Real-world data on ALK rearrangement test in Chinese advanced non-small cell lung cancer (RATICAL): a nationwide multicenter retrospective study
Lin Li, Wencai Li, Chunyan Wu, Yanfeng Xi, Lei Guo, Yuan Ji, Lili Jiang, Ji Li, Jingping Yun, Gang Chen, and Yuan Li contributed equally to this work as co-first authors.
Abstract
Background
Anaplastic lymphoma kinase (ALK) test in advanced non-small cell lung cancer (NSCLC) can help physicians provide target therapies for patients harboring ALK gene rearrangement. This study aimed to investigate the real-world test patterns and positive rates of ALK gene rearrangements in advanced NSCLC.
Methods
In this real-world study (ChiCTR2000030266), patients with advanced NSCLC who underwent an ALK rearrangement test in 30 medical centers in China between October 1, 2018 and December 31, 2019 were retrospectively analyzed. Interpretation training was conducted before the study was initiated. Quality controls were performed at participating centers using immunohistochemistry (IHC)-VENTANA-D5F3. The positive ALK gene rearrangement rate and consistency rate were calculated. The associated clinicopathological characteristics of ALK gene rearrangement were investigated as well.
Results
The overall ALK gene rearrangement rate was 6.7% in 23,689 patients with advanced NSCLC and 8.2% in 17,436 patients with advanced lung adenocarcinoma. The quality control analysis of IHC-VENTANA-D5F3 revealed an intra-hospital consistency rate of 98.2% (879/895) and an inter-hospital consistency rate of 99.2% (646/651). IHC-VENTANA-D5F3 was used in 53.6%, real-time polymerase chain reaction (RT-PCR) in 25.4%, next-generation sequencing (NGS) in 18.3%, and fluorescence in-situ hybridization (FISH) in 15.9% in the adenocarcinoma subgroup. For specimens tested with multiple methods, the consistency rates confirmed by IHC-VENTANA-D5F3 were 98.0% (822/839) for FISH, 98.7% (1,222/1,238) for NGS, and 91.3% (146/160) for RT-PCR. The overall ALK gene rearrangement rates were higher in females, patients of ≤ 35 years old, never smokers, tumor cellularity of > 50, and metastatic specimens used for testing in the total NSCLC population and adenocarcinoma subgroup (all P < 0.05).
Conclusions
This study highlights the real-world variability and challenges of ALK test in advanced NSCLC, demonstrating a predominant use of IHC-VENTANA-D5F3 with high consistency and distinct clinicopathological features in ALK-positive patients. These findings underscore the need for a consensus on optimal test practices and support the development of refined ALK test strategies to enhance diagnostic accuracy and therapeutic decision-making in NSCLC.
Abbreviations
-
- ALK
-
- anaplastic lymphoma kinase
-
- EMR
-
- electronic medical record
-
- FISH
-
- fluorescence in-situ hybridization
-
- IHC
-
- immunohistochemistry
-
- NGS
-
- next-generation sequencing
-
- NSCLC
-
- non-small cell lung cancer
-
- NMPA
-
- national medical products administration;
-
- RT-PCR
-
- real-time polymerase chain reaction
1 BACKGROUND
Primary lung cancer (hereafter refers to as lung cancer) that begins in the lungs is one of the most common malignant tumors in China [1] and worldwide [2]. According to the World Health Organization, the Global Cancer Observatory (GLOBOCAN) 2020 and the National Cancer Center database, 815,600 new lung cancer patients and 714,700 deaths caused by lung cancer were seen in 2020 in China, ranking first among all malignant tumors causing mortality [3]. More than 80% of lung cancer patients have non-small cell lung cancer (NSCLC) [1, 2].
In the past decade, discovering predictive biomarkers has paved the way for new treatment opportunities using targeted therapy and immunotherapy in the NSCLC [4-6]. Anaplastic lymphoma kinase (ALK) gene rearrangement is the third most common driving mutation in the development of NSCLC after mutations in epidermal growth factor receptor (EGFR) and Kirsten rat sarcoma viral oncogene homolog (KRAS) [7, 8], and previous studies from China [9] and America [10] showing that ALK gene rearrangement occurs in 3%-7% of NSCLC cases, with a high incidence among adenocarcinoma and non-smoking patients. The echinoderm microtubule-associated protein-like 4 (EML4)-ALK fusion is the primary type of ALK rearrangement. In addition to EML4, other genes fused with the ALK gene in lung cancer include kinesin family 5B (KIF5B), kinesin light chain (KLC), trafficking from ER to golgi regulator (TFG), and others [11-13].
Currently, the molecular target drugs in the market (e.g., crizotinib and alectinib) for lung cancer with ALK gene rearrangement and other genetic variations have significantly improved the patients' outcome [14, 15], even for squamous cell carcinoma [16]. Therefore, detecting the ALK gene rearrangements in an accurate, standardized, and cost-effective manner is critical for selecting drugs to manage lung cancer. Presently, China's National Medical Products Administration (NMPA) has approved ALK gene rearrangement diagnostics in four technical platforms, such as immunohistochemistry (IHC)-VENTANA⁃D5F3, fluorescence in-situ hybridization (FISH), real-time polymerase chain reaction (RT-PCR), and next-generation sequencing (NGS). The FISH technique requires a small amount of tumor sample, but the procedure is technologically demanding and has atypical negative signals. RT-PCR has high specificity and sensitivity but can only detect currently known variants. IHC-VENTANA-D5F3 is simple, inexpensive, and can be performed in almost all locations, but the criteria for positive response are not applicable in all cases, leading to ambiguous results and the need for other platforms. NGS can detect most variants, but it is expensive and requires a test run. To address this situation, experts from the RATICAL study and the Chinese Society of Clinical Oncology issued guidelines for the ALK fusion test in NSCLC in 2021 to adopt in clinical practice [17].
However, selecting an appropriate diagnostic method with good quality control for detecting ALK rearrangements has been challenging, impacting the clinical benefit. To gain knowledge on the current status and quality control of ALK gene test in lung adenocarcinoma, we planned a nationwide multicenter clinical study (the RATICAL study) including NSCLC patients who underwent ALK gene test from 30 medical centers. This study aimed to investigate the real-world test patterns of ALK gene rearrangements in advanced NSCLC, including the detection methods and quality control, and to investigate the positive rates, the associated clinicopathological characteristics, and the presence of other mutations.
2 METHODS
2.1 Study design and participants
In this real-world data on ALK Test In Chinese Advanced Lung cancer (i.e., the RATICAL study) (registered at https://www.chictr.org.cn/, with identifier ChiCTR2000030266) conducted across 30 nationwide medical centers, data from patients with advanced NSCLC who underwent an ALK test between October 1, 2018 and December 31, 2019 were retrospectively collected (28 centers provided data). This retrospective study was approved by the Ethics Committee of the Cancer Hospital of the Chinese Academy of Medical Sciences (approval no. 2018091013463902) and reported to the Ethics Committee of all study centers.
The inclusion criteria were 1) patients histologically or cytologically diagnosed with NSCLC and 2) patients with advanced NSCLC (IIIB, IIIC, or IV), as defined by the American Joint Committee on Cancer Staging Manual, 8th edition, who underwent ALK test. The exclusion criteria were 1) patients who went for a third-party ALK assay outside the participating hospitals, 2) patients who underwent ALK test using different types of specimens, where each sample type used a different test method, 3) patients with no tumor cells in hydrothorax, as the absence of tumor cells may compromise the reliability of ALK test results, 4) patients who had the RT-PCR detection method with targeted multiple genes/locations but no ALK gene rearrangements, 5) patients with missing information such as age, sex or ALK results, and 6) patients who did not have conclusive ALK test results that could be analyzed.
2.2 Data collection
The baseline information of the patients, such as age, sex, smoking status (current, past, or never), type of cancer (adenocarcinoma, squamous, or others), tumor site (non-metastatic or metastatic), and cancer stage, was collected from the electronic medical record (EMR) system.
This study was a real-world study. Therefore, ALK test was based on the clinical reality of each participating hospital. In order to detect ALK and other driver genes, particulars on the type of sample tested (samples from tissue, body fluid, and cells), the test methods (FISH, IHC, NGS, and RT-PCR), and the results (positive or negative) were collected. Specifically, for IHC testing, data regarding the use of different antibody variants such as 5A4, and 1A4 were also gathered. Information on other driver genes or biomarkers was also recorded, including B-Raf proto-oncogene serine/threonine-protein kinase (BRAF), human epidermal growth factor receptor 2 (ERBB2), KRAS, mesenchymal-epithelial transition factor (MET), neuroblastoma rat sarcoma viral oncogene homolog (NRAS), rearranged during transfection (RET), c-ros oncogene 1 (ROS1), and programmed death ligand-1 (PD-L1).
2.3 Quality control
Supplementary Figure S1 presents the technological workflow. We included 109 cases of lung adenocarcinoma diagnosed using the automated ALK IHC-VENTANA-D5F3 test for the analysis of consistency between local pathologists who reported initial results at each participating cancer and study group pathologists who initiated this RATICAL study. These cases included 49 positive and 60 negative diagnoses, all re-assessed by 31 pathologists from the RATICAL study group (including 13 chief pathologists, 11 associate chief pathologists, and 7 attending pathologists). The concordance rate achieved was 98.2%, with only two cases showing inconsistency between local pathologists and pathologists from the RATICAL study group, as previously reported [18].
To further ensure the accuracy of the IHC-VENTANA-D5F3 results, an intra-hospital quality control procedure was implemented across 22 participating hospitals. In addition to the intra-hospital quality control, the inter-hospital concordance rate of IHC-VENTANA-D5F3 was assessed by evaluating 10 ALK gene rearrangement cases and 20 ALK-negative cases across different hospitals within the same city or region, using multifocal microscopes to ensure the reproducibility of results across nearby facilities. For samples initially tested using RT-PCR or NGS, a sample of cases (both positive and negative) that had results available for ALK test through IHC, RT-PCR, or NGS were selected for inter-hospital consistency analysis. Each of 11 hospitals randomly selected cases in a recommended ratio of RT-PCR: NGS = 6:4, or 10 RT-PCR cases if NGS was not available. These samples were sent to the central laboratory at Department of Pathology at the Cancer Hospital of the Chinese Academy of Medical Sciences, Beijing, China for retesting using ALK IHC-VENTANA-D5F3 to compare and validate the findings across different diagnostic platforms.
2.4 Statistical analysis
The continuous data were tested for normal distribution using the Shapiro-Wilk method. Continuous data with a normal distribution are expressed as mean ± standard deviation; otherwise, they are expressed as median (upper and lower quartiles). Categorical data are expressed as percentages, and the chi-square test was used to compare groups. The differences were statistically significant when the P value was <0.05. R programming language version 4.1.1 (R Foundation for Statistical Computing, Vienna, Austria) was used for all statistical analyses.
3 RESULTS
3.1 Quality control outcomes
Using the IHC-VENTANA-D5F3 protocol, nationwide clinical centers tested 895 cases for intra-hospital quality control. Among these, 288 were initially reported as positive, 601 were negative, and 6 were classified as suspiciously positive. Sixteen patients exhibited inconsistent results compared to initial evaluations, resulting in an overall concordance rate of 98.2%. The inconsistencies were primarily due to several factors: squamous carcinoma with inconclusive IHC results, positive control failure, incorrect interpretations, and excessive non-specific background staining.
In the inter-hospital consistency analysis of IHC-VENTANA-D5F3, a total of 651 samples (216 with positive ALK gene rearrangement and 435 samples without) were tested. Upon retesting these samples in the central laboratory (Department of Pathology at the Cancer Hospital of the Chinese Academy of Medical Sciences, Beijing, China), 5 of these results were inconsistent with the initial findings, with a concordance rate of 99.2%. These inconsistencies were primarily attributed to being misclassified as positive due to intracytoplasmic patch-like staining in squamous carcinoma, weak staining, and positive control failure.
In order to determine quality control using other protocols, 119 samples were retested, and 2 were not included in the analyses due to the absence of tumor cells on received IHC-VENTANA-D5F3 slides. Eight of the 117 patients had inconsistent results compared with the initial results, reaching a concordance rate of 93.2%. Among these inconsistencies, 5 cases initially diagnosed as positive by RT-PCR at local laboratories at each participating center were tested negative by IHC-VENTANA-D5F3 at the central laboratory. Upon retesting at the local laboratories, 3 of these cases were confirmed as negative by both IHC-VENTANA-D5F3 and 2 by FISH, suggesting initial false positives. One case, initially diagnosed as weakly positive by IHC-VENTANA-D5F3, showed positive results by FISH upon retesting, and another case had no tumor cells available at the time of retesting. One positive case by initial FISH was tested negative by IHC-VENTANA-D5F3 at the center laboratory, and no tumor cells were found upon retesting at the local laboratories. Two cases were were tested negative by FISH at local centers but tested positive by IHC-VENTANA-D5F3 at the center laboratory; both were confirmed as positive upon retesting at the local laboratories at each participating center.
3.2 Patient characteristics
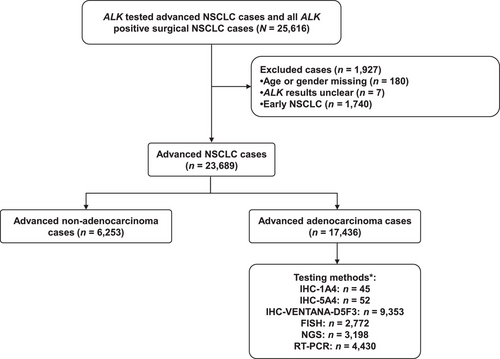
A total of 25,616 patients were screened, 187 patients were excluded due to missing data and unclear ALK gene rearrangement results, and 1,740 patients were excluded because they had early-stage NSCLC. Therefore, 23,689 samples were analyzed, including 17,436 patients with adenocarcinoma (Figure 1). In the present study, 19.6% of patients were tested with samples obtained from metastatic foci and 81.7% of ALK tests were performed on histological samples in clinical practice, followed by cytological samples (15.9%). The overall ALK gene rearrangement rate was 6.7% in advanced NSCLC. ALK rearrangements were more common in adenocarcinoma (8.2%) than in squamous (1.5%) and other types (4.2%) (P < 0.001). The characteristics of the patients with NSCLC and adenocarcinoma are shown in Table 1.
| Advanced NSCLC (n = 23,689) | Advanced adenocarcinoma (n = 17,436) | |||||
|---|---|---|---|---|---|---|
| Variable | ALK-positive | ALK-negative | P | ALK-positive | ALK-negative | P |
| Total, n (%) | 1,590 (6.7) | 22,099 (93.3) | 1,422 (8.2) | 16,014 (91.8) | ||
| Sex, n (%) | <0.001 | <0.001 | ||||
| Male | 757 (5.2) | 13,725 (94.8) | 658 (7.1) | 8,669 (92.9) | ||
| Female | 833 (9.0) | 8,374 (91.0) | 764 (9.4) | 7,345 (90.6) | ||
| Age, n (%) | <0.001 | <0.001 | ||||
| ≤ 35 | 110 (31.9) | 235 (68.1) | 102 (34.6) | 193 (65.4) | ||
| 36-65 | 1,217 (8.4) | 13,320 (91.6) | 1,090 (9.8) | 10,001 (90.2) | ||
| > 65 | 263 (3.0) | 8,544 (97.0) | 230 (3.8) | 5,820 (96.2) | ||
| Smokers, n (%) | <0.001 | <0.001 | ||||
| Current | 280 (4.2) | 6,381 (95.8) | 238 (5.6) | 3,985 (94.4) | ||
| Never | 853 (8.3) | 9,439 (91.7) | 782 (9.3) | 7,650 (90.7) | ||
| Unknown | 457 (6.8) | 6,279 (93.2) | 402 (8.4) | 4,379 (91.6) | ||
| Tumor cellularity, n (%) | 0.024 | 0.033 | ||||
| ≤ 30 | 227 (6.7) | 3,181 (93.3) | 215 (7.9) | 2,494 (92.1) | ||
| 31-50 | 214 (7.2) | 2,756 (92.8) | 196 (8.3) | 2,155 (91.7) | ||
| > 50 | 325 (8.3) | 3,590 (91.7) | 295 (9.8) | 2,720 (90.2) | ||
| Tumor location, n (%) | <0.001 | <0.001 | ||||
| Right upper lobe | 173 (4.4) | 3,760 (95.6) | 155 (5.2) | 2,807 (94.8) | ||
| Right lower lobe | 196 (7.8) | 2,317 (92.2) | 177 (9.5) | 1,696 (90.5) | ||
| Right middle lobe | 132 (9.8) | 1,213 (90.2) | 121 (11.9) | 898 (88.1) | ||
| Left upper lobe | 226 (6.8) | 3,103 (93.2) | 204 (8.4) | 2,230 (91.6) | ||
| Left lower lobe | 179 (8.7) | 1,887 (91.3) | 160 (10.5) | 1,371 (89.5) | ||
| Other | 684 (6.5) | 9,819 (93.5) | 605 (7.9) | 7,012 (92.1) | ||
| Sampling lesion, n (%) | <0.001 | <0.001 | ||||
| Primary | 1,043 (5.6) | 17,443 (94.4) | 934 (7.1) | 12,247 (92.9) | ||
| Metastatic | 498 (10.7) | 4,151 (89.3) | 452 (11.5) | 3,469 (88.5) | ||
| Unknown | 49 (8.8) | 505 (91.2) | 36 (10.8) | 298 (89.2) | ||
| Pathology, n (%) | <0.001 | |||||
| Adenocarcinoma | 1,422 (8.2) | 16,014 (91.8) | ||||
| Squamous | 50 (1.5) | 3,392 (98.5) | ||||
| Other | 118 (4.2) | 2,693 (95.8) | ||||
| TNM stage, n (%) | 0.001 | 0.003 | ||||
| III | 331 (5.7) | 5,512 (94.3) | 293 (7.8) | 3,463 (92.2) | ||
| IV | 874 (6.9) | 11,752 (93.1) | 780 (7.8) | 9,240 (92.2) | ||
| Unknown | 385 (7.4) | 4,835 (92.6) | 349 (9.5) | 3,311 (90.5) | ||
| Sample type, n (%) | 0.003 | 0.003 | ||||
| Histological specimen | 1,340 (6.9) | 18,017 (93.1) | 1,204 (8.3) | 13,224 (91.7) | ||
| Cytological specimen | 226 (6.0) | 3,536 (94.0) | 196 (7.7) | 2,337 (92.3) | ||
| Body fluid specimens | 15 (3.2) | 448 (96.8) | 13 (3.4) | 375 (96.6) | ||
| Other | 9 (8.4) | 98 (91.6) | 9 (10.3) | 78 (89.7) | ||
- Abbreviation: NSCLC, non-small cell lung cancer.
In the total NSCLC patients, the overall ALK gene rearrangement rates were higher in females (P < 0.001), ≤ 35 years of age (P < 0.001), never smokers (P < 0.001), with tumor cellularity of >50 (P = 0.024), samples from metastatic tumors (P < 0.001), and diagnosed using histological specimens (P = 0.003), similarly in the adenocarcinoma subgroup (Table 1). Supplementary Figure S2 shows the distribution of non-adenocarcinoma cases from the participating centers.
3.3 Frequency and positive ALK gene rearrangement rate by different detection methods
Table 2 presents the data of ALK gene rearrangement determined by different detection methods in the adenocarcinoma subgroup. The most common detection method was IHC-VENTANA-D5F3 (53.6%), followed by RT-PCR (25.4%), NGS (18.3%, all DNA-based), and FISH (15.9%). The frequency of detection methods in each center differed (Supplementary Table S1). In the meantime, the ALK gene rearrangement rates determined by RT-PCR, NGS, and FISH were 9.1%, 8.2%, and 5.3%, respectively. The ALK gene rearrangement rate varied from 4.3% to 11.0% in advanced NSCLC and from 4.7% to 11.9% in advanced adenocarcinoma across hospitals (Supplementary Table S2). For specimens tested with multiple methods, the agreement of detection was high (>95%) among the methods, except between IHC-VENTANA-D5F3 and PCR (91.3%), NGS and PCR (90.9%), and IHC-5A4 and IHC-VENTANA-D5F3 (75.0%) (Table 3).
| Advanced adenocarcinoma, n (%) | ||||
|---|---|---|---|---|
| Detection method | Overalla | ALK-positive | ALK-negative | Uncertain |
| IHC-VENTANA-D5F3 | 9,353 (53.6) | 847 (9.1) | 8,494 (90.8) | 12 (0.1)b |
| IHC-1A4 | 45 (0.3) | 3 (6.7) | 41 (91.1) | 1 (2.2)b |
| IHC-5A4 | 52 (0.3) | 2 (3.8) | 49 (94.2) | 1 (1.9)b |
| RT-PCR | 4,430 (25.4) | 404 (9.1) | 4,026 (90.9) | 0 |
| NGS | 3,198 (18.3) | 261 (8.2) | 2,937 (91.8) | 0 |
| FISH | 2,772 (15.9) | 146 (5.3) | 2,626 (94.7) | 0 |
- Abbreviations: FISH, fluorescence in-situ hybridization; IHC, Immunohistochemistry; NGS, next-generation sequencing; RT-PCR, real-time polymerase chain reaction.
- a The percentage represents the proportion of patients using each method among 17,436 patients.
- b The 12 samples with uncertain results using IHC-VENTANA-D5F3 were confirmed using FISH (11 negative and 1 positive); 1 sample with uncertain result using IHC-1A4 and 1 using IHC-5A4 were confirmed using FISH (both negative).
| Detection methods | IHC-D5F3 | FISH | NGS | PCR | |
|---|---|---|---|---|---|
| IHC | 1A4 | 100% (7/7) | 96.9% (31/32) | ||
| 5A4 | 75.0% (3/4) | 96.6% (28/29) | |||
| IHC-VENTANA-D5F3 | — | 98.0% (822/839) | 98.7% (1,222/1,238) | 91.3% (146/160) | |
| FISH | — | 96.1% (123/128) | 100% (1/1) | ||
| NGS | — | 90.9% (20/22) | |||
| PCR | — | ||||
- Abbreviations: FISH, fluorescence in-situ hybridization; IHC, Immunohistochemistry; NGS, next-generation sequencing; PCR, polymerase chain reaction.
3.4 Common mutations in patients with advanced NSCLC and adenocarcinoma
As shown in Table 4, the positive rates of EGFR, KRAS, ERBB2, ROS1, MET, BRAF, and RET gene alterations in patients with advanced NSCLC were 44.9% (5,730/12,763), 9.0% (912/10,181), 2.2% (177/7,933), 2.0% (276/13,748), 1.8% (143/7,733), 1.3% (134/9,978), and 1.2% (104/8,932), respectively.
| Advanced NSCLC | Advanced adenocarcinoma | |||
|---|---|---|---|---|
| Genotype | N |
ALK-positive n (%) |
N |
ALK-positive n (%) |
| BRAF | ||||
| Positive | 134 | 1 (0.7) | 113 | 1 (0.9) |
| Negative | 9,844 | 692 (7.0) | 7,467 | 608 (8.1) |
| ERBB2 | ||||
| Positive | 177 | 3 (1.7) | 156 | 3 (1.9) |
| Negative | 7,756 | 487 (6.3) | 5,838 | 431 (7.4) |
| EGFR | ||||
| Positive | 5,730 | 32 (0.6) | 5,309 | 30 (0.6) |
| Negative | 7,033 | 917 (13.0) | 4,769 | 820 (17.2) |
| KRAS | ||||
| Positive | 912 | 10 (1.1) | 745 | 9 (1.2) |
| Negative | 9,269 | 707 (7.6) | 7,011 | 624 (8.9) |
| MET | ||||
| Positive | 143 | 3 (2.1) | 122 | 3 (2.5) |
| Negative | 7,590 | 488 (6.4) | 5,752 | 434 (7.5) |
| RET | ||||
| Positive | 104 | 2 (1.9) | 92 | 2 (2.2) |
| Negative | 8,828 | 540 (6.1) | 6,731 | 478 (7.1) |
| ROS1 | ||||
| Positive | 276 | 10 (3.6) | 234 | 10 (4.3) |
| Negative | 13,472 | 852 (6.3) | 9,827 | 755 (7.7) |
| PD-L1 [22C3] | ||||
| Positive | 779 | 75 (9.6) | 597 | 70 (11.7) |
| Negative | 1,042 | 60 (5.8) | 822 | 56 (6.8) |
| PD-L1 [288] | ||||
| Positive | 510 | 24 (4.7) | 348 | 19 (5.5) |
| Negative | 824 | 34 (4.1) | 672 | 28 (4.1) |
| PD-L1 [SP142] | ||||
| Positive | 1,009 | 42 (4.2) | 602 | 35 (5.8) |
| Negative | 1,709 | 83 (4.9) | 1,292 | 94 (7.3) |
| PD-L1 [SP263] | ||||
| Positive | 186 | 21 (11.3) | 129 | 18 (14.0) |
| Negative | 120 | 5 (4.2) | 79 | 5 (6.3) |
- Abbreviation: NSCLC, non-small cell lung cancer.
In patients with advanced adenocarcinoma, compared with various positive mutations, EGFR-negative (17.2%), BRAF-negative (8.1%), ROS1-negative (7.7%), and MET-negative (7.5%) tumors had more frequent ALK rearrangements, but PD-L1-negative tumors had fewest ALK rearrangements. The ALK gene rearrangement was more frequently seen with ROS1 (4.3%), MET (2.5%), RET (2.2%), ERBB2 (1.9%), KRAS (1.2%), BRAF (0.9%), and EGFR mutations (0.6%) than other mutations in advanced adenocarcinoma.
3.5 Mutation-positive rates in subgroups of patients with advanced adenocarcinoma
Table 5 presents the positive rates of driver genes in different subgroups of patients with advanced adenocarcinoma. The ALK-positiveALK gene rearrangement rate was higher in females (9.4%) than in males (6.8%). The EGFR-positive tumors were higher in females (66.5%) and mainly in never smokers of different age groups. Three hundred and seventy-five males out of 2,609 (14.4%) had KRAS mutation, and 99 females out of 2,492 (4.0%) were KRAS mutation, predominantly current smokers aged > 35 years. Additionally, the BRAF, ERBB2, MET, RET, and ROS1 positive rates were 1.7%, 3.0%, 3.3%, 1.5%, and 2.0% in males and 1.6%, 3.1%, 2.0%, 1.2%, and 3.0% in females. The results showed that 74.3% of male patients and 90.8% of female patients had mutations in at least one of these eight driver genes, making them potential candidates for targeted drugs.
| Age (years) and smoking status | |||||||
|---|---|---|---|---|---|---|---|
| ≤ 35 | 36-65 | > 65 | |||||
| Mutated gene | Mutated rate | Never | Current | Never | Current | Never | Current |
| ALK | |||||||
| Male | 6.8% (456/6,681) | 37.7% (23/61) | 46.2% (18/39) | 10.3% (169/1,635) | 6.7% (171/2,541) | 4.0% (38/942) | 2.5% (37/1,463) |
| Female | 9.4% (564/5,974) | 33.9% (42/124) | 0 (0/2) | 11.3% (430/3,796) | 8.2% (8/98) | 4.3% (80/1,874) | 5.0% (4/80) |
| BRAF | |||||||
| Male | 1.7% (42/2,525) | 0 (0/21) | 0 (0/14) | 1.6% (10/626) | 1.4% (13/961) | 1.2% (4/332) | 2.6% (15/571) |
| Female | 1.6% (39/2,431) | 4.3% (2/47) | 0 (0/0) | 1.3% (21/1,559) | 2.1% (1/47) | 2.0% (15/755) | 0 (0/23) |
| EGFR | |||||||
| Male | 41.6% (1,521/3,659) | 34.5% (10/29) | 30.4% (7/23) | 51.3% (434/846) | 37.2% (547/1,471) | 50.4% (231/458) | 35.1% (292/832) |
| Female | 66.5% (2,332/3,505) | 35.8% (24/67) | 0 (0/1) | 66.1% (1,504/2,275) | 61.4% (35/57) | 70.1% (751/1,071) | 52.9% (18/34) |
| ERBB2 | |||||||
| Male | 3.0% (54/1,817) | 7.7% (1/13) | 0 (0/11) | 2.6% (11/422) | 2.7% (20/731) | 3.7% (9/224) | 3.1% (13/416) |
| Female | 3.1% (54/1,734) | 0 (0/30) | 0 (0/0) | 2.6% (29/1,112) | 2.8% (1/36) | 4.5% (24/537) | 0 (0/19) |
| KRAS | |||||||
| Male | 14.4% (375/2,609) | 4.8% (1/21) | 0 (0/16) | 9.4% (60/640) | 18.5% (186/1,008) | 10.1% (34/338) | 16.0% (94/586) |
| Female | 4.0% (99/2,492) | 0 (0/47) | 0 (0/0) | 3.9% (63/1,600) | 6.0% (3/50) | 3.8% (29/772) | 17.4% (4/23) |
| MET | |||||||
| Male | 3.3% (59/1,797) | 0 (0/13) | 8.3% (1/12) | 3.7% (15/410) | 2.9% (21/731) | 3.9% (8/203) | 3.3% (14/428) |
| Female | 2.0% (33/1,684) | 3.1% (1/32) | 0 (0/0) | 1.8% (19/1,062) | 0 (0/37) | 2.3% (12/530) | 4.3% (1/23) |
| RET | |||||||
| Male | 1.5% (33/2,180) | 0 (0/16) | 7.7% (1/13) | 3.4% (18/536) | 1.1% (9/805) | 0.6% (2/311) | 0.6% (3/499) |
| Female | 1.2% (25/2,019) | 0 (0/31) | 0 (0/1) | 1.6% (20/1,267) | 2.6% (1/39) | 0.6% (4/659) | 0 (0/22) |
| ROS1 | |||||||
| Male | 2.0% (74/3,659) | 11.1% (4/36) | 0 (0/20) | 2.6% (24/928) | 2.5% (33/1,322) | 1.5% (8/551) | 0.6% (5/802) |
| Female | 3.0% (103/3,385) | 7.7% (5/65) | 100% (1/1) | 3.0% (64/2,103) | 8.0% (4/50) | 2.6% (29/1,132) | 0 (0/34) |
- Note: *Patients with unknown history of smoking (n = 4781) were excluded from this analysis.
4 DISCUSSION
This retrospective study aimed to comprehensively understand the current status and quality control of ALK test for lung cancer in China (the RATICAL study). The analyses included 23,689 patients with advanced NSCLC who underwent ALK test in 30 medical centers across China. The overall ALK gene rearrangement rate was 6.7% in advanced NSCLC, with higher ALK gene rearrangement rates in patients aged ≤ 35 years, females, never smokers, and patients with adenocarcinoma. The IHC-VENTANA-D5F3 was used to test the ALK gene rearrangements most frequently. The largest number of samples tested were histological samples.
The overall ALK gene rearrangement rate was 6.7% in advanced NSCLC in the present study. This outcome was consistent with that of the previous studies from China (6.6% [9] and 4%-11.6% [19]). Notably, these rates are higher compared to those observed in Western populations, underscoring potential ethnic and regional variations in genetic susceptibility to ALK rearrangements. For example, a study reported that Asian patients exhibited a significantly higher prevalence of ALK gene rearrangement disease at 22.0%, compared to New Zealand Europeans at 4.4% [20]. This suggests that genetic factors may contribute to the higher incidence of ALK rearrangements in Asian populations. The ALK gene rearrangement rates were 31.9%, 8.4%, and 3.0% for patients aged ≤ 35, 36-65, and > 65 years in advanced NSCLC in the present study, respectively. The ALK gene rearrangement rate was 8.3% for advanced NSCLC patients who never smoked, which was higher than for patients who smoked or were smoking (4.2%). The positive detection rate among women was 9.0%, higher than 5.2% among men in the present study. These results were consistent with those from a previous study [10]. The ALK gene rearrangement rate was 8.2% for patients with advanced lung adenocarcinoma, numerically higher than that in patients with advanced NSCLC, indicating a lower positive rate in patients with non-adenocarcinoma. Despite this outcome, several squamous cell carcinoma patients with ALK gene rearrangement have benefitted from ALK inhibitors [16]. Therefore, Chinese clinical experts recommend ALK test for patients with advanced NSCLC (non-adenocarcinoma) to make better decisions on treatment options [21].
The diagnostic guidelines strongly recommend the IHC-VENTANA-D5F3, FISH, RT-PCR, and NGS methods for detecting the ALK fusion gene [17]. Each method has distinct advantages: FISH is highly specific but may have false-negative issues; IHC-VENTANA-D5F3 offers quick screening with high sensitivity; RT-PCR excels in sensitivity for known variants; and NGS not only detects known ALK rearrangements but also identifies concurrent genomic alterations that may impact therapy outcomes [17]. For instance, NGS has shown potential in predicting better disease control and longer progression-free survival in crizotinib-treated patients [22]. This highlights the importance of selecting appropriate diagnostic techniques based on clinical needs and available resources. The IHC methods are commonly used in clinical practice to determine ALK fusions because they are simple, fast, and cost-effective. Four ALK antibodies, such as IHC-VENTANA-D5F3, ALK1, 5A4, and 1A4, are presently available. Among them, ALK1 is less sensitive, 1A4 is less specific, and 5A4 has an unclear cut-off value [23]. Therefore, the IHC-VENTANA-D5F3 is the preferred choice for IHC due to its high sensitivity and specificity [24], and it was used in more than half of the samples analyzed in the present study. However, this test can give false-positive results when non-adenocarcinoma samples are used, warranting experienced pathologists for interpretation [24]. In the previously published first quality control analysis of the RATICAL study, 31 pathologists from 31 nationwide medical centers were selected to read 109 IHC sections. Only 19.4% of the pathologists were 100% accurate in the diagnosis. The false negative rate was 26.5%, positive cases were read as negative by at least one pathologist, and 41.7% of the 60 negative cases were read as positive (false positive) by at least one pathologist. The inconclusive interpretation was 31.2% [18]. These results highly emphasized the heterogeneity in viewing and interpretation among the pathologists on IHC slides, contributing to the variations in ALK gene rearrangement results observed in the present study. Consequently, guidelines have been issued in China to standardize the ALK test and recommend other retesting methods when the IHC-VENTANA-D5F3 results are inconclusive [17, 21].
In this study, 651 patients whose sample underwent testing using the IHC-VENTANA-D5F3 protocol had a concordance rate of 99.2% in the inter-hospital agreement analysis, and 119 samples who underwent testing using other protocols (PCR, NGS, FISH, and IHC) were reviewed by the IHC-VENTANA-D5F3 and resulted in a concordance rate of 93.2%. In addition, the agreement of detection of different methods specifically for those validated cases confirmed by FISH were 98.0% for IHC-VENTANA-D5F3, 98.7% for NGS, and 91.3% for RT-PCR, showing a high concordance rate with the initial tests, but some inconsistencies remained. Indeed, the concordance rate between IHC and RT-PCR was relatively low at 91.3%. This may be attributed to the specificity of RT-PCR, which can lead to false negatives if the specific ALK fusion sequences are not present. Additionally, the potential for IHC to produce false positive results [17], contributes to these discordances. Furthermore, due to the small sample size of discordances, the detailed analysis to track possible reasons is infeasible. In a previous study of ALK test in over 3,000 NSCLC patients, FISH and IHC-VENTANA-D5F3 were used. As a result, 14 patients were read as negative by FISH and positive by IHC but further validated as ALK gene rearrangement by NGS [12]. Therefore, when inconsistencies arise with different test methods, clinicians and pathologists should discuss them to reach a consensus [17].
Regarding selecting specimens for downstream test, tumor tissue samples are highly recommended. When tumor tissue specimens do not meet the requirements or are unavailable, cytology samples can be used. In a few circumstances of advanced lung cancer, when tissue or cytology samples are objectively unavailable, blood or cerebrospinal fluid was used for the diagnosis [21]. In this study, most (more than 80%) of ALK tests were performed on histological samples in clinical practice, followed by cytological samples. Only a few evaluations were performed using body fluid samples. However, a previous study found that the ALK test on cytological specimens was reliable and sensitive [25]. In the present study, about 20% of patients were tested with samples obtained from metastatic foci. While our study observed a higher ALK gene rearrangement rate in metastatic tumors compared to primary lesions, it is important to recognize that the underlying ALK gene rearrangement status remains consistent between primary and metastatic sites. This consistency supports the reliability of using either primary or metastatic tissue samples for detecting ALK rearrangements. The literature corroborates this, showing high concordance of ALK gene rearrangements in both tumor sites, which validates the use of metastatic samples in clinical diagnostic settings when primary tumor tissue is not available [26, 27].
We also analyzed for other driver genes or biomarkers concurrently with ALK gene rearrangement detection and found a small number of cases of mutations, except for high co-positivity with PD-L1. The co-occurrence of ALK gene rearrangement and mutations in other driver genes was rare, as supported by the literature [28], except for the TP53 mutation [29], which was not assessed in the present study. RT-PCR or NGS test is recommended when ALK gene rearrangement and other driver gene mutations are considered [17]. Since co-occurrence of mutations is rare, the treatment options are uncertain [30-32]. Notably, EGFR mutation was more common in women, and KRAS mutation was more common in males, as supported by the literature [33, 34].
ALK test is available in China by IHC and FISH (these methods only target the ALK gene or protein, and therefore, not all patients have EGFR test), but also by RT-PCR and NGS assays that include ALK and multiple other genes (as listed in Tables 4–5). Therefore, the results for these genes are also available when NGS and RT-PCR assays are performed. The results were generally consistent with the mutation rates of these driver genes reported in the literature without new signals [35-37].
In managing advanced NSCLC, comprehensive molecular profiling that includes not only ALK but also ROS1, RET, NTRK, and other actionable gene fusions is critical. However, the practical limitations of conventional methods like ALK IHC necessitate the use of more advanced techniques. At our institution, we have established a versatile diagnostic model that integrates multiple test platforms to accommodate the variable quality of clinical biopsy specimens. When tissue NGS is not feasible due to sample quality issues, we employ ARMS-PCR+IHC/FISH or switch to blood ctDNA NGS test, which allows us to continue detecting driver gene variants despite the challenges [38]. This strategic approach ensures that comprehensive genomic profiling can be achieved, supporting the use of targeted therapies based on a broad spectrum of detected mutations, including common and rare gene loci. Our experiences emphasize the importance of a flexible, multi-platform test strategy to overcome the limitations of tissue sample quality and ensure that molecular diagnostics can inform effective treatment planning for all patients.
This study had some limitations. First, ALK test was based on the clinical reality of each participating hospital. Each hospital might have different test protocols to match their local equipment, hardware, software, and the pathologists' preferences. Nevertheless, the quality control principles are national and were uniformly applied by all centers. Second, it was a cross-sectional study, and the treatment of patients with ALK gene rearrangement (e.g., treatment with ALK inhibitors) was not followed. Third, missing data might have occurred due to the large sample size. Fourth, our study included data from 30 centers across the country, but the standards of ALK test and the testers' experience might have varied from center to center. Fifth, only nine of the 30 centers were performing NGS. Since biopsies provided limited specimens used primarily for histology, immunohistochemistry, and basic genetic test, insufficient material was often left for NGS. Nevertheless, multiple quality controls were performed at all centers to minimize the impact of this bias on the outcomes. Besides, while this study provides comprehensive insights into the test patterns and positivity rates of ALK rearrangements, it does not include clinical outcome data such as overall survival.
In conclusion, we investigated ALK gene rearrangement rates and test patterns in China through this real-world study. The overall ALK gene rearrangement rate among advanced NSCLC in China was 6.7%, while advanced NSCLC patients with ALK gene rearrangement had specific clinicopathological characteristics. The IHC-VENTANA-D5F3 was used most frequently for detecting the ALK gene rearrangement with high consistency. This study provides a basis for detecting ALK gene rearrangements in patients with NSCLC, aiming to optimize future ALK gene rearrangement test patterns in patients with NSCLC.
AUTHOR CONTRIBUTIONS
Lin Li: Data curation; formal analysis; writing—original draft. Lei Guo: Data curation; project administration. Wencai Li, Chunyan Wu, Yanfeng Xi, Yuan Ji, Lili Jiang, Ji Li, Jingping Yun, Gang Chen, Yuan Li, Yueping Liu, Dianbin Mu, Yuchen Han, Leina Sun, Qingxin Xia, Xiaodong Teng, Nanying Che, Wei Wu, Xueshan Qiu, Chao Liu, Xiaochu Yan, Daiqiang Li, Zhihong Zhang, Zhe Wang, Yujun Li, Zheng Wang, Lingchuan Guo, Xiu Nie, Jingshu Geng and Jianhua Zhou: Data curation; project administration; supervision. Jianming Ying: Conceptualization; methodology; funding acquisition; supervision; formal analysis; writing—review & editing.
ACKNOWLEDGEMENTS
We would like to thank Genowis (Beijing) Gene Technology Co., Ltd. for undertaking data analysis tasks in the project. This work was supported by CAMS Innovation Fund for Medical Sciences (CIFMS) (grant no. 2021-1-I2M-012 and 2022-I2M-C&T-B-078), Beijing Hope Run Special Fund of Cancer Foundation of the People's Republic of China (grant no. LC2019L04), National Key Research and Development Program (grant no. 2022YFC2409902) and Capital's Funds for Health Improvement and Research (grant no. 2020-2Z-4028). They had no role in study design, in the collection, analysis, and interpretation of data; in the report's writing; or in the decision to submit the article for publication.
CONFLICT OF INTEREST STATEMENT
All authors declare no competing interests.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This retrospective study was approved by the Ethics Committee of the Cancer Hospital of the Chinese Academy of Medical Sciences and reported to the Ethics Committee of all study centers. Informed consent was obtained from all patients.
PATIENT CONSENT STATEMENT
Informed consent was obtained from the patients.
Open Research
DATA AVAILABILITY STATEMENT
The data supporting the findings of this study are available within the article and its Supplementary Materials.




