry (IHC) staining were collected by the Department of Radiation and Medical Oncology, Zhongnan Hospital of Wuhan University (Wuhan, Hubei, China). Plasma and tumor tissue samples were obtained from NPC patients diagnosed at stages I to IV according to the American Joint Committee on Cancer 8th edition, who had never undergone any immunotherapy. Approval for conducting this study, involving the analysis of anonymized data, was obtained from the Medical Ethics Committee at Zhongnan Hospital of Wuhan University (ZN2021073). Written informed consent was obtained from all subjects in compliance with the principles of the Declaration of Helsinki. The clinical characteristics of the patients are detailed in Supplementary Table S1.
2.2 Multiplex immunofluorescence (mIF) and IHC staining
The NPC tissue microarray for mIF staining was purchased from Outdo Biotech, Shanghai, China. The sections were first deparaffinized and rehydrated through a graded series of xylene and ethanol immersions. Antigen retrieval was achieved using a citric acid-based solution (pH 9.0), with the sections then subjected to heat-induced epitope retrieval in a preheated oven for 15 min. For mIF, the Tyramide Signal Amplification (TSA) system (Baiqiandu, Wuhan, Hubei, China) was employed. The slides underwent a 3% hydrogen peroxide treatment to inhibit the activity of endogenous peroxidase and were blocked with 10% goat serum to minimize non-specific binding; both steps were conducted at ambient temperature for 25 and 30 min, respectively. The sections were then incubated with a series of primary antibodies, followed by appropriate secondary antibodies and TSA reagents each tagged with distinct fluorophores. After each fluorophore development, the previous antibodies were stripped from the sections to allow for subsequent rounds of antibody binding. Nuclear counterstaining was performed using 4′,6-diamidino-2-phenylindole (DAPI) for 10 min, and the sections were ultimately preserved with an anti-fade mounting medium.
For IHC, tissue sections were blocked with 3% bovine serum albumin for 30 min at room temperature. Primary antibodies were applied and allowed to bind overnight at 4°C. The sections were then exposed to horseradish peroxidase-conjugated secondary antibodies for 30 min at room temperature. Visualization of the antigens was conducted using 3,3'-diaminobenzidine chromogens, followed by nuclear counterstaining with Mayer's hematoxylin.
The method for assessing protein expression is summarized as follows: For each patient, 3 fields of view from tumor tissue are randomly selected. The average fluorescence intensity of all cells in each field is calculated, and the average of the 3 fields is taken as the protein expression level for that patient. Subsequently, patients were stratified into ‘low’ or ‘high’ expression groups based on the median IHC scores of LMP1 and ALIX proteins. For spatial dot analysis, cell subpopulation identification and spatial analysis, including intercellular distance, were performed using R package “Phenoptr” and “phenoptrReports” (R, version 4.1.1; https://github.com/PerkinElmer/phenoptr and https://github.com/PerkinElmer/phenoptrReports). The distance between two cell subtypes was calculated using the x and y coordinates from the raw inForm data, and the nearest neighbor distance for each cell was identified through phenoptr.
2.3 Cell lines and culture
The human NPC 5-8F and 6-10B cells were donated by Sun Yat-sen University Cancer Center (Guangzhou, Guangdong, China). To make stable cell lines expressing LMP1, 5-8F or 6-10B cells were initially transduced with lentiviral particles containing LMP1-GFP or control-GFP plasmid (Genechem, Shanghai, China) followed by puromycin selection (2 µg/mL, BS111, Biosharp, Hefei, Anhui, China). These NPC cells were cultured at 37°C and 5% CO2 in RPMI-1640 medium (HyClone, Logan, Utah, USA) supplemented with 100 units/mL penicillin, 0.1 mg/mL streptomycin, and 10% fetal bovine serum (FBS). HEK293T cells were obtained from the American Type Culture Collection and cultured at 37°C and 5% CO2 in DMEM medium (HyClone) supplemented with 100 units/mL penicillin, 0.1 mg/mL streptomycin and 10% FBS. All the cells were subject to regular authentication through analysis of short tandem repeats and testing to confirm the absence of mycoplasma contamination.
2.4 Plasmids, RNA interference and antibodies
GFP-tagged LMP1 wild type and mutants: transmembrane domain deletion (ΔTM), aa 1-351, aa 1-233, aa 1-193; MYC-tagged ALIX wild type and mutants: aa 1-703 (ΔPRR), aa 351-873 (ΔBro1), aa 365-703 deletion (ΔV); FLAG-tagged PD-L1 were constructed by Genechem Technology (Shanghai, China). PLV3-CMV-LMP1 (P38232) was obtained from MiaoLingBio (Wuhan, Hubei, China). siRNAs targeting ALIX and nontargeting control siRNA were purchased from Tsingke Biotech (Beijing, China). siRNA sequences against human ALIX: 5ʹ-GCATCTCGCTATGATGAATAT-3ʹ (#1); 5ʹ-CCTGAATTACTGCAACGAAAT-3ʹ (#2). Transfections were conducted using Lipofectamine 3000 (L3000001, Thermo Fisher, Waltham, MA, USA) following the manufacturer's guidelines, and the transfection efficiency was assessed through Real-time quantitative PCR (RT-qPCR) and Western blotting within 24-48 h after transfection. The RT-qPCR was performed under conditions of 95°C for 3 min, and 45 cycles of 95°C for 5 s and 60°C for 30 s, using Taq Pro Universa l SYBR qPCR Master Mix (Q712-03, Vazyme, Nanjing, Jiangsu, China) on the iCycler iQ system (Bio-Rad, Hercules, CA, USA). The antibodies used in the article and their detailed information are summarized in Supplementary Table S2.
2.5 RNA sequencing (RNA-seq) and bioinformatic analysis
For RNA-seq, LMP1-overexpressing and control 6-10B cells' total RNA were extracted using TRIzol reagent (R401-01, Vazyme). After quality control, the RNA-seq library was sequenced with paired-end reads on the BGISEQ platform (https://en.genomics.cn/sequecplatform.html) at the Beijing Genomics Institute.
For analysis of RNA-seq results, RNA-seq reads quality was estimated using FastQC (v0.11.9, https://www.bioinformatics. babraham.ac.uk/projects/fastqc/) and matched with the human genome GRCh38 by HISAT2 (v2.2.1, https://daehwankimlab. github.io/hisat2/). We use Feature Counts (v2.0.1, http://subread.sourceforge.net/) to quantitate the transcriptome with the General Transfer Format annotation files. After standardized data processing, the R package DEGSeq2 (v1.28.1, https://bioconductor.org/packages/ release/bioc/html/DESeq2.html) was employed to conduct differential analysis of gene expression data. Genes with a P < 0.05 and an absolute value of log2foldchange > 2 were identified as differentially expressed genes (DEGs), which were visualized using volcano plots generated with the R package ggplot2 (v3.3.5, https://cran.r-project.org/web/packages/ggplot2/index.html). The R package clusterProfiler (v3.18.1, https://bioconductor. org/packages/release/bioc/html/clusterProfiler.html) was utilized for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis.
The single-cell RNA-sequencing data for analysis of ALIX mRNA expression and features of tumor microenvironment including cell compositions in human NPC was downloaded from Gene Expression Omnibus (GEO) (GSE150430).
2.6 Purification and characterization of sEVs
For cell culture supernatant samples, sEVs were purified by sequential centrifugation, following the MISEV2023 guidelines [22]. Briefly, at 90% confluency, the cells underwent a phosphate-buffered saline (PBS) wash and were then incubated in a serum-free culture medium for 48 h to allow sEV release. Subsequently, the individual supernatants from cells were sequentially centrifuged at 300 ×g, 4°C for 10 min to remove cells, followed by 3,000 g for 10 min and 10,000 g for 30 min at 4°C to remove microvesicles. Subsequently, the supernatants were filtered through a 0.22 µm syringe filter and centrifuged at 4°C, 120,000 ×g (Beckman Type 90 Ti) for 2 h. Ultimately, the sEV pellet was reconstituted in either PBS or lysis buffer for further analysis. For plasma samples, we extracted plasma-derived sEVs according to the instructions provided in the Total sEVs Isolation Reagent (4484453, Thermo Fisher). The quantification of total sEV protein concentrations was performed utilizing the Bicinchoninic Acid (BCA) Protein Assay kit (23227, Thermo Fisher).
To detect EBV DNA loads in plasma sEVs, we followed the EBV nucleic acid detection kit (JL-T1729, JONLNBIO, Shanghai, China) protocol and extracted sEVs from the plasma to investigate the correlation between EBV DNA loads and sEV PD-L1 expression levels. For the purpose of transmission electron microscopy (TEM) verification, purified sEVs suspended in PBS were deposited onto formvar carbon-coated nickel grids. Subsequently, the grids were stained with 2% uranyl acetate and quickly rinsed with a drop of PBS. After being air-dried, the grids were finally observed under a JEM-1011 TEM (Hitachi, Tokyo, Japan). We conducted nanoparticle tracking analysis (NTA) to verify the concentration and size distribution of isolated sEVs, which were re-suspended in filtered PBS, using the NTA instrument (NS500, NanoSight, Salisbury, Wiltshire, UK) with a 488 nm laser.
2.7 Co-IP and Western blotting
The NPC 5-8F and 6-10B cells were collected and washed with PBS, then lysed with either protein lysis buffer (P0013B, Beyotime, Shanghai, China) or IP lysis buffer (87787, Thermo Fisher), containing a protease inhibitor cocktail (HY-K0011, MedChemExpress, Monmouth Junction, NJ, USA), on ice for 30 min. After centrifugation at 12,000 ×g for 20 min at 4°C, total protein concentrations were measured using BCA Protein Assay Kits (23227, Thermo Fisher).
For co-IP assays, 1-2 mg of whole-cell lysate protein was incubated with 1 µg of specific antibodies (anti-FLAG/anti-GFP/anti-MYC or others as specified) overnight at 4°C. Magnetic protein A/G beads (HY-K0202, MedChemExpress) were added and incubated with the lysate for 4 h at 4°C. The beads were washed four times with IP buffer at 4°C, and immuno-complexes were eluted through boiling in 2× sodium dodecyl sulfate (SDS) loading buffer for subsequent Western blotting analysis.
In Western blotting, protein samples underwent SDS-polyacrylamide gel electrophoresis (PAGE) and electro-transfer to a polyvinylidene difluoride membrane. The membrane underwent blocking with 5% nonfat milk for 1 h at room temperature, probed with specific primary antibodies at 4°C overnight, and then washed 3 times with PBST (PBS with 0.05% Tween 20) for 5 min each. Details of the antibodies used are listed in Supplementary Table S2. ALIX, tumor susceptibility 101 (TSG101), and CD63 were used as marker proteins to characterize sEV. After incubation with peroxidase-conjugated secondary antibodies for 1 h at room temperature, the membrane was washed 5 more times with PBST for 8 min each. Blots were visualized using the ChemiDoc XRS system (Bio-Rad) and Image Lab Software (Bio-Rad).
2.8 Immunofluorescence
After treatment, cells were washed with PBS and fixed with 4% neutral formaldehyde for 20 min at room temperature. They were then permeabilized with 0.5% Triton X-100 in PBS for 15 min. The cells were incubated with a blocking buffer (10% FBS in PBS) for 1 h. The above steps were performed at room temperature. Following 3 washes with PBS, the primary antibodies PD-L1 (1:200, 13684, CST, Danvers, MA, USA), CD63 (1:200, 25682-1-AP, Proteintech, Wuhan, Hubei, China), and ALIX (1:200, 2171, CST) were added and incubated overnight at 4°C. The next day, after 3 washes with PBST, the cells were incubated with anti-mouse Cy-3 (1:200, SA00009-1, Proteintech), anti-rabbit Cy-5 (1:400, L138A, ABP Biosciences, Wuhan, Hubei, China), and anti-rabbit Alexa Fluor-647 (1:400, A0468, Beyotime) for 2 h at room temperature. Subsequently, the cells were washed and stained with DAPI (100 ng/mL). Finally, an anti-fluorescence quencher was used to seal the slides, and the NPC cells treated as specified were analyzed by confocal microscopy (ZEISS LSM880, 63× oil lens, Oberkochen, Baden-Württemberg, Germany). The entire process was conducted under light protection.
2.9 Silver staining and MS analysis
Immunocomplexes were resolved on SDS-PAGE and visualized using a Fast Silver Stain Kit (BL620A, Biosharp), adhering strictly to the protocols recommended by the manufacturer. Distinct protein bands were excised from the gel for further analysis. Proteomic characterization was facilitated by LC-MS/MS, undertaken at SpecAlly Life (Wuhan, Hubei, China). This was conducted on a high-resolution Q Exactive HF-X MS integrated with an UltiMate 3000 RSLCnano system (Thermo Scientific), in strict accordance with established methodologies. For data interrogation, the MaxQuant software (version 1.6.6) was utilized, leveraging the Andromeda database search engine. The search parameters were set against the UniProt human proteome reference database. A stringent false discovery rate of 1% was enforced for both protein and peptide validation to ensure the reliability of the identifications.
2.10 Protein half-life analysis
To assess PD-L1 protein stability, LMP1-overexpressing cells were treated with cycloheximide (CHX, 200 µg/mL, HY-12320, MedChemExpress) at specified time points. Subsequently, the cells were harvested for Western blotting using the designated antibodies. The intensities of the PD-L1 protein bands were quantified using ImageJ software and then normalized to vinculin.
2.11 Molecular docking
The possible binding models of the indicated proteins was determined using molecular docking. The 3D protein structure of LMP1 and ALIX was predicted from AlphaFold Protein Structure Database (https://alphafold.com/), while that of PD-L1 was obtained from RCSB PDB database (https://www.rcsb.org/structure/8AOM). HDOCK (http://hdock.phys.hust.edu.cn/) enables comprehensive analysis of protein-protein docking, including the evaluation of diverse conformations, binding affinities across different conformational states, and identification of amino acid residues within a 5Å interaction distance. PyMOL (v4.3.0, https://pymol.org/2/) and Ligplus (https://www.ebi.ac.uk/thornton-srv/software/LIGPLOT/) software were employed to visualize the interacting amino acid residues between the two proteins in both three-dimensional and two-dimensional representations.
2.12 Enzyme-linked immunosorbent assay (ELISA)
To detect PD-L1 on sEVs obtained from NPC cells, we employed a PD-L1 ELISA kit (KE00074, Proteintech). Each step was meticulously performed in strict accordance with the manufacturer's instructions provided with the kit.
2.13 TEM
For fixation, the cells were first treated with 2.5% glutaraldehyde and then with 1% osmium tetroxide, each step conducted on ice for 2 h and washed with PBS (3 times, every time 15 min). Dehydration of the cell aggregates was achieved using a sequential ethanol and propylene oxide series, after which they were embedded in an Epon 812 mixture and polymerized at 60°C for 48 h. The polymerized blocks were cut into 60-80 nm sections by a slicer and then counterstained with uranyl acetate and lead citrate. A FEI TECNAI G2 20 TWIN electron microscope (Thermo Fisher) was employed for the examination of all samples.
2.14 PBMC isolation and treatment with sEVs
To elucidate the immunomodulatory impact of sEV PD-L1 on T cell function, PBMCs were harvested from the blood of healthy donors [29]. These PBMCs were then cultured in conjunction with sEVs. Specifically, PBMCs were plated at a density of 2 × 106 cells per well in 24-well plates and activated with anti-CD3 (2 µg/mL, BE0231, Bioxcell, Lebanon, NH, USA) and anti-CD28 (2 µg/mL, BE0015-1, Bioxcell) antibodies for 48 h. Following this activation phase, the cells were treated with sEVs (10 µg/mL) derived from human NPC cells for an additional 48 h in the continued presence of anti-CD3/CD28 antibodies. To neutralize the PD-L1 sites on the sEVs, PD-L1 blocking antibodies (329746, Biolegend, London, UK) were employed. To specifically monitor CD8+ T cell proliferation, we utilized carboxyfluorescein succinimidyl ester (CFSE), a cell-permeable dye that facilitates the tracking of cellular division (65-0850-84, Thermo Fisher). A 1 µmol/L solution of CFSE was used to label 1 × 106 CD8+ T cells, which were then incubated at 37°C for 20 min. Termination of the staining reaction was achieved by the addition of a quenching volume of ice-cold medium supplemented with 10% FBS. The cells were then processed further for flow cytometric analysis as previously described. In brief, live cells were distinguished using a live/dead dye. Cell surface staining was performed for 30 min at 4°C, followed by intracellular staining for 50 min on ice using a fixation/permeabilization kit (554714, BD, San Jose, CA, USA).
2.15 PBMC-humanized mouse tumor model establishment
Mice (NOD-SCID, 6-8 weeks old, male) were obtained from the Laboratory Animal Center of Bestcell Model Biology Center (Wuhan, Hubei, China) and maintained under specific-pathogen-free conditions. All experiments involving mice were carried out following animal ethical standards, with the approval of the Ethics Committee of the Bestcell Model Biology Center. Then 1×107 LMP1-overexpressing 6-10B cells were subcutaneously injected into the right thigh root of each mouse. After 7 days, when the tumor length reached approximately 5 mm, 5×106 human PBMCs (harvested from the blood of healthy donors) were injected via the tail vein. To ensure the successful construction of the model, we routinely assessed the proportion of T cells in the mice's peripheral blood via flow cytometry and monitored their body weight. The subsequent 2 weeks involved monitoring tumor growth, graft-versus-host disease, drug administration, and other experimental procedures until the mice were euthanized for tissue collection. Mice were euthanized employing carbon dioxide asphyxiation 14 days after tumor cell inoculation or if the longest dimension of the tumors reached 2.0 cm before 14 days.
For live-cell imaging of T cells, we used a CD8 MicroBeads kit (130-045-201, Miltenyi, Bergisch Gladbach, North Rhine-Westphalia, Germany) to isolate CD8+ T cells from PBMCs and subsequently labeled the CD8+ T cells with CFSE fluorescence, then injected into tumor cell-derived xenograft NOD-SCID mice via the tail vein. Following gas anesthesia in mice, a small animal live imaging system (IVISLuminaXRMS, Waltham, MA, USA) was utilized to dynamically monitor the in vivo distribution and intensity alterations of PBMCs. To ensure accurate signal values, background fluorescence was subtracted using the small animal live imaging system. We calculated the relative fluorescence intensity for the tumor region by dividing the signal value of the tumor region by that of the thoracic region.
To evaluate the experimental treatment of ALIX inhibition combined with PD-1 monoclonal antibody in PBMC-humanized mouse model, tumor-bearing mice were randomly divided into 4 groups based on 2 treatment factors: ALIX knockdown or not, and treatment with PD-1 monoclonal antibody or IgG. Anti-PD-1 antibody was given every 3 days with a dosage of 100 mg per mouse for a total of 5 treatments.
2.16 Statistics and reproducibility
Experimental results are expressed as mean ± standard deviation. Statistical analyses were conducted using Prism (Version 8, GraphPad, San Diego, CA, USA). If the samples follow a normal distribution, the significance of differences between two groups was assessed by the Student's t-test; if the samples do not follow a normal distribution, non-parametric tests such as the Mann-Whitney test will be employed for comparing the 2 groups. The correlation analysis between 2 variables will be conducted using the Pearson analysis. Two-way ANOVA was employed to evaluate the statistical differences in efficacy between combined ALIX suppression and immunotherapy treatments. Survival outcomes, both overall and disease-free, were estimated via the Kaplan-Meier method, with comparisons drawn using the log-rank test. Survival time was calculated from the date of diagnosis for the patient. P < 0.05 was considered to indicate statistical significance. Representative results were replicated independently at least 3 times with similar results.
3 RESULTS
3.1 EBV-encoded LMP1 restricted CD8+ T cell infiltration in human NPC specimens and PBMC-humanized mouse model
EBV influences host immune surveillance through its encoded products such as proteins, nucleic acids, etc [32, 33]. To elucidate the impact of LMP1 expression on tumor-infiltrating lymphocytes, we performed immunofluorescence to detect the expression of LMP1, CD8 and PD-1 in tissue samples from NPC patients. The results showed that tumors with high LMP1 expression had fewer infiltrating CD8+ T cells and PD-1+CD8+ T cells (Figure 1A-B). Further, we performed spatial dot plot analysis on the tissue immunofluorescence images. The results showed that in the tissues with high LMP1 expression, there were significantly fewer CD8+ T cells within a close distance (less than 30 µm) to tumor cells compared to the low LMP1 expression group (Figure 1C). Moreover, our previous studies explored the correlation of PD-L1 expression with patients' EBV DNA loads and prognosis [34]. Considering the role of circulating sEVs containing PD-L1 in mediating tumor local and peripheral immune suppression, here we detected the EBV DNA loads and sEV PD-L1 content in the plasma of NPC patients and explored the correlation between them. The results of Western blotting and ELISA both indicated that patients with high EBV DNA loads had higher levels of sEV PD-L1 content, and there was a significant positive correlation between them (Supplementary Figure S1A-C).

As LMP1 plays an important role in remodeling the tumor microenvironment, RNA-seq was performed on LMP1-overexpressing and control 6-10B cells. We performed DEG analysis on the 2 groups of data, revealing a total of 1,451 DEGs, with 755 upregulated and 696 downregulated (Figure 1D). Then, we performed GO and KEGG enrichment analysis on DEGs. The results showed that LMP1 could affect the leukocyte migration pathway (Supplementary Figure S1D) and PD-L1 expression and PD-1 checkpoint pathway in cancer (Figure 1E). We subsequently examined the association between LMP1 expression and tumor-infiltrating immune cells in PBMC-humanized tumor-bearing NOD-SCID mice (Figure 1F) and NPC tissue samples. The success of constructing the immunologically reconstituted human xenograft mouse model was verified by routinely assessing the proportion of T cells in the mice's peripheral blood via flow cytometry and monitoring their body weight (Supplementary Figure S1E-F). The analysis of tumor growth curves indicated that LMP1 promoted NPC cell growth in vivo (Figure 1G), and the results of weighing tumors after complete removal also demonstrated that tumors in the LMP1 overexpression group were larger (Figure 1H). Next, we measured the proportion of tumor-infiltrating CD8+ T cells in mice by flow cytometry. Representative contour plots presenting the general gating strategy are shown (Supplementary Figure S1G). Flow cytometry of tumor tissues revealed a significant decrease in the number and cytotoxicity of infiltrating CD8+ T cells in tumors overexpressing LMP1 (Figure 1I). The in vivo imaging results tracking the fate of CD8+ T cells showed that the infiltration of CD8+ T cells at the tumor site peaked around one day post-injection, with the fluorescent signal of immune cells within the tumor gradually diminishing over the following week. Notably, LMP1 significantly restricted the infiltration of CD8+ T cells within the tumor compared to the control group lacking LMP1 expression (Figure 1J-K). Integrating the above results, it can be concluded that LMP1 restricts the infiltration of CD8+ T cells in tumors in both human NPC specimens and PBMC-humanized mouse tumor model.
3.2 LMP1 selectively enriched PD-L1 in sEVs, thereby inhibiting CD8+ T cell activity
LMP1 regulates the contents of host cell-derived sEVs that exert influence on the tumor microenvironment [35, 36]. We hypothesized that LMP1 could mediate immunosuppression via promoting the secretion of PD-L1-containing sEVs. In accordance with the MISEV2023 guidelines [22], we utilized ultracentrifugation to separate sEVs from the supernatants of LMP1-overexpressing and control 5-8F and 6-10B cells. The successful purification was validated by NTA and TEM, revealing that the majority of isolated vesicles fell within the 30-200 nm size range, aligning with typical sEV dimensions (Figure 2A-B). LMP1 overexpression led to a 3-fold increase in PD-L1 levels in sEVs (Figure 2C). We performed immunofluorescence to visualize the co-localization of endogenous PD-L1 with CD63, a marker for late endosomes and MVBs [37]. This analysis showed a marked increase in PD-L1 localized to CD63+ MVBs in the LMP1 overexpression group (Figure 2D-E; Supplementary Figure S2A-C). Through TEM, we observed MVBs and ILVs within cells, with the results showing that LMP1 significantly enlarged the MVBs containing ILVs (Figure 2F). In addition, the quantification results of sEV PD-L1by ELISA were consistent with Western blotting, indicating higher sEV PD-L1 levels in the LMP1 overexpression group (Figure 2G), providing robust evidence of LMP1's role in enhancing PD-L1 loading into sEVs in NPC cells.
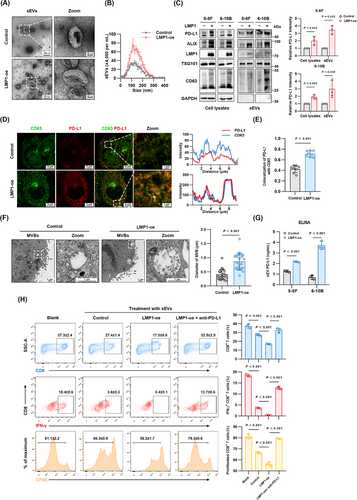
sEVs are emerging as critical mediators in oncogenesis, notably in facilitating immune evasion by tumor cells [38, 39]. Next, we aimed to validate whether LMP1-promoted sEV PD-L1 could inhibit CD8+ T cell proliferation and activity in vitro through the PD-1/PD-L1 axis. We treated PBMCs with sEVs from specified sources for 3 days, and analyzed CD8+ T cell proliferation and cytotoxicity through flow cytometry (Supplementary Figure S2D). Our results indicated that LMP1-induced sEVs significantly impeded CD8+ T cell proliferation and cytotoxicity. This was manifested as a reduction in CFSE intensity due to diminished cell division, alongside lowered cell counts and attenuated interferon-γ (IFN-γ) production (Figure 2H). Importantly, neutralization of sEV PD-L1 using blocking antibodies markedly reversed these inhibitory effects, highlighting the predominant role of PD-L1 in the LMP1-mediated suppression of CD8+ T cell activity. Collectively, these findings demonstrated that LMP1 facilitated the immune escape of NPC cells via enhancing PD-L1 expression within sEVs.
3.3 LMP1 hijacked ALIX to form the PD-L1/LMP1/ALIX trimolecular complex
Next, we aimed to unravel the molecular mechanisms underlying LMP1's facilitation of PD-L1 loading into sEVs. We conducted IP to isolate LMP1-bound protein complexes from NPC cell lysates. Subsequent SDS-PAGE and silver staining visualized these proteins (Figure 3A). MS identified ALIX, a known participant in sEV biogenesis, as an LMP1-binding protein (Figure 3B). To verify this interaction, we performed co-IP in HEK293 cells overexpressing ALIX-MYC and LMP1-GFP. Western blotting revealed the mutual interaction between ALIX and LMP1 (Figure 3C). This specific interaction between endogenous ALIX and LMP1 was further validated in NPC cells (Figure 3D).
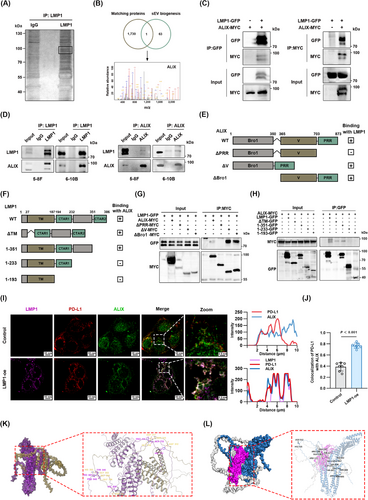
To further elucidate the specific domain of ALIX that interacts with LMP1, we constructed 3 ALIX mutants: the N-terminal Bro1 domain deletion (ΔBro1), the central V domain deletion (ΔV), and the malleable C-terminal proline-rich region (PRR) deletion (ΔPRR, Figure 3E). We also engineered 4 LMP1 mutants: transmembrane domain deletion (ΔTM), aa 1-351, aa 1-233 and aa 1-193 (Figure 3F). The co-IP results identified the PRR as the specific binding partner of LMP1 (Figure 3G). As shown in Figure 3H, LMP1 residues 1-233 did not bind to ALIX, while residues 1-351 showed binding to ALIX, indicating that the intracellular 234-351 residues of LMP1 was the critical domain for ALIX interaction. These findings suggest that LMP1 interacts with the PRR domain of ALIX through its intracellular 234-351 domain. CTAR1 and CTAR2 are 2 important functional domains located in the intracellular region of LMP1 [12, 13]. To investigate whether LMP1-induced sEV PD-L1 upregulation is independent of CTAR2, we overexpressed the 1-351 residue in 6-10B cells and assessed the level of secreted sEV PD-L1 using Western blotting and ELISA. The results showed that LMP1 1-351, which lacks CTAR2, still enhanced sEV PD-L1 secretion in 6-10B cells (Supplementary Figure S3A-B). These findings indicate that LMP1 upregulates sEV PD-L1 by hijacking ALIX, independent of CTAR2.
ALIX is involved in the ESCRT pathway, regulating the formation of ILVs in MVBs [26]. In our research model, the role of ALIX is the gateway for LMP1 to control the contents of the host cell-derived sEVs [16]. We employed laser confocal microscopy to assess the co-localization of LMP1, PD-L1 and ALIX in NPC cells. The results of immunofluorescence showed the vesicles simultaneously positive for LMP1, ALIX, and PD-L1 (Figure 3I). In control cells lacking LMP1 expression, PD-L1 and ALIX displayed negligible co-localization, whereas cells overexpressing LMP1 exhibited a significant increase in the co-localization coefficient between PD-L1 and ALIX (Figure 3J).
Besides, we also predicted the structure of the trimolecular complex by protein docking. The molecular docking results of LMP1 and ALIX showed that they bound stably (Figure 3K), further confirming the interaction between LMP1 and ALIX. We then simulated the docking of LMP1, PD-L1 and ALIX molecules, and the results showed that LMP1 could stably bind to ALIX and PD-L1 respectively, and it was these 2 combinations that enabled these 3 molecules to form stable complexes. (Figure 3L). In summary, these findings bolstered our hypothesis that LMP1 hijacked ALIX to form the PD-L1/LMP1/ALIX trimolecular complex that drives the augmented loading of PD-L1 into sEVs.
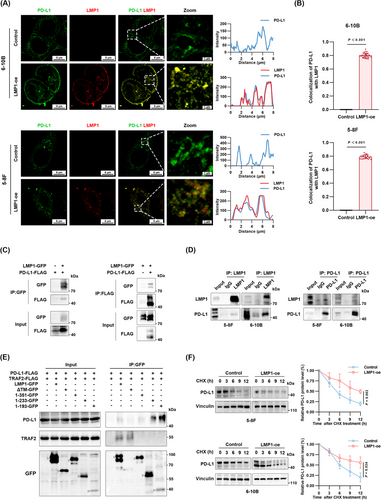
3.4 LMP1 specifically interacted with PD-L1 via its transmembrane domain, while engaging with ALIX through the intracellular domain
Through immunofluorescence, we observed co-localization between LMP1 and PD-L1, with a co-localization coefficient of 0.88 (Figure 4A-B), leading us to hypothesize the potential for protein interaction between LMP1 and PD-L1. The results of molecular docking between LMP1 and PD-L1 also showed that they formed a stable interaction (Supplementary Figure S3C). To validate the LMP1-PD-L1 interaction, co-IP was conducted in HEK293 cells transfected with PD-L1-FLAG and LMP1-GFP. Co-IP results indicated that the FLAG signal was detectable in the anti-GFP immunoprecipitated complexes, and conversely, the GFP signal was detectable in the anti-FLAG immunoprecipitated complexes, confirming the interaction between LMP1 and PD-L1 (Figure 4C). This interaction was further validated in both 5-8F and 6-10B cells endogenously expressing PD-L1 (Figure 4D). Then we sought to pinpoint the specific region in the LMP1 protein responsible for its interaction with PD-L1. Tumor necrosis factor receptor (TNFR)-associated factor 2 (TRAF2) was used as a positive control for LMP1 interaction [13, 40]. While the interaction between TRAF2 and LMP1 lacking CTAR2 was rarely detectable, our co-IP results localized the transmembrane domain of LMP1 as the specific binding site for PD-L1 (Figure 4E). CHX chase assays suggested that LMP1 overexpression prolonged the half-life of PD-L1 proteins (Figure 4F). Simultaneously, we also verified the upregulation of PD-L1 membrane expression by LMP1 through flow cytometry (Supplementary Figure S3D). Summarily, these findings illuminated that LMP1 engaged with PD-L1 through its transmembrane domain, stabilizing PD-L1. Considering the earlier findings, we believe that the concurrent binding of LMP1 with PD-L1 and ALIX respectively is crucial for the formation of the PD-L1/LMP1/ALIX trimolecular complex, resulting in enhanced secretion of sEV PD-L1.
3.5 ALIX was required for the LMP1-induced sEV PD-L1 secretion and CD8+ T cell disfunction in vitro
The crucial role of ALIX in promoting the assembly of the PD-L1/LMP1/ALIX complex prompted us to investigate its impact on the secretion of sEV PD-L1 and subsequent impact on anti-tumor immune responses. We downregulated ALIX in NPC cells stably expressing LMP1 and assessed the levels of sEV PD-L1 by Western blotting and immunofluorescence. ALIX deficiency significantly decreased sEV PD-L1 levels, whereas the PD-L1 levels in whole cell lysates remained unchanged (Figure 5A-B). This finding highlighted ALIX's specific involvement in directing PD-L1 into sEVs without affecting its expression. In addition, immunofluorescence revealed a significant reduction in PD-L1 signals within CD63+ MVBs in ALIX-deficient cells (Figure 5C-D, Supplementary Figure S4). Considering ALIX's established function in the ESCRT-mediated formation of ILVs and sEV cargo selection [26, 27], we employed TEM to evaluate how ALIX suppression impacted MVB and ILV morphology and quantity. Notably, the size of ILV-containing MVBs was considerably reduced (Figure 5E-F). These results demonstrated that ALIX was required for the LMP1-induced sEV PD-L1 secretion.
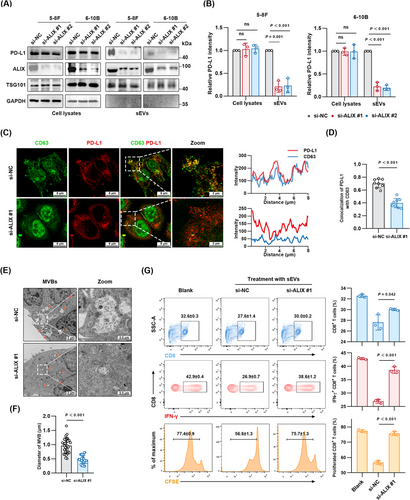
To further understand the impact of ALIX inhibition on anti-tumor immunity, especially T cell responses, we assessed the effects of sEVs on PBMCs. The results demonstrated a significant increase in the proliferation and cytolytic activity of CD8+ T cells following ALIX knockdown in NPC cells. This enhancement was evidenced by an increased fraction of CFSE-labeled cells, indicative of higher cell proliferation rates, and elevated production of IFN-γ in the ALIX knockdown (si-ALIX) group relative to the siRNA control (si-NC) group (Figure 5G). Overall, these findings highlighted that ALIX inhibition diminished the release of sEV PD-L1 and augmented the anti-tumor efficacy of CD8+ T cells.
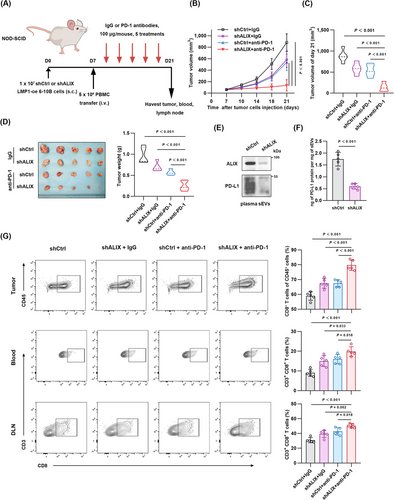
3.6 Targeting ALIX improved ICB therapy responses in vivo
It has been reported that sEVs containing PD-L1 could weaken the effectiveness of immune therapy via neutralizing PD-1 monoclonal antibodies [29]. In this study, ALIX deficiency suppressed the loading and secretion of sEV PD-L1, providing a theoretical basis for combining ALIX inhibition with immune checkpoint blockade (ICB)-based immunotherapy. Schematic diagram of ALIX inhibition combined with PD-1 monoclonal antibody therapy is shown in Figure 6A. Analysis of tumor growth curves showed that the combination therapy group significantly suppressed tumor growth compared to other groups (Figure 6B-C). Similarly, the size and weight of tumors in the combination therapy group were significantly smaller than those in other groups (Figure 6D). Extraction of sEVs from mouse plasma and analysis of PD-L1 content by Western blotting and ELISA showed that inhibition of ALIX suppressed the secretion of sEV PD-L1 in vivo (Figure 6E-F). Flow cytometry of tumor tissues revealed that the combination therapy group significantly increased the number of tumor-infiltrating CD8+ T cells (Figure 6G). Considering the ability of sEVs to inhibit peripheral immunity, we collected draining lymph nodes and peripheral blood for flow cytometry. The results showed that ALIX inhibition combined with anti-PD-1 therapy increased the number of CD8+ T cells in both peripheral and draining lymph nodes (Figure 6G). These results demonstrate that ALIX inhibition can effectively enhance the efficacy of immunotherapy in vivo.
3.7 LMP1-ALIX axis correlated with immunosuppressive microenvironment and NPC patient outcomes
To assess the clinical importance of the LMP1-ALIX axis, we examined the levels of LMP1, ALIX, CD8 and PD-1 in tumor tissue microarray by mIF and assessed the correlation between LMP1/ALIX and the clinical outcomes of patients. Patients were divided into the high and low LMP1/ALIX expression groups based on the median values of their fluorescence intensity. The clinical characteristics of enrolled NPC patients are listed in Supplementary Table S1. The mIF and Pearson correlation analysis demonstrated that ALIX was positively correlated with LMP1 levels, which was consistent with our immunofluorescence results in NPC cells (Figure 7A-B). NPC patients with elevated ALIX expression presented a reduced density of CD8+ T cells and PD-1+CD8+ T cells within the tumor milieu (Figure 7C-D). The results of IHC also suggested that LMP1 and ALIX shared similar localization and expression characteristics in NPC tissues (Figure 7E). To assess the effect of ALIX expression on the distribution of immune cells in the tumor immune microenvironment, we analyzed single-cell RNA-sequencing data for NPC from a GEO public dataset (GSE150430), which showed that high ALIX expression was associated with a low degree of CD8+ T cell infiltration (Figure 7F-G), which was also in accordance with our mIF results.
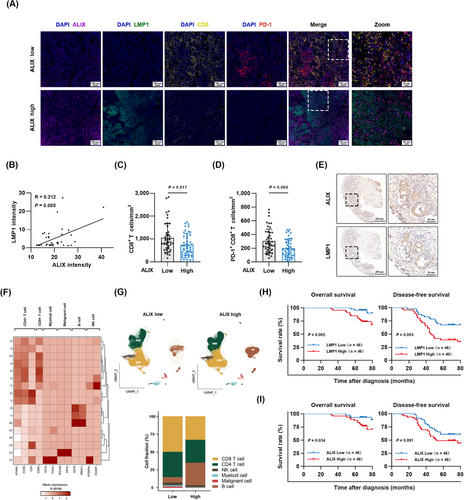
Furthermore, the NPC patients with a high level of LMP1 exhibited shorter median overall survival (69 months vs. 72 months) and disease-free survival (46 months vs. 68 months) than the patients with lower levels of LMP1 (Figure 7H). Meanwhile, a high level of ALIX in NPC patients was also associated with shorter median overall survival (69 months vs. 72 months) and disease-free survival (49.5 months vs. 66.5 months) than a low level of ALIX according to results from the NPC tissue microarray (Figure 7I). In summary, our findings suggested that the upregulated LMP1-ALIX axis might foster an immunosuppressive tumor microenvironment, shedding light on potential prognostic markers and therapeutic targets for immunotherapy in NPC.
4 DISCUSSION
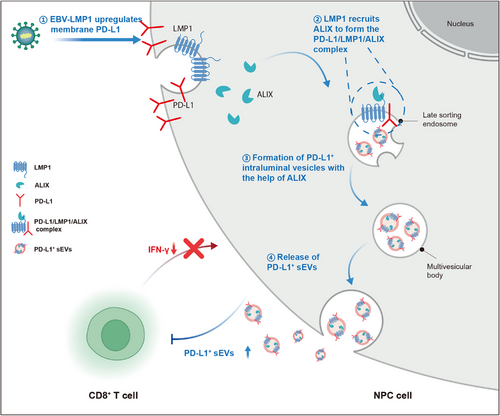
In this study, we explored the impact of LMP1 on the tumor immune microenvironment and identified the underlying mechanisms. Using a PBMC-humanized mouse tumor model and human NPC tissue samples, we found that LMP1 is linked to an immunosuppressive tumor microenvironment, partly due to its ability to enhance sEV PD-L1 expression. Through co-IP and MS, we identified ALIX as essential for LMP1's upregulation of sEV PD-L1. Mechanistically, LMP1 interacts with ALIX and PD-L1 to form a trimolecular complex, facilitating ALIX-dependent PD-L1+ sEV secretion (Figure 8). Targeting ALIX, combined with PD-1 therapy, significantly enhances anti-tumor immune responses. Additionally, mIF in NPC tissue microarrays showed that the LMP1-ALIX axis is associated with an immunosuppressive microenvironment and poorer patient prognosis.
LMP1, an oncoprotein of EBV in the latency program, plays a crucial role in mediating EBV-induced malignant transformation [10, 13]. However, the role of LMP1 in the interaction between EBV and host immunity remains controversial [41]. Previous studies indicated that immune evasion signaling mediated through LMP1 might immunize EBV-infected cells from host immune surveillance during the latent infection [11]. In contrast, Choi et al. [42] argued that LMP1 expression in B cells resulted in the overexpression of multiple cellular antigens, which subsequently led to the induction of robust cytotoxic responses by CD4+ and CD8+ T cells. These findings suggested that the role of LMP1 in the interaction between the virus and the host immune system still requires further investigation. In our research, we have established the correlation between LMP1 and an immunosuppressive microenvironment in both PBMC-humanized mouse tumor models and human NPC tissue samples. Kase et al. [43] also reported a correlation between LMP1 expression and elevated serum PD-L1 levels in NPC patients. It is well-established that serum PD-L1 is prone to degradation in circulation and faces challenges in penetrating blood vessels or stroma. In contrast, PD-L1+ sEV, with its double lipid membrane structure, offers superior penetrability and more effectively mediates both local and peripheral immune suppression. Our findings further reveal that LMP1 enhances the secretion of sEV PD-L1 both in vivo and in vitro, which mediates the dysfunction of CD8+ T cells. While this study focused on the impact of LMP1 on tumor infiltrating lymphocytes, the impact of LMP1 on other types of immune cells within the tumor microenvironment remains to be investigated.
The role of sEV PD-L1 in regulating immune responses and mediating resistance to immunotherapy is receiving increasing attention [31, 44]. Therefore, understanding the mechanism of PD-L1 sorting into sEVs is crucial for overcoming immunotherapy resistance. In this study, we uncovered that the process of loading PD-L1 into sEVs was manipulated by EBV-encoded LMP1 in EBV+ NPC. The mechanism by which LMP1 upregulates sEV PD-L1 is partly due to the interaction between LMP1 and PD-L1. Previous studies primarily focused on the upregulation of PD-L1 by LMP1 through the NF-κB pathway at the transcriptional level [18, 45], without addressing the direct interaction between LMP1 and PD-L1. It is noteworthy that LMP1 can bind to PD-L1 through its transmembrane domains. In addition, our studies revealed that the interaction between LMP1 and PD-L1 enhanced the stability of PD-L1, with the underlying mechanisms remaining an area for future research. Moreover, given the structural and functional similarities between sEVs and enveloped viral particles [46], it is fascinating to explore whether similar mechanisms exist in other types of viruses rather than EBV to regulate the selection and sorting of sEVs. Overall, our findings highlighted a novel pathway regulating PD-L1 loading into sEVs hijacked by EBV. Considering the recognized prognostic value of sEV PD-L1 in predicting responses to immunotherapy and cancer progression [29, 47], sEV PD-L1 holds promise as a future biomarker for immunotherapy efficacy in NPC.
Our co-IP and MS results demonstrated that LMP1-promoted sEV PD-L1 secretion depended on ALIX, also known as programmed cell death 6-interacting protein, which is recognized as an ESCRT-associated protein [27]. In prior studies, ALIX was shown to regulate ILV formation and control the sEV cargo sorting. Interestingly, Baietti et al. [28] identified that the trimolecular complex syndecan/syntenin/ALIX regulates the biogenesis of sEVs. This discovery led us to consider whether PD-L1 and LMP1, as well as ALIX, could form a trimolecular complex and thus promote PD-L1 packaging into sEVs. Rider et al. [48] further supported this hypothesis by using BioID combined with MS to identify over 1,000 proteins interacting with LMP1, including CD63, syntenin-1, ALIX, TSG101, hepatocyte growth factor-regulated tyrosine kinase substrate, and charged multivesicular body proteins. This suggested a potential interaction between LMP1 and ALIX. To validate this hypothesis, we confirmed that the intracellular domain of LMP1 binds directly to the PRR domain of ALIX, and that the transmembrane domain of LMP1 binds to PD-L1, conforming a trimolecular complex for sEV cargo sorting. Interestingly, as a known binding partner of LMP1, TRAF2's interaction with LMP1 was interfered by the deletion of aa 351-382 including CTAR2 domain, which was not consistent with Devergne's work [49]. While the interaction of aa 187-231 LMP1 with TRAF2 was detected in the cell-free GST-pulldown assay, our co-IP assays were performed in HEK293T cells, which might better preserve the tertiary structure of the proteins. The detailed functions of specific domains in the LMP1 protein with its binding partners are required to be investigated.
Moreover, several reports demonstrated that retroviruses, including human immunodeficiency virus type 1 [50] and simian immunodeficiency virus [51], bound the V domain of ALIX through YPXnL late-domain motifs and recruited ESCRT-III subunits to support their virus budding. Combined with our findings, we suspected that ALIX might be a vital gateway that viruses exploit to achieve their own goals in virus-host interactions. Compared to previous studies, our research innovates in showing that ALIX's role is not limited to being utilized by viruses for the assembly of viral particles; it can also be exploited by viruses to alter the contents and properties of sEVs derived from tumor cells. Additionally, our results indicated that LMP1 binds to the PRR domain of ALIX rather than the V domain. These findings might provide novel evidence that PRR interaction with LMP1 was essential for EBV-mediated sEV cargo sorting. As for whether PRR is responsible for EBV assembly and budding in host cells, further investigations are still required. Furthermore, in our PBMC-humanized mouse tumor model, we have shown for the first time that the combination of ALIX inhibition and immunotherapy can enhance anti-tumor immune responses. Our mIF results of NPC tissue microarray indicated that ALIX expression is associated with an immunosuppressive microenvironment and its high expression correlates with poor patient prognosis. These findings consistently suggest that ALIX is a promising candidate target for enhancing immunotherapy efficacy, with its effectiveness in other cancer types yet to be verified.
This study revealed that EBV-encoded LMP1 upregulates PD-L1 on sEVs by interacting with PD-L1 and ALIX, leading to immune suppression of CD8+ T cells. However, due to the complex interactions among various immune cells in the tumor microenvironment, it remains to be seen whether LMP1 also affects other immune cells, such as B cells and natural killer cells. While our in vivo and in vitro studies demonstrate that inhibiting ALIX improves the tumor immune microenvironment, and multiplex immunofluorescence staining of NPC patient samples links ALIX to an inhibitory immune environment, further validation is needed to determine if these findings apply to other cancer types.
5 CONCLUSIONS
In summary, our work makes 3 contributions. First, we identified an essential role of LMP1 in sEV PD-L1 sorting and EBV-mediated immune evasion in NPC. Second, we elucidated the mechanism by which LMP1 acted as a scaffolding protein linking PD-L1 and ALIX, thereby enabling PD-L1 loading into sEVs. Third, we provided original evidence that the PRR domain of ALIX served as a gateway to EBV-controlled host cellular sEV cargo sorting and that targeting ALIX helps enhancing immunotherapy efficacy. Together, we offer novel insights into EBV-mediated immune escape in NPC through the LMP1/ALIX/sEV PD-L1 axis and highlight ALIX as a promising therapeutic target for enhancing anti-tumor immunity.
DECLARATIONS
AUTHOR CONTRIBUTIONS
C.X., Y.Z., Y.G. and F.H. conceive the study and design experiments. F.H., G.T., J.Z., Y.C., C.W., L.H., Y.G., J.H., R.B., P.S., and X.Y. carried out the experiments and analyzed data. G.T., Y.T. and J.Y. helped with animal experiments. Q.W. and Z.W. provided critical suggestions and technical support. F.H. wrote the draft. C.X., Y.Z. and Y.G. revised the manuscript. All authors read and approved the final manuscript.
ACKNOWLEDGEMENTS
We thank all members of the Xie laboratory for helpful discussions and technical assistance. We would like to thank Professor Melvin L. K. Chua from the National Cancer Centre Singapore for his professional advice during the manuscript revision process. We also appreciate Professor Gang Chen from School and Hospital of Stomatology of Wuhan University for technique support. This study was supported by the National Natural Science Foundation of China (81803065 and 81972852), Research Projects of Biomedical Center of Hubei Cancer Hospital (2022SWZX12), Key Research & Development Project of Hubei Province (2020BCA069), Health Commission of Hubei Province Medical Leading Talent Project, and Translational Medicine and Interdisciplinary Research Joint Fund of Zhongnan Hospital of Wuhan University (ZNJC201922 and ZNJC202007).
CONFLICT OF INTEREST STATEMENT
The authors declare no conflict of interests.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The collection of plasma samples from NPC patients was approved by the Medical Ethics Committee at Zhongnan Hospital of Wuhan University (ZN2021073). Written informed consent was obtained from all patients. The NPC tissue array was purchased from Outdo Biotech, Shanghai, with approval from the Ethics Committee of Shanghai Outdo Biotech Company (YB M-05-02). All mouse experiments were conducted in compliance with animal ethical standards and approved by the Ethics Committee of the Bestcell Model Biology Center (ZN2024012).
Open Research
DATA AVAILABILITY STATEMENT
The RNA-seq results of NPC 6-10B cells with or without LMP1 overexpression is deposited in the National Genomics Data Center (NGDC) under the accession number: PRJCA028411. Raw data for Figures 1-7 and Supplementary Figures S1-S4 were available in the Supplementary files. Any additional information required to reanalyze the data in this paper can be requested from the corresponding author upon reasonable request.




