Exposure of benzo[a]pyrene induces HCC exosome-circular RNA to activate lung fibroblasts and trigger organotropic metastasis
Wei Mu and Pengfei Gu contributed equally to this work.
Abstract
Background
Benzo[a]pyrene (B[a]P), a carcinogen pollutant produced by combustion processes, is present in the western diet with grilled meats. Chronic exposure of B[a]P in hepatocellular carcinoma (HCC) cells promotes metastasis rather than primary proliferation, implying an unknown mechanism of B[a]P-induced malignancy. Given that exosomes carry bioactive molecules to distant sites, we investigated whether and how exosomes mediate cancer-stroma communications for a toxicologically associated microenvironment.
Method
Exosomes were isolated from B[a]P stimulated BEL7404 HCC cells (7404-100Bap Exo) at an environmental relevant dose (100 nmol/L). Lung pre-education animal model was prepared via injection of exosomes and cytokines. The inflammatory genes of educated lungs were evaluated using quantitative reverse transcription PCR array. HCC LM3 cells transfected with firefly luciferase were next injected to monitor tumor burdens and organotropic metastasis. Profile of B[a]P-exposed exosomes were determined by ceRNA microarray. Interactions between circular RNA (circRNA) and microRNAs (miRNAs) were detected using RNA pull-down in target lung fibroblasts. Fluorescence in situ hybridization and RNA immunoprecipitation assay was used to evaluate the “on-off” interaction of circRNA-miRNA pairs. We further developed an adeno-associated virus inhalation model to examine mRNA expression specific in lung, thereby exploring the mRNA targets of B[a]P induced circRNA-miRNA cascade.
Results
Lung fibroblasts exert activation phenotypes, including focal adhesion and motility were altered by 7404-100Bap Exo. In the exosome-educated in vivo model, fibrosis factors and pro-inflammatory molecules of are up-regulated when injected with exosomes. Compared to non-exposed 7404 cells, circ_0011496 was up-regulated following B[a]P treatment and was mainly packaged into 7404-100Bap Exo. Exosomal circ_0011496 were delivered and competitively bound to miR-486-5p in recipient fibroblasts. The down-regulation of miR-486-5p converted fibroblast to cancer-associated fibroblast via regulating the downstream of Twinfilin-1 (TWF1) and matrix metalloproteinase-9 (MMP9) cascade. Additionally, increased TWF1, specifically in exosomal circ_0011496 educated lungs, could promote cancer-stroma crosstalk via activating vascular endothelial growth factor (VEGF). These modulated fibroblasts promoted endothelial cells angiogenesis and recruited primary HCC cells invasion, as a consequence of a pre-metastatic niche formation.
Conclusion
We demonstrated that B[a]P-induced tumor exosomes can deliver circ_0011496 to activate miR-486-5p/TWF1/MMP9 cascade in the lung fibroblasts, generating a feedback loop that promoted HCC metastasis.
List of abbreviations
-
- B[a]P
-
- Benzo[a]pyrene
-
- HCC
-
- hepatocellular carcinoma
-
- CAF
-
- cancer-associated fibroblast
-
- AAV
-
- adeno-associated virus
-
- HLF
-
- human derived lung fibroblast
-
- Exo
-
- exosome
-
- i.v.
-
- intravenous injection
-
- qRT-PCR
-
- quantitative reverse transcription-PCR
-
- 3’ UTR
-
- 3' untranslated regions
-
- CCK8
-
- cell counting kit-8
-
- FAP
-
- fibroblast activation protein
-
- TGF-β
-
- transforming growth factor-β
-
- α-SMA
-
- α-smooth muscle actin
-
- H&E
-
- hematoxylin and eosin
-
- IHC
-
- immunohistochemistry
-
- TNF
-
- tumor necrosis factor
-
- VEGF
-
- vascular endothelial growth factor
-
- BMP
-
- bone morphogenetic protein
-
- CCL
-
- chemokine C-C motif ligand
-
- IL
-
- interleukin
-
- circRNA
-
- circular RNA
-
- miRNA
-
- microRNAs
-
- siRNA
-
- small interfering RNA
-
- FISH
-
- fluorescence in situ hybridization
-
- IF
-
- Immunofluorescence
-
- RIP
-
- RNA immunoprecipitation
-
- AGO2
-
- argonaute 2
-
- TWF1
-
- twinfilin actin binding protein 1
-
- STAT3
-
- signal transducer and activator of transcription 3
-
- MMP9
-
- matrix metalloproteinase 9
-
- HUVEC
-
- human umbilical vein endothelial cell
-
- ceRNA
-
- competitive endogenous RNA
1 BACKGROUND
In addition to genetic mutations and hepatitis viral infections, unhealthy behaviors linked to lifestyle “westernization”, particularly fried food preference diets, have been postulated to promote the malignant of liver cancers [1-3]. Benzo[a]pyrene (B[a]P) is a major pro-carcinogen formed in the process of deep-fried and smoked meats [4], making humans are inevitably exposed by foods [5]. Epidemiological studies reported the populations who prefer “Western diet” is associate with an elevated risk of hepatocellular carcinoma (HCC) [3, 6, 7]. The ingested B[a]P can be transported and deposited in liver, as a consequence to the liver burden and inflammation [8]. However, the dietary intake of B[a]P in health populations has been overlooked because of its imperceptible toxicity. Public health issues stemming from B[a]P exposure focus on patients with HCC, who are more susceptible to B[a]P exposure [9]. Thus, biomarkers and therapeutic targets of foodborne B[a]P in populations with liver disease are more emphatically investigated. Clinically, lung metastasis is the most common outcome of HCC progression and one of the leading causes of cancer-related deaths [10, 11]. Our previous study also reported that sustained B[a]P exposure could not alter tumor cell growth but significantly enhanced lung-specific metastasis in cell and animal models [12-14]. Based on these results, we speculated an unknown mechanism exists for facilitating organotropic invasion and administrating foodborne B[a]P exposure.
Metastasis is a temporal and spatial development that begins with forming favorable distant tissues before tumor cells arrive [15]. The supportive pre-metastatic niche is characterized by cancer-associated properties, such as inflammation, angiogenesis preparation, and extracellular matrix remodeling [16]. One of the most abundant and notable populations of local stroma cells is that of fibroblasts [17, 18]. Activated fibroblasts are regarded as plastic and resilient cells interacting with tumors and other cell types [19]. In this framework, tumor derived exosomes emerge as master regulators to alter cellular signal transduction via autocrine and paracrine functions [20]. Exosomes are membrane vesicles of endocytic origin ranging in size from 30 to 150 nm, that deliver bioactive molecules from donor to recipient cells [21, 22]. Within exosomes, non-coding RNAs are protected from degradation via nucleases and pH fluctuations in the extracellular space, thereby shutting not only to nearby cells but also to distant organs [23]. Circular RNA (circRNA), contains a particular circular form as a covalently closed-loop without 5’ caps or 3’ tails, representing an abundant and stable class with regulatory functions in exosomes [24]. circRNA selectively conserves microRNA (miRNA) target sites and exerts regulatory effects by regulating miRNA gene expression as sponges, which mechanisms are known as the competitive endogenous RNA (ceRNA) theory [25, 26]. Given that the circRNAs of tumor-derived exosome could be transported to adjacent or distant cells, a recent study demonstrated the exosome-mediate transfer of circRNAs engages various circRNA-miRNA-mRNA axes between tumor and target cells [27].
Environmental and foodborne factors can affect circRNA production [28, 29], and dysregulated circRNAs have been identified as novel carcinogen biomarkers [30]. Our previous study also developed ceRNA networks for B[a]P-induced carcinogenicity through integrated bioinformatics analysis in tumor cell models [31]. Indeed, exosomes most likely participate in the adaptive response of an organism to external stimuli. However, the biological mechanisms underlying environment-driven alterations and exosome-mediated communications are poorly understood. To characterize the adverse outcome pathways of real-life scenarios of pollutants in cancer-stroma communications in this study, a novel RNA pull-down assay in target cells rather than donor cells was developed in vitro, and the adeno-associated virus (AAV)-inhalation model for lung-specific activation was developed in vivo. Our purpose was to determine whether the contents of exosomes were altered by B[a]P exposure and how exosomal circRNAs were transferred to recipient cells.
2 MATERIALS AND METHODS
2.1 Cell lines and cell culture
Human-derived HCC cell lines, HepG2, HCC-LM3 (LM3) and BEL-7404 (7404), and human-derived lung fibroblast (HLF) cells were obtained from Cell Bank of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China), and cultured in RPMI1640 or DMEM medium (Cytiva, Marlborough, MS, USA) supplemented with 10% fetal bovine serum (FBS; Sage Creation Science Co. Ltd, Beijing, China), 100 μg/mL penicillin (Gibco, Waltham, MA, USA), and 100 μg/mL streptomycin (Gibco), and maintained in an incubator with 5% CO2 at 37°C. For B[a]P exposure procedure, 7404 cells were co-cultured with 100 nmol/L B[a]P (Macklin, Shanghai, China) or 0.1% dimethylsulfoxide (DMSO; Beyotime, Haimen, Jiangsu, China) for up to 4 weeks.
2.2 Exosome collection, isolation, and purification
For exosome collection, when cultured cells were covered 80% confluent, the culture medium was changed to RPMI1640 without FBS for 48 h. The cell culture mediums were collected and filtrated filtered through a 0.22 μm filters (Millipore, Burlington, MA, USA). Exosomes were isolated at 4°C by density gradient ultracentrifugation as follows: 500 × g and 2000 × g for 20 min, respectively to remove residual cell debris, and 10,000 × g for 60 min to remove micro-vesicles, finally, 100,000 × g for 2 h to collect exosomes in pellets. Meanwhile, the supernatant without pellet was also collected and named with exosome-non supernatant. For exosomes purification, the exosomes were overlaid by a continuous sucrose gradient (0.25 mol/L-2 mol/L) and ultracentrifuge for 100,000 × g for 15 h, at 4°C. The exosomes solutions with sucrose were divided as 12 fractions (0.2 mL/fractions), and the 2-8 fractions were purified exosomes. Ultracentrifugation was performed with using a high-speed centrifuge and the ultracentrifuge (Beckman Coulter, Jersey City, NJ, USA).
2.3 Transmission electron microscopy (TEM) and nanoparticle tracking analysis (NTA)
For exosome observation, exosome was suspended in 4% paraformaldehyde (PFA), and images were captured using the HT7700 TEM system (Hitachi, Tokyo, Japan) at a voltage of 80 kV. For NTA evaluation, the sizes and numbers of exosomes were tracked via the NanoSight NS300 system (NanoSight, Malvern, UK). Exosomes were resuspended in phosphate buffer saline (PBS) at a concentration of 5 μg/mL and further diluted 100- to 500-fold to achieve between 20 and 100 objects per frame. Subsequently, samples were injected into sample chambers at room temperature, each of which was configured with a 488 nm laser. All the data were analyzed by the NTA analytical software (NanoSight).
2.4 Cell counting kit-8 (CCK-8) proliferation and tube formation assays
For CCK-8 assays, 3 × 103 cells/well from different groups were seeded in 96-well plates. CCK-8 (Meilunbio, Dalian, China) reagent was added to each well at a fixed time each day for 3 days, and the cells were incubated at 37°C for 2 h. Then, the absorbance was measured in the single-wavelength mode (450nm) by microplate reader (Biotek, Synergy H1, Vermont, USA).
For tube formation assays, 250 μL matrigel (Invitrogen, Carlsbad, CA, USA) was pre-coated to 24-well plates for 30 min at 37°C incubator before seeding cells. 2 × 104 cells/well were cultured with supernatant from indicated cultured tumor cells and exosomes. After 24 h, tube formation on each well was examined under bright field using an inverted microscopy (Olympus IX83, Tokyo, Japan).
2.5 Cell migration assays
The wound healing and transwell assays were used to detect the migration ability of tumor and fibroblast cells. For wound healing assay, 5 × 105 cells were cultured in 6-well plates and scratched with a sterile pipette tip when each well was filled with cells. Cells were cultured in serum-free DMEM medium and the wound size was measured at 0 and 24 h. For transwell assay, 3 × 104 cell suspending in 200 μL serum-free DMEM medium were cultured in upper chambers (8 μm), and 500 μL 10% FBS-medium was filled in the lower chamber. After 24 h incubation, 4% PFA (Sangon Biotech, Shanghai, China) and 0.1% crystal violet (beyotime) were used to fix and stain the cells covered in the bottom chamber. After removing the cells in the upper chamber, the residual cells adhered to the bottom chambers were photographed.
2.6 Sanger sequencing
Total circRNA was extracted from 7404-100Bap Exo and reversely transcribed into cDNA. PCR amplification products were purified by Qiaquick Gel Extraction Kit (Qiagen, Hilden, Germany) and subjected to Quick Shot Sanger Sequencing by using BioSune (Shanghai, China) to confirm the head-to-tail splicing.
2.7 RNA isolation, real-time qPCR, RNase R treatment, and Actinomycin D assay
Total RNAs were extracted from tissues and cells using TRIzol (Invitrogen) and Isopropanol (Aladdin, Shanghai, China), and reversely transcribed into cDNA using the PrimeScriptTM RT reagent kit (Takara, Dalian, Liaoning, China). Exosomal RNA from the culture media was extracted with an exoRNeasy Midi Kit (Qiagen, Valencia, CA, USA) following the manufacturer's instructions. Small RNA from HLF cells was extracted using RNAiso for small RNA kit (Takara). Real-time qPCR was performed using TB Green Premix ExTaq Ⅱ (Takara). The primers are listed in Supplementary Table S1.
To achieve lentivirus-mediated circRNA overexpression, the lentivirus vector pLC5-ciR control and pLC5-ciR harboring circRNA was purchased from Ribo. siRNAs were transfected into HLF cells using Lipofectamine 3000 reagent (Invitrogen) according to the manufacturer's protocol. The sequences were listed in the Supplementary Table S1.
For RNase R treatment, 2 mg total RNA was incubated with tumor cells for 15 min at 37°C with 3 units/mg RNase R (Epicentre Technologies, Madison, WI, USA). Reverse transcription (RT) was then performed using random hexamers or oligo (dT) and SuperScript III (Invitrogen) and quantitative PCR (qPCR) was performed using TaqMan master or SYBR Green master mix (Applied Biosystems, Carlsbad, CA, USA).
For Actinomycin D assay, 2 mg/L actinomycin D (Sigma, Shanghai, China) was added into cultured cells last to 4, 8, 12 and 24 h. The cells were harvested according to the time of treatment. Then qRT-PCR was performed to analyze the stability of mRNA and circRNA. Three independent experiments were applied in triplicate. The primer sequences are listed in Supplementary Table S1.
2.8 Agilent methods for circRNA array
Total RNA was isolated using TRIzol (Invitrogen) according to the manufacturer's instructions, and purified by using RNeasy Mini Kit (Qiagen). The SBC human ceRNA microarray v1.0 (Shanghai Biotechnology Corporation, China) was used for analysis of differentially expressed circRNAs in exosome samples collected from cultured 7404 cells and 7404-100Bap cells (7404Exo and 7404-100Bap Exo). The labeled RNA targets were then hybridized with the slides. After hybridization, slides were scanned on the Agilent Microarray Scanner (Agilent technologies, Santa Clara, CA, US). Data were extracted with Feature Extraction software 10.7 (Agilent technologies). Raw data were normalized by Quantile algorithm, limma package the R program. The microarray experiments were performed by after the protocol of Agilent technologies Inc. at Shanghai Biotechnology Corporation. Abnormally expressed genes were added to the Database for Annotation, Visualization and Integrated Discovery v6.7.
2.9 Exosomes labeling with lipid and protein-binding dyes for cell uptake
Uptake of dye-labeled exosomes into the individual recipient cells. 1 mg of exosomes was labeled with 200 μL of SP-DioC18 in PBS (1:10,000; Invitrogen) for 25 min. Labeled exosomes were washed twice and incubated with exosome-sized vesicles removed FBS (Exo non-FBS, Thermo Fisher, CA, USA) in the dark for 30 min so that the free dye could be removed. The labeled exosomes were purified using 40% sucrose solution as described previously. Meanwhile, recipient HLF cells were fixed with 4% PFA (Sangon Biotech, Shanghai, China) for 15 min and permeabilized with 0.5% TritonX-100 (Macklin) for 20 min. The lipophilic Dil dyes (1:10,000; Invitrogen) was used to label HLF cells likewise. The SP-DioC18 labeled exosomes were co-incubated with Dil labeled cells for 12h at 4°C, and images were collected via a confocal microscope ((Leica, Wetzlar, Germany).
For staining cell membrane proteins, these fixed cells were staining with phalloidin (Abcam, Cambridge, MA, USA), overnight at 4°C and subsequently incubated with fluorescently conjugated Alexa Fluor 488 and Alexa Fluor 555 secondary antibodies (ThermoFisher).
2.10 Establishment of education animal model and in vivo tumorigenesis assay
The animal experiments required the demands of laboratory animal welfare and ethics, and approved by the Institutional Animal Care and Use Committee of Shanghai Jiao Tong University (Shanghai, China). The nude male BALB/c mice (4-6 weeks old) were purchased from Jiesijie (Shanghai, China). They were bred and housed in specific pathogen-free (SPF) animal facilities.
Lung pre-metastatic model was generated via exosome “education”, each group was received intravenously injected (i.v.) of 200 μg exosomes (dilute with 100 μL PBS, exosomes derived from 7404 and 7404Bap-100 cells) or PBS control once every 3 days for 15 days. BALB/c nude mice were randomly divided into the PBS group (n = 8), 7404Exo group (i.v. with 7404Exo/mice; n = 8), and 7404-100Bap group (i.v. with 7404-100Bap Exo/mice; n = 8). The exosome-modulated lungs from PBS, 7404Exo, and 7404-100Bap (n = 3/group) were evaluated using RT2 Profiler™ PCR Array (mouse cytokines and chemokines; Catalog No. 330401, Qiagen; Supplementary Table S2). After generation of lung-educated model, luciferase-labeled LM3 cells (5 ×105) were i.v. injected in these educated mice, and then received an intraperitoneal injection of 200 μL of D-luciferin (100 mg/kg, Genomeditech, Shanghai, China) on a weekly basis. Luciferase activity was measured within 10 min after injection using the Xenogen IVIS bioluminescence imaging system (Xenogen, Alameda, CA, USA) to real-time monitor tumor motility. At the end of the experiment, mice were sacrificed with CO2. Their tumor-bearing lungs were collected and fixed in Bouin fixative at 4°C for 7 h (Ricca Chemicals, Arlington, TX, USA), colonies numbers of lungs were counted. Then the lungs were cut in 5 μm sections for subsequent staining with Hematoxylin and Eosin (H&E, Beyotime, Jiangsu, China) and Masson's Trichrome (Beyotime) for histopathologic evaluation.
The model of twinfilin-1 (TWF1)-overexpressed specific on lung was generated by adeno-associated virus (AAV). Linearized expression vector (AOV002: Paav-CAG-MCS-EGFP-3FLAG) was purchased from OBiO (OBiO Tech, Inc. Shanghai, China). The TWF1 was recombined into the linearized vector by Geneart seamless cloning and assembly kit (Thermo Fisher Scientific). The TWF1-transfected vectors were introduced into the lungs of mice via inhalation every 5-6 days for 5 times. In the middle of the 14 days of TWF1-AAV inhalation, LM3 cells were i.v. injected. At the end of 40 days, the mice were sacrificed and these tumor-bearing lungs were analyzed.
2.11 Western blotting
Western blotting experiments were conducted as we previously described [15]. Briefly, proteins were extracted with sodium dodecyl sulfate (SDS) lysis buffer (Beyotime, Shanghai, China) containing proteinase and phosphatase inhibitors (Roche Applied Science, Indianapolis, IN, USA). Then, the proteins were separated in 10% polyacrylamide gels and transferred to polyvinylidene difluoride (PVDF) membranes (Merck Millipore, Darmstadt, Germany). After blocking with 5% bovine serum albumin and incubation with antibodies, the blotting bands were analyzed by an Odyssey Infrared Imaging System (Biosciences, Lincoln, NE, USA). GAPDH was utilized as the control protein, and the antibodies used are shown in Supplementary Table S3.
2.12 Biotin-labeled RNA pull-down assay and northern blotting
Biotin-labeled RNA pull-down assay was performed using Pierce™ Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific, CA, USA) following the manufacturer's manual. Upstream and downstream RNA fragments of circ_0011496 were synthesized by Ribo biotech (Guangzhou, Guangdong, China). Cells were crosslinked by formaldehyde, equilibrated in glycine buffer, and scraped with lysis buffer. Firstly, HLF cells and 7404-100Bap Exo-educated HLF cells were sonicated and then centrifuged. The supernatants were transferred to a 2 mL tubes and separately saved 50 μL as input analysis. Cell lysates were incubated by the biotin-tagged specific circ RNA-probe for 3 h. Approximately 1 × 107 cells were harvested and these lysates were incubated with labeled RNA and streptavidin magnetic beads at 4°C for 1 h, and the magnetic beads were washed and the pull-down complexes were boiled with the loading buffer. Northern blot with 50 μg RNA isolated from HLF cells transfected with circ_0011496 probe or random probe. These blots were hybridized with probes specifically recognizing miR-486-5p. The probe sequences are listed in Supplementary Table S1.
2.13 RNA binding protein immunoprecipitation (RIP) assay
The RIP assay was performed in strict accordance with the instructions of the Millipore kit (Millipore). After incubation in a shaker overnight at 4°C, protein G magnetic beads were added to capture the complex. RNA was extracted after washing with the buffer. The extracted RNA was reverse transcribed, and the RNA levels were detected by qRT-PCR. The primer sequences are listed in Supplementary Table S1.
2.14 Fluorescence in situ hybridization (FISH), immunofluorescence (IF), and immunohistochemistry (IHC) in vitro and in vivo
FISH assay was used to find the intracellular location of circRNA. Probes of FISH assay for circ_0011496, human U6, and human18S (listed in Supplementary Table S1) were synthesized by Ribo. These FISH samples were analyzed by IF, briefly, cells were rinsed with PBS and fixed in 4% formaldehyde solution for 10 min, and incubated with 0.5% Triton X-100 solution (Beyotime, Haimen, Jiangsu, China) for 5 min at 4°C. After pre-hybridization for 30 min, the cells were hybridized with fluorescence-hybridization probes overnight in the dark at 37°C, and laser scanning confocal microscopy (Leica) was used to visualize the images at 400 × magnification. Furthermore, FITC-labeled circ_0011496 and Cy3-labeled miRNA probes were designed and synthesized (listed in Supplementary Table S1). The probe signals were detected with a fluorescent in situ hybridization kit (Ribo) according to the manufacturer's instructions.
For IHC, Specimens the were deparaffinized using dimethylbenzene and alcohol (Sigma) with corresponding concentration gradient, rehydrated and subsequently washed by using PBS. After retrieving antigens by using citrate buffer (Sigma) for 30 min and blocking endogenous enzymes, the slides were incubated with corresponding antibodies. The slides were then stained and analyzed under microscope (Leica). The antibodies used are shown in Supplementary Table S3.
To evaluate the uptaken and co-localization of circRNA in lung fibroblasts in vivo, we performed in situ hybridization using the ViewRNA™ ISH Cell Assay Kit (Thermo fisher) for circRNA in situ hybridization with custom-made probes targeting the circRNA exon junctions. Slices of mice lungs were dissected and fixed overnight in 4% PFA, and then incubated with the anti-vimentin.
2.15 Prediction and construction of the circRNA-miRNA-mRNA network
Interactions between circRNA and miRNA were analyzed by online databases of circInteractome (https://circinteractome.nia.nih.gov/) and circBase (http://circbase.org/). Meanwhile, mRNAs that regulated by miRNAs were integrated predicted using the TargetScan (https://www.targetscan.org/), miRTarBase (https://mirtarbase.cuhk.edu.cn/), miRDB (http://www.mirdb.org/) and miRwalk (http://mirwalk.umm.uni-heidelberg.de/) databases. Only when mRNAs are overlapped in the 4 databases of miRNA tragets can be were considered as candidate target mRNAs. Moreover, intersections with the different expression mRNAs (DEmRNAs) were identified to screen out the hub genes targeted by the different expression miRNAs (DEmiRNAs). The circRNA-miRNA-mRNA regulatory network was constructed by using a combination of circRNA-miRNA pairs and miRNA-mRNA pairs and visualized by Cytoscape 3.6.1 (The Cytoscape Consoritum, New York, NY, USA).
2.16 Subcellular fractionation and localization
Using total RNA as input, Cytoplasmic & Nuclear RNA Purification Kit (Norgen, Belmont, CA, USA) was used to isolate and purify cytoplasmic and nuclear RNA. After which, the nuclei were pelleted by 5000×g centrifugation at 4°C, and supernatants were taken as cytoplasmic fraction. The cellular localization of circRNA was determined by qRT-PCR analysis of the cytoplasmic RNAs, nuclear RNAs, and cell culture-medium RNAs. The primer sequences are listed in Supplementary Table S1.
2.17 Dual-luciferase reporter assay
The promoter-luciferase of wildtype and mutant genes were constructed into pRL-TK Renilla luciferase vectors (GenePharma Co.), and these plasmids were transfected into HLF or 7404 cells. The luciferase activity was assessed with a Dual-Luciferase Reporter Assay kit (Beyotime) 48 h after transfection following on the manufacturer's protocol.
2.18 Statistical analysis
Statistical data were analyzed using GraphPad Prism 9.0 (GraphPad Software Inc., San Diego, CA, USA). Each experiment was repeated at least three times, and each group contains at least 3 triplicates. Data were shown as mean ± standard error of the mean in the bar charts. The differences among multiple groups were performed with the unpaired Student's t-test or one-way ANOVA with Dunnett's multiple comparisons test. P values of <0.05 were considered statistically significant.
3 RESULTS
3.1 Exosome secreted from B[a]P-exposed HCC cells regulate lung fibroblast activation in vitro
Chronic B[a]P exposure at an environmental-relevant dose (100 nmol/L) promotes 7404 HCC cells rapidly metastasis to lungs [9, 14]. To explore the messenger functions of B[a]P-exposed HCC exosomes between tumor cells and the lung niche of recipients, we isolated HCC cell-secreted exosomes (7404 Exo and 7404-100Bap Exo) and co-incubated with HLF cells (Figure 1A). The cup-shaped structures of vesicles were captured and identified using TEM (Figure 1B). The size distribution of 7404 Exo and 7404-100Bap Exo displayed mostly diameters were 80-140 nm via NTA, which is the typical size for exosomes (Figure 1C). The characteristics of these vesicles were verified by the detection of exosomal biomarkers (CD81, CD63, and TSG101; Figure 1D). To further visualize the delivery of exosomes into HLF in vitro, 7404 Exo and 7404-100Bap Exo were labeled with SP-DioC18 (green) and HLF cells were labeled with Dil (red), respectively. Confocal imaging revealed the presence of green spots in recipient fibroblasts, suggesting the uptake of tumor-derived exosomes (Figure 1E).
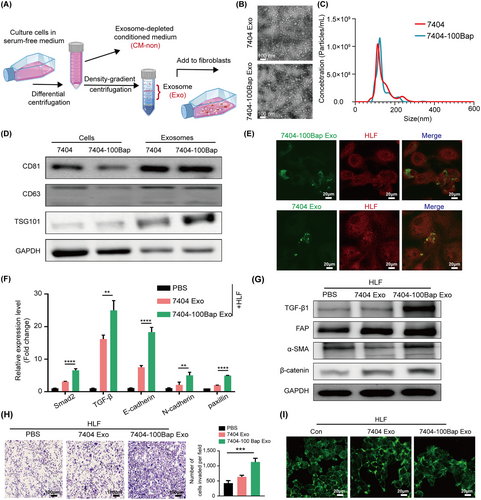
B[a]P induced HCC-derived exosomes activate lung fibroblasts.
(A) Workflow diagram of the procedure for exosomes extraction and pellet-free supernatant collection. The exosomes were further co-culture with fibroblasts for 48 h. (B) Exosomes released by B[a]P treated (7404-100Bap Exo) and control cells (7404 Exo) were detected by TEM. Scale bar, 100 nm. (C) The size range of exosomes from 7404-100Bap and 7404 were checked by nanosight particle tracking analysis. (D) Western blotting assay of indicated proteins in exosomes and donor cells. (E) Confocal imaging showed the delivery of SP-DioC18-labeled exosomes (green) to Dil-labeled fibroblasts (red), scale bar, 20μm. (F-G) qRT-PCR (F) and Western blotting (G) for the indicated proteins in the HLF cells pre-treated with 7404 Exo and 7404-100Bap Exo. (H) Migration transwell assays for HLF treated with equal quantities of exosomes (7404-100Bap Exo and 7404 Exo) or blank control (PBS). Scale bar, 100 μm. (I) Phalloidin staining of actins in HLF cells pre-treated with exosomes (7404 Exo and 7404-100 Bap Exo). Scale bar, 20 μm. **P < 0.01; ***P < 0.001, ****P < 0.0001. Data in F and H were analyzed using one-way ANOVA with Dunnett's multiple comparisons test. Abbreviations: B[a]P, Benzo(a)pyrene; Con, control; Exo, exosome; HCC, hepatocellular carcinoma; HLF, human derived lung fibroblast; qRT-PCR,quantitative reverse transcription-PCR; TEM, transmission electron microscope.
Activated fibroblasts or cancer-associated fibroblasts (CAFs) sense external cues and respond by switching to their activated states [32]. After co-cultured with different exosomes, we found higher expression levels of fibrosis (FAP, α-SMA, and β-catenin) and pro-inflammatory (Smad2, TGF-β, E-cadherin, N-cadherin and paxillin) markers in the 7404-100Bap Exo pre-treated fibroblasts, suggested the acquisition of specialized functions of motility and matrix deposition (Figure 1F-G). To evaluate the activation phenotypes of HLF, supernatants with depletion/free of exosomes (CM-non) were obtained and used as a control (Supplementary Figure S1A). Although HLF cell proliferation was neither altered by exosome or CM-non (Supplementary Figure S1B), increasing motility cells was promoted by exosomes rather than exosome-depleted supernatants (Figure 1H, Supplementary Figure S1C). Additionally, the morphology of focal adhesions in 7404-100Bap Exo-treated fibroblasts also showed more sprouts via the FITC-phalloidin staining (Figure 1I). All these results determined B[a]P induced exosomes promote changes in cytoskeletons of HLF cells, thereby creating a positive status that leads to the fibrotic cycle.
3.2 B[a]P contributes to lung pre-metastatic niche formation in vivo
Lung education model was established to evaluate the contribution of B[a]P-exposed exosomes on organotropic metastasis (Figure 2A). 7404 Exo and 7404-100Bap Exo were intravenously injected into mice (n = 8/group) every 3 days for 15days to prepare exosome-modulated lungs. These pre-exosome-educated lungs were randomly selected and dissected (n = 3/group), their expression of cytokines was examined using RT2 Profiler™ PCR Array (Figure 2B). Of the 81 analyzed genes, 38 (46.9%) genes showed more than 2-fold upregulation in both of exosome injection groups, whereas 15 genes (18.5%) were only up-regulated in the 7404-100Bap Exo group (33 important cytokines of pre-metastatic niche are shown in Figure 2B). The classification of cytokines, such as the BMP family members, CCL family members, growth factors, and pro-inflammatory molecules in metastatic tissues could confirm the contribution of B[a]P-induced exosomes to pre-metastatic niche formation (Figure 2C, Supplementary Table S2). Next, the remaining of lung-educated mice were i.v. injected with 5 × 105 LM3 cells that with stably expressing firefly luciferase (n = 5/group). The metastatic tumors were monitored in real-time using in vivo imaging system, as shown in the 7404-100Bap Exo injected group, which trigger the spread of HCC LM3 cells (Figure 2D). All mice were euthanized on day 35, and the resected lungs were stained for H&E and evaluated for the presence of tumor metastases (Figure 2E). Masson's trichrome staining also showed increased collagen deposition in exosome-modulated lungs (Figure 2E). In addition, higher expression of α-SMA demonstrated the differentiation of activated fibroblasts in the 7404-100Bap Exo educated-lungs (Figure 2E). Finally, metastatic foci were observed and photographed after lungs were immersed in Bouin's fixative. After LM3 injection, lungs pre-treated by 7404-100Bap Exo showed more tumor nidus (arrows) than that pre-treated with PBS control and 7404Exo (Figure 2F). Taken together, compared to 7404 Exo pre-injected lungs, lungs pre-treated by 7404-100Bap Exo could stimulate higher expression of pro-inflammatory molecules as well as enhanced tumor-bearing in the lungs.
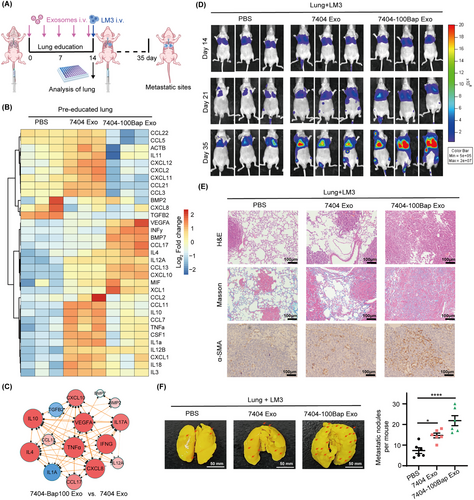
The contribution of 7404-100Bap Exo in lung metastasis in vivo.
(A) Workflow of lung education by tumor derived exosome in vivo. 7404 Exo and 7404-100Bap Exo were i.v. injected into mice (n = 8/group) every 3 days for 5 times, the educated lungs were evaluated by PCR array before tumor cells injection. Luciferase-labeled LM3 cells were next i.v. injected to these educated mice, and the growth and spread of tumor cells were monitored every 7 days till 35 days. (B) Heat map of PCR array for genes encoding cytokines and chemokines were analyzed in the exosome-educated lung tissues (n = 3/group) after 5 times injection of PBS, 7404 Exo and 7404-100Bap Exo. Expression values were normalized over the expression of GAPDH and presented as log2 of relative changes. As shown, genes with higher expression are depicted in red, genes with lower expression are depicted in blue. The cytokines and chemokines molecules were grouped according to the pathway in which they participate. (C) The Top 15 dysregulated gene expression network in the pre-educated lung tissues was analyzed and visualized by String and Cytoscape3.6.1 software. (D) Mice were i.v. injected with 5 × 105 LM3 cells that with stably expressing firefly luciferase (n = 5/group). Representative images and quantitative analysis of lung metastasis of mice using luciferase-based bioluminescence imaging. (E) The mice in D were sacrificed after 35 days of LM3 injection. H&E and Masson's trichrome staining were detected in the lung tissues harvested from the mice and the IHC results showed the expression levels of α-SMA. Scale bar, 100μm. (F) Lung metastasis nidus of tumor-bearing mice after injection of PBS, 7404 Exo and 7404-100Bap Exo were counted after fixed and stained with bouin's solution. *P < 0.05, ****P < 0.0001. Data in F were analyzed using one-way ANOVA with Dunnett's multiple comparisons test. Abbreviations: αSMA, alpha smooth muscle actin; GAPDH, Glyceraldehyde-3-Phosphate Dehydrogenase; IHC, immunohistochemistry; HCC, hepatocellular carcinoma; H&E, Hematoxylin and eosin; i.v. intravenous.
3.3 B[a]P-regulated exosome circRNAs in cancer-stroma communications
The profile of 7404-Exo and 7404-100Bap Exo were evaluated using ceRNA microarray. The differentially expressed circRNAs (DEcricRNAs) and miRNAs (DEmiRNAs) were identified (fold change >2 or <0.5 and P < 0.05, Figure 3A). The 30 most significantly up-regulated circRNAs in 7404-100Bap Exo were firstly selected, and 342 potential miRNA targets via circInteractome and circBase databases were predicted. We next wonder whether these predicted miRNAs were also matched in 7404-100Bap Exo. In line with the “on-off” characteristics of circRNA-miRNA pairs [33, 34], the down-regulated miRNAs in exosomes were then intersected with predicted miRNAs to acquire 60 overlapped circRNA (up-regulated)-miRNA (down-regulated) pairs. Meanwhile, through taking advantages of miRwalk, miRDB, miRTarBase, and TargetScan databases, we predicted target miRNA-mRNA paris from the intersection. By using 4 selected circRNA as center, and their regulatory miRNA and mRNA as the targets, the B[a]P induced circRNA-miRNA-mRNA network were visualized (Figure 3B). We have confirmed the delivery of these B[a]P-induced circRNAs from tumor exosomes to HLF cells using qRT-PCR (Figure 3C). To further evaluate the effects of circRNAs on cells, HLF were transfected with vectors overexpressing different circRNAs. Overexpression of circ_0011496 promoted HLF migration (Figure 3D), and altered morphology of focal adhesions (Figure 3E). HLF transfected with circ_0011496 also showed higher expression of fibrosis (FAP and E-cadherin) and pro-inflammatory (IL-6 and VEGF) genes (Figure 3F). To demonstrate circ_0011496 were not only stemmed from B[a]P stimulation of tumor cells, but also can be delivered into these 7404-100Bap Exo, we isolated the nucleus, cytoplasm, and cultured mediums of 7404-100Bap cells. The significantly higher expression of circ_0011496 was detected in the culture medium of 7404-100Bap cells than in the nucleus and cytoplasm, suggesting the paracrine of circRNA and they were packaged into exosomes (Figure 3G). Taken together, B[a]P stimulated circRNAs encapsulated in exosomes and can be transmitted to and then act as regulatory in target HLF cells.
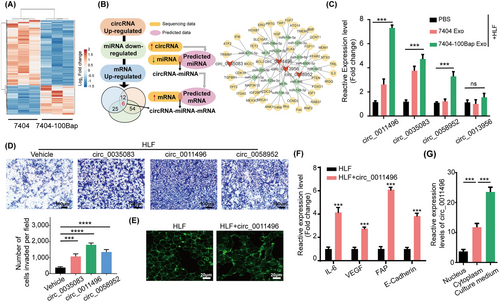
Identification of exosomal circ_0011496.
(A) ceRNA microarray analysis of the indicated circRNAs in 7404 Exo and 7404-100Bap Exo. Red and blue represent upregulated and downregulated circRNAs, respectively. (B) Flowchart of cicrRNA-miRNA-mRNA network analysis. B[a]P stimulated circRNAs were selected and their sponge miRNAs were predicted 2 databases (circInteractome and circBase). These prediction miRNAs were second screened in the 7404-100Bap Exo, the down-regulated miRNAs were selected according to “on-off” characteristics of circRNA-miRNA pairs. Interaction of miRNAs and mRNAs were predicted from the intersection of 4 databases (miRwalk, miRDB, TargetScan and miRTarbase). (C) qRT-PCR analysis of the indicated circRNAs in the control HLF and the HLF pre-treated with 7404 Exo or 7404-100Bap Exo. (D) Migration transwell assay of the HLF transfected with the indicated circRNAs. Scale bar, 100 μm. (E) Phalloidin staining showed the adherent morphology of the control HLF and the HLF transfected with circ_0011496. Scale bar, 20 μm. (F) qRT-PCR analysis of CAF activation-related genes in the HLF transfected with circ_0011496 cells. (G) The expressions of circ_0011496 in the nuclear, cytoplasmic and cultured medium of 7404-100Bap cells were detected by qRT-PCR analysis. ***P < 0.001, ****P < 0.0001. Data in C, D, and G were analyzed using one-way ANOVA tests with Dunnett's multiple comparisons test. Data in F were analyzed using Student's t-tests. Abbreviations: BMP, bone morphogenetic protein; CAF, cancer-associated fibroblast; CCL, chemokine C-C motif ligand; ceRNA, competing endogenous RNAs; circRNA, circularRNA; HLF, human derived lung fibroblast; IL6, interleukin 6; qRT-PCR, quantitative reverse transcription PCR; TNF, tumor necrosis factor; VEGF, vascular endothelial growth factor.
3.4 Characterization the effect of circ_0011496 in fibroblast activation
Circ_0011496, identified as circCSMD2, was evaluated by head-to-tail splicing of the RT-PCR product of circCSMD2 using the expected size of Sanger sequencing (Figure 4A). RNase R digestion (Figure 4B) and actinomycin D treatment (Figure 4C) were performed to verify the circular nature and stability of circ_0011496, which suggested that it is an abundant, circular, and stable candidate circRNA transcript. To further investigate whether circ_0011496 regulates recipient cells through its circular characteristics, we detected the subcellular localization of circRNAs in recipient HLF cells rather than in donor tumor cells. FISH assay through confocal microscopy demonstrated that exosomal circ_0011496 probe (red) was localized to the cytoplasm of Exo-pretreated HLF cells (Figure 4D). Small interfering RNAs (siRNAs) were designed that specifically targeted the back-splice junction sequence of circ_0011496 in the 7404-100Bap cells. Circ_0011496 expressions decreased in the si-circ_0011496-01 and si-circ_0011496-02 groups compared to that in the control (siNC) group, but did not affect CSMD2 gene (Figure 4E). In line with this, the expression levels of circ_0011496 of exosomes that isolated from si-circ_0011496 transfected cells also decreased, implying the transfection of si-circRNA in donor cells could inhibit circ_0011496 expressed in derived exosomes (Figure 4F).
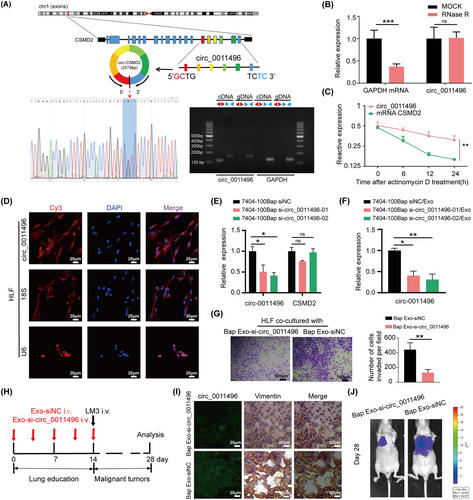
The circular characteristic of circ_0011496 in the CAF activation.
(A) Sketch map and Sanger sequencing to confirm the specific back splicing site of circ_0011496 (circCSMD2). (B) qRT-PCR analysis of circ_0011496 and GAPDH mRNA after RNase R treatment. (C) qRT-PCR analysis of circ_0011496 and linear CSMD2 mRNA in the tumor cells treated with actinomycin D. (D) The cytoplasmic distribution and location of circ_0011496 in the 7404-100Bap Exo pre-treated HLF cells via FISH assay. Scale bar, 20μm. (E) 7404-100Bap cells were transfected with siRNA-control (siNC) and 2 circ_0011496 siRNAs (si- circ_0011496-01 and si- circ_0011496-02). Silencing efficiency of two circ_0011496 siRNAs in these 7404-100Bap cells were detected by qRT-PCR. (F) Exosomes isolated from 7404-100Bap cells that transfected with siNC or circ_0011496 siRNAs (7404-100Bap siNC/Exo and 7404-100Bap si-circ_0011496-01/02/Exo), and expressions of exosomal circ_0011496 were evaluated by qRT-PCR. (G) Migration transwell assay of HLF cells treated with the exosomes derived from 7404-100Bap cells (Bap Exo-siNC) and 7404-100Bap/si-circ_0011496-02 cells (Bap Exo-si- circ_0011496). (H) Exosomes, Bap Exo-siNC and Bap Exo-si-circ_0011496, were collected (200μg/mL) and then i.v. injected to mice every 3days for 5 times to generate circ_0011496-deficient lungs (n = 8/group). LM3 cells (5 × 105) were further i.v. injected and the tumor metastasis were monitored till 28 days. (I) Representative FISH and IHC images for circ_0011496 and fibroblast markers (vimentin) in the same areas of these lungs after 14 days of Bap Exo-siNC and Bap Exo-si-circ_0011496 education. Scale bar, 20 μm. (J) Luciferase-based bioluminescence imaging of metastatic LM3 cells after 28 days i.v. injection in the exosome-educated models to determine the key role of circ_0011496 in promoting lung metastasis. *P < 0.05; **P < 0.01, ***P < 0.001. Data in E and F were analyzed using one-way ANOVA tests with Dunnett's multiple comparisons test. Data in B, C, and G were analyzed using Student's t-tests. Abbreviations: CSMD2, CUB and Sushi multiple domains 2; FISH, fluorescence in situ hybridization; IHC, immunohistochemistry; i.v., intravenous injection; qRT-PCR, quantitative reverse transcription-PCR; siRNA, small interfering RNA.
To explore the effect of circ_0011496 in vitro and in vivo, exosomes were secreted from 7404-100Bap cells transfected with vector-control (Bap Exo-siNC) or vector-si-circ_0011496-01 (Bap Exo-si-circ-0011496). We confirmed the reduced number of migrating cells in circ_0011496-depleted exosomes (Figure 4G). in vivo assay was designed to confirmed the effect of circ_0011496 in lung metastasis. As shown, Bap Exo-siNC and Bap Exo-si-circ-0011496 were i.v. injected to generate lung education models, and the HCC LM3 cells were next injected for evaluating tumor spread (Figure 4H). After 14 days of exosome injection, we performed in situ hybridization of circ_0011496, and the same areas were stained with fibroblast marker (vimentin) by IHC assay. The fluorescence signals of circ_0011496 probes (green) were also present in the vimentin-positive fibroblasts in Exo-siNC injected groups (Figure 4I). Until the end of 28 days, the visualization of LM3 spread and metastasize also demonstrated a significant reduction of lung metastatic foci when inhibition of exosomal circ_0011496 expressions (Figure 4J).
To further confirm the important role of circ_0011496 secreted from B[a]P-exposed HCC cells, we designed the exposure experiments in two liver cancer cell lines, a high-metastatic line LM3 and a low-metastatic cell line HepG2. These two cell lines were treated with 100 nmol/L B[a]P for 30 days (LM3-Bap and HepG2-Bap; Supplementary Figure S2A). Wound healing assay demonstrated an increasing motility capacity of LM3-Bap cells and HepG2-Bap cells than control groups (Supplementary Figure S2B). Next, tumor derived exosomes were isolated from LM3-Bap cells and HepG2-Bap cells, and the expression of circ_0011496 of exosomes were up-regulated after B[a]P exposure (Supplementary Figure S2C). The HLF cells were pre-treated with LM3-Bap Exo and HepG2-Bap Exo also showed enhanced motility phenotype (Supplementary Figure S2D). These results indicated that, after B[a]P exposure, HCC cells exhibited increased expression of circ_0011496, which can be delivered to fibroblasts, and then regulate fibroblast activation.
3.5 Exosomal circ_0011496 directly regulates miR-486-5p in lung fibroblasts
Given that circ_0011496 can regulate fibroblast activation, we performed a biotin-labeled RNA pull-down to screen for sponges of circ_0011496 (circ-probe) in recipient HLF cells. Specific probes were designed to against circ_0011496, and precipitated miRNAs were obtained and compared in the 7404-100Bap Exo-treated HLF and HLF cells (Figure 5A). Next, in order to confirm these dysregulated miRNAs are original from endogenous HLF cells and then were pulled-down by circ_0011496-probe, rather than delivered by exosomal circRNA-miRNA axis. We selected molecules bind to circ_0011496-probe from the lysis total RNA input (HLF-IP) and small RNA (HLF-P) from HLF cells, respectively. Overlapped miRNAs including miR-486-5p were identified as sponges that theoretically bind to and directly regulated by circ_0011496 (Figure 5B). The probe complementary to circ_0011496 pulled down more miR-486-5p than mutant-random control, suggesting the positive correlation (Figure 5C). Additionally, knockdown or overexpression of circ_0011496 in tumor derived exosomes could influence the expression of miR-486-5p in recipient HLF cells (Figure 5D). Dual-luciferase reporter assay also indicated the consistent result that miR-486-5p overexpression could inhibit the luciferase activity of wide-type circ_0011496 vector (WT) as opposed to mutant vector (MUT) (Figure 5E). Considering that miRNAs silence the expression of target genes by binding with Argonaute 2 (Ago2) [35, 36], we performed an RIP assay on Ago2 in exosome-modulated HLF cells. circ_0011496 was significantly enriched in complexes with Ago2 compared to that in the control IgG group, suggesting that circ_0011496 can act as a platform for the binding between miRNAs and Ago2 (Figure 5F). Importantly, after incubation of HLF cells with the 7404-100Bap Exo, we found that miR-486-5p were gradually decreased along with treatment time (Figure 5G), implying the exosomes were continued uptaken by target HLF cell and interacted with HLF miRNA. Colocalization of circ_0011496 and miR-486-5p was observed by immunofluorescence using RNA FISH (FISH-IF), which revealed that the interaction occurred in the cytoplasm of HLF cells (Figure 5H). These results demonstrated the direct regulation of circ_0011496 and miR-486-5p.
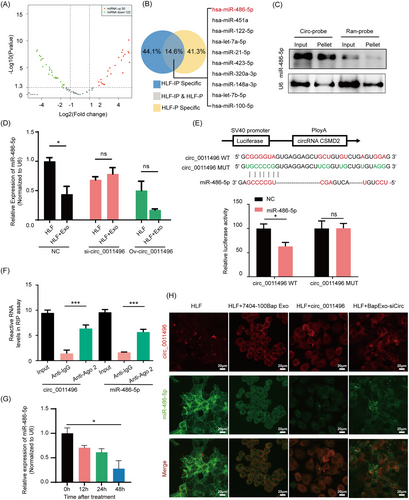
circ_0011496 served as a molecular sponge for miR-486-5p.
(A) Volcano plot illustrates the profile of sponges’ miRNAs obtained by Biotin-labeled RNA pull-down assay of circ_0011496 probe in the 7404-100Bap Exo educated-HLF cells comparing with HLF cells. |log2(Fold change)| ≥1 and P < 0.05). (B)The sponges’ miRNAs were screened by pull down of circ_0011496 specific probe in the total RNA input of HLF cells (HLF-IP) and the small RNA of HLF cells (HLF-P). The top 10 expression miRNAs in the intersection (HLF-IP & HLF-P) that were identified as specifically sponges bind to circ_0011496. (C) Northern blotting performed to detect interaction between circ_0011496 and miR-486-5p. miRNA-486-5p was pulled down by the circ_0011496 probe (circ-probe) but not its mutant random probe (ran-probe). (D) 7404-100Bap cells were transfected with si-NC, si- circ_0011496 and the overexpression plasmid of circ_0011496 (Ov-circ_0011496), and their derived exosomes were further collected, respectively. HLF cells were co-cultured with these different exosomes (HLF + Exo-si-circ_0011496, HLF + Exo-ov-circ_0011496,) for 48h, and the expression levels of mir-486-5p were evaluated by qRT-PCR assay. (E) A schematic of wild-type (WT) and mutant (MUT) circ_0011496 luciferase reporter vectors. Dual-luciferase reporter assay was performed to assess the interaction between circ_0011496 WT or circ_0011496 MUT of circ_0011496 and miR-486-5p. (F) Anti-AGO2 RIP assays were used in 7404-100Bap Exo modulated fibroblasts to determine the enrichment of circ_0011496 and miR-486-5p. Anti-IgG was used as control. (G) qRT-PCR analysis of the expression levels of miR-486-5p in the lung fibroblasts treated with7404-100Bap Exo at indicated time points. (H) HLF control, HLF treated with 7404-100Bap Exo (HLF + 7404-100Bap Exo), HLF transfected with circ_0011496 (HLF + circ_0011496), and HLF treated with exosomes derived form 7404-100Bap cells-transfected si-circ_0011496 (HLF + BapExo-siCirc) were used as cell models. FISH indicates the colocation of circ_0011496 and miR-486-5p in these cell models, circ_0011496 probe was labeled red with biotin. Scale bar, 20 μm. *P < 0.05; ***P < 0.001; ns, not significant. Data in F and G were analyzed using one-way ANOVA tests with Dunnett's multiple comparisons test. Data in D and E were analyzed using Student's t-tests. Abbreviations: FISH, fluorescence in situ hybridization; HLF, human derived lung fibroblast; IHC, immunohistochemistry; i.v., intravenous injection; qRT-PCR, quantitative reverse transcription-PCR; RIP, RNA immunoprecipitation; siRNA, small interfering RNA.
3.6 Twinfilin-1 (TWF1) is a direct target of miR-486-5p in vitro and in vivo
Bioinformatic tools including miRDB, TargetScan, miRWalk, and miRTarBase, were used to predict potential targets of miR486-5p, of that 13 genes were identified at the intersection of these databases (Figure 6A). We next evaluated the expression of these predicted candidate mRNAs using in vitro and in vivo models. TWF1 is not only up-regulated in the HLF cells treated with 7404-100Bap Exo (in vitro model, HLF + Exo), but also increased in the 7404-100Bap Exo pre-metastatic lungs (in vivo model, Lung + Exo) (Figure 6B). TWF1 expression was further inhibited when HLF cells transfected with miR-486-5p mimic (Figure 6C). Considering that TWF1is regarded as a highly conserved actin monomer-binding protein that regulates cytoskeletal dynamics [37], we speculated that dysregulation of TWF1 also play an important role in activated fibroblasts. Altogether, TWF1 is the potential target of miR-486-5p. To explore the regulation of miR-486-5p and TWF1, WT and mutated miR-486-5p binding sites were cloned into luciferase vectors. Activities of luciferases with mutated 3’-UTRs were no longer suppressed by corresponding miRNAs (Figure 6D). HLF cells treated with 7404-100Bap displayed increased expression of TWF1, as well as the expression of matrix and transmembrane associating molecules such as interleukin-11 (IL11), signal transducer activator transcription 3 (STAT3) and metallopeptidase 9 (MMP9) than that treated with 7404 Exo (Figure 6E). To further explore the regulatory mechanisms of the circRNA-miRNA-mRNA network, HLF cells were transfected with miRNA mimic, circRNA vector, or siRNA-TWF1. The expression levels of TWF1 and MMP9 were downregulated by miR-486-5p but were rescued by circ_0011496 expressions (Figure 6F). The knockdown of TWF1 only decreased expression of MMP9, all these data suggested that TWF1 is a direct downstream target that mediates exosomal circ_0011496 and HLF miR-486-5p axis. Functionally, the overexpression of miR-486-5p mimic and inhibition of TWF1 enhanced HLF cell migration in vitro (Figure 6G). Finally, it is important to determine whether TWF1 expression could promote organotropic metastasis by activating the lung microenvironment before tumor cell arrival. We established a TWF1 overexpression model specific in the lungs via AAV inhalation (Figure 6H). The injury and fibrosis in the lung along with the increasing numbers of metastatic nidus in the AAV-TWF1 model (Figure 6I-J), suggesting that TWF1 affects HLF transformation in the pre-metastatic niche.
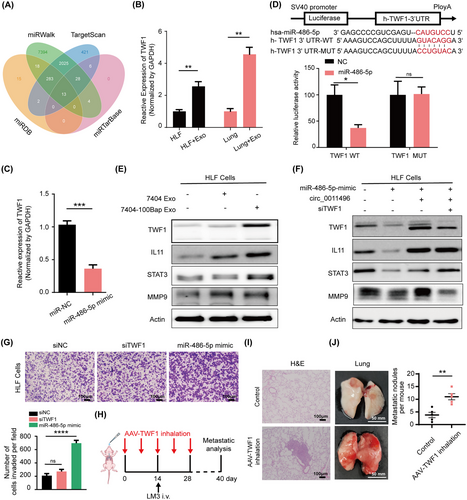
TWF1 is a direct downstream target of miR-486-5p.
(A) Target gene prediction of miR-486-5p with four bioinformatics tools. (B) TWF1 expressions were evaluated in the control HLF cells and 7404-100Bap Exo pre-treated HLF cells (HLF + Exo). For in vivo assessment, i. v. injection of PBS control (Lung) and 7404-100Bap Exo (Lung + Exo) to mice for 15 days to generated an education model as described, and TWF1 expressions of these lungs were evaluated by qRT-PCR. (C) qRT-PCR analysis of TWF1 expression in the HLF cells transfected with miR-486-5p mimic or control miRNA (miR-NC), respectively. (D) Relative luciferase activity of the wild-type (WT) and mutant type (MUT) of miR-486-5p binding site in TWF1 3’UTR. (E) Western blotting of indicated proteins in 7404 Exo or 7404-100Bap Exo treated HLF cells. (F) Western-blotting of indicated proteins in HLF cells transfected with miR-486-5p mimic, circ_0011496 overexpression plasmid, and TWF1 siRNA (siTWF1). (G) Migration transwell assay for HLF transfected with siTWF1 and miR-486-5p mimic. Scale bar, 100 μm. (H) Schematic illustration for the in vivo delivery of AAV-mediated TWF1 overexpression in lung and lung metastasis analysis. TWF1 transfected into the AAV vectors (50μg/mL) and introduced to mice via inhalation every 5-6 days for 5 times to specific express TWF1 in lungs. LM3 cells (5 × 105) were i.v. injected by the 14 days of TWF1-AAV inhalation, and the tumor growth and metastasis were analyzed at the end of 40 days. (I-J) Mice along with lung metastasis mention in H were sacrificed 40 days after LM3 injection. H&E staining of lung tissues (I) and the images of HCC tumor-bearing lung (J) were shown. *P < 0.05; **P < 0.01, ***P < 0.001, ****P < 0.0001. Scale bar, 100 μm.Data in G were analyzed using one-way ANOVA tests with Dunnett's multiple comparisons test. Data in B, C, and D were analyzed using Student's t-tests. Abbreviations: 3’UTR, 3' untranslated regions; AAV, adeno-associated virus; HLF, human derived lung fibroblast; IL11, interleukin; i.v., intravenous; MMP9, matrix metalloproteinase 9; STAT3, signal transducer and activator of transcription 3; TWF1, twinfilin-1.
3.7 Activated fibroblasts generate a feedback loop for distant ograntropic metastasis
Activated fibroblasts have strong tumor-modulating effects on stromal cell activation and distant tumor cell recruitment [38]. We herein developed a set of experiments to simulate the effect of activated fibroblasts on other cells in pre-metastatic niche. After 48h treatment with exosomes, these activated HLF cells (con/HLF, 7404 Exo/HLF, and 7404-100Bap Exo/HLF) were set on the bottom chamber, meanwhile the tumor cells or surrounding stroma cells were set on the upper chamber (Figure 7A). The homologous 7404 or high metastatic LM3 tumor cells were selected to evaluate the recruitment of activated HLF on tumor cells (upper). We found both of 2 HCC cells co-cultured with 7404-100Bap Exo/HLF cells showed enhanced invading capacities (Figure 7B). Similarly, tube formation assay for human umbilical vein endothelial cells (HUVEC) also displayed the most enhanced angiogenetic ability when co-cultured with 7404-100Bap Exo/HLF group (Figure 7A and C).
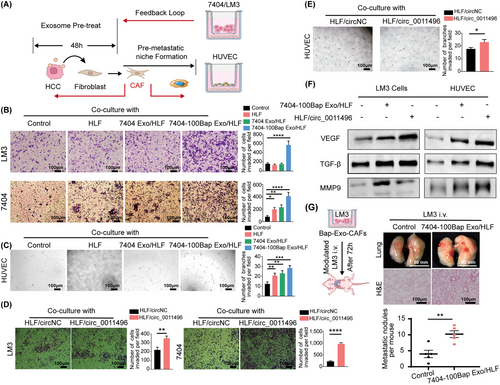
CAFs promote angiogenesis of HUVEC and migration of LM3 exoaomal circRNA.
(A) Schematic diagram of Exo-modulated fibroblast cells regulated surrounding stroma cells, and generated a feedback loop for distant tumor cells. The HLF cells were pre-treated with 7404 Exo or 7404-100Bap Exo for 48h for activation (7404Exo/HLF, 7404-100Bap Exo/HLF), these activated-HLF cells were next co-cultured with 7404 and LM3 cells for transwell assay, and with HUVEC cells for angiogenesis tube formation. (B) Migration of HCC 7404 and LM3 cells co-cultured with PBS control, HLF cells, 7404Exo/HLF cells, 7404-100Bap Exo/HLF cells were accessed by transwell assay. Scale bar, 100 μm. (C) Angiogenesis ability of HUVEC cells co-cultured with PBS control, HLF cells, 7404Exo/HLF cells, 7404-100Bap Exo/HLF cells were accessed by matrigel-based tube formation. Scale bar, 100 μm. (D) Migration of HCC cells treated with HLF-transfected circ_0011496 vector was accessed by transwell assay. Scale bar, 100 μm. (E) Tube formation ability of HUVEC treated with HLF-transfected circ_0011496 vector. Scale bar, 100 μm. (F) Western blotting of indicated proteins in the HLF cells co-cultured 7404-100Bap Exo, as well as co-cultured with HLF-transfected circ_0011496 vector. (G) We developed an in vivo experiment to explore the cancer-stroma crosstalk, LM3 cells and 7404-100Bap Exo/HLF cells were co-cultured using 0.4μm-transwell chamber for 72h, so that LM3 cells could not pass through the pore but effected by signal transduction. These co-cultured LM3 cells were i.v. injected into mice, and the lungs after 35 days LM3 injection were scarfed and evaluated by H&E staining. Scale bar, 100 μm for H&E and 50mm for lung. *P < 0.05; **P < 0.01, ***P < 0.001, ****P < 0.0001. Data in B and C were analyzed using one-way ANOVA tests with Dunnett's multiple comparisons test. Data in E and D were analyzed using Student's t-tests. Abbreviations: CAF, cancer-associated fibroblast; HCC, hepatocellular carcinoma; H&E, hematoxylin and eosin; HUVEC, human umbilical vein endothelial cell; MMP9, matrix metalloproteinases 9; STAT3, signal transducer and activator of transcription 3; TGF-β, transforming growth factor-β.
Next, we evaluated whether circRNAs paly essential roles in converting activated myofibroblasts. HLF cells were transfected with circ_0011496 (HLF/circ_0011496) or circNC (HLF/circNC) in advance, and these transfected HLF cells were further co-incubated with LM3, 7404 and HUVEC in the transwell chambers. Both LM3 and 7404 significantly migrated and invaded when co-cultured with circ_0011496 overexpressed HLF (Figure 7D). Similarly, HUVEC showed increased tube formation after co-cultured with circ_0011496 transfected HLF (Figure 7E). To confirm whether these activated fibroblasts could reprogram surrounding tumor and stroma cells in the microenvironment by regulating MMP9 pathway through exosomal circ_0011496. LM3 and HUVEC were co-cultured either with 7404-100Bap Exo/HLF, or HLF/circ_0011496 for 48 h, respectively, and MMP9-associated tumor vasculature and inflammatory signals were evaluated. We found significant higher expressions of VEGF and MMP9 in HUVEC/LM3 cells that treated with 7404-100Bap Exo/HLF and HLF/circ_0011496, and the slightly upregulation of TGF-β in these samples (Figure 7F). in vivo, LM3 cells were pre-treated with exosome-modulated CAFs for 72h, and these pre-treated LM3 cells were then i.v. into mice. Increased metastatic nodes in the tumor-bearing lungs are shown in Figure 7G. Therefore, we demonstrated that exosomal circ_0011496 contributes to fibroblast-dependent niche formation, as a consequence to transform endothelial cells and recruit tumor cells.
4 DISCUSSION
In this study, we investigated the adverse outcome of B[a]P from the perspectives of tumor-host crosstalk and pre-metastatic niche formation. We introduced tumor derived exosomes as regulatory carriers for intracellular signal transduction in vitro, and established an exosome-educated lung model in vivo. Our results showed that exposure to B[a]P could convert fibroblasts to activated fibroblasts or CAFs via paracrine of exosome-mediated oncogenes in the lungs before HCC cell arrive. Compared to non-exposed HCC cells, circ_0011496 was up-regulated following B[a]P treatment and was mainly packaged into HCC-derived exosomes Pull-down assay was conducted to screen for circ_0011496 sponge miRNAs in recipient fibroblasts rather than in their donor HCC cells. We herein demonstrated exosomal circ_0011496 were delivered into fibroblasts and competitively bound to miR-486-5p, leading to fibroblast transformation via regulation of the TWF1 cascade. Additionally, we established AAV-inhalation model, confirming that specially overexpression of TWF1 in the lungs could trigger pre-metastatic niche formation via activating cytokines, such as IL11, MMP9 and VEGF. These cytokines stimulated by activated fibroblasts not only promote endothelial cell angiogenesis, but also recruit the distant HCC cells migrate to lungs (Figure 8). Therefore, the hepatotoxicity feedback-loop of B[a]P accelerated organotropic metastasis by delivering the tumor derived exo-circ_0011496 to activate the miR-486-5p/TWF1/MMP9 cascade.
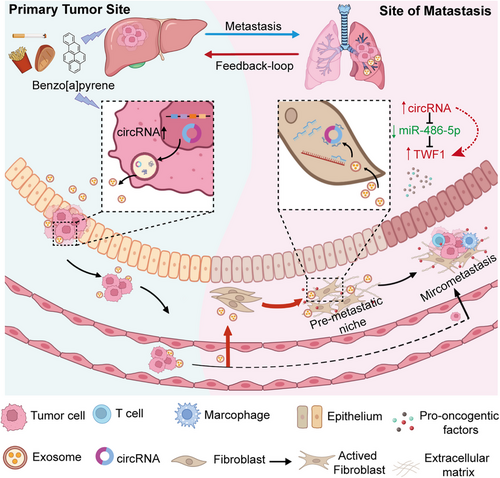
Schematic representation of the role of B[a]P in HCC lung-specific metastasis.
The schematic shows the effect of B[a]P in HCC lung-specific metastasis through exosomal circRNA. Before tumor cell arriving, exposure of B[a]P trigger an increased in the amount of exosomal circRNA to target lung fibroblasts, contributing to the transformation of fibroblasts in the metastatic organ. These Exo-activated fibroblasts further generate a feedback loop either for epithelial cell angiogenesis or recruitment of primary HCC invasion. Abbreviations: B[a]P, Benzo[a]pyrene; HCC, hepatocellular carcinoma.
Liver tissue has the highest capacity for toxic metabolites of B[a]P, making it sensitive and inevitably exposed [39, 40]. “Western diet” becomes a kind of food preference that accounting the main source of B[a]P exposure, but foodborne intake concentrations are too low to be taken seriously in healthy populations [41]. Our previous study found that low-dose B[a]P exposure promoted lung-specific metastasis but did not alter primary tumor growth, implying an unexplored toxicological mechanism of B[a]P-induced malignancy [13, 14]. We herein established an “exosome-educated model” to report that B[a]P-exposed HCC cells converted lung fibroblasts to activated phenotypes through paracrine of exosome-mediated oncogenes. These activated fibroblasts are believed to reprogram microenvironment by expressing pro-inflammatory genes. Our results also confirmed the Exo-educated lungs could secret cytokines, such as IL-11, VEGF, and MMP9 to promote tube formation in HUVEC, leading to pre-metastatic niche generation before primary tumor cell arrival.
Adverse outcome pathway is a chemical-independent description of a linear or branching path from a molecular initiating event (MIE) to an eventual adverse outcome at the organism or population level [42, 43]. In our study, we identified the dysregulation of exosomal oncogenes as a novel type of MIE, and the distant lung metastasis as final outcome. TWF1 can affect tumor progress by modulating the phenotype of tumor cells to influence immune cell function, or by affecting inflammation-related signaling pathways [44]. Our data provide a novel concept and paradigm to support the effect of B[a]P on exosome-mediated metastasis to distal organs. Therapeutic interference of this pathway, especially the circRNA-loaded exosomes, suggested an attractive approach for treating HCC patients with unhealthy living habits. However, the lack of studies on the clinical significance of exosomal circRNA in the serum of B[a]P-exposed HCC patients is one of the major limitations. In addition, it is difficult to simulate real-life scenarios on exosome changes, much of our knowledge discussing the regulatory roles of exosomal circRNAs obtained from cell and animal models, but few studies conducted in humans generally involved, making it difficult to simulate real-life scenarios of exosome changes. Furthermore, single cell analysis on the B[a]P-educated lung tissues would be necessary for further investigation, providing an overview profile of cancer communications.
5 CONCLUSIONS
In conclusion, our findings demonstrated that exosomes derived from B[a]P stimulated HCC cells converted lung fibroblasts to CAFs through paracrine of circRNAs. We herein reported a new mechanism by which exosomes transfer circ_0011496 to lung fibroblasts, and then regulate miR-486-5p/TWF1/MMP9 cascades of activation of target cells, contributing new evidence for the crosstalk between primary tumor and distant host cells.
AUTHOR CONTRIBUTIONS
Conceptualization: Yang Ge and Wei Mu. Methodology: Pengfei Gu, Wei Mu and Jinjin Zhou. Bioinformatics and validation: Wei Mu and Pengfei Gu. Formal analysis and investigation: Huating Li and Yulun Jian. Data curation and visualization: Wei Mu and Weiping Jia. Original draft preparation: Wei Mu and Pengfei Gu. Review and editing: Wei Mu and Yang Ge. Supervior: Yang Ge and Weiping Jia. Project administration: Weiping Jia and Wei Mu. All authors have read and agreed to the published version of the manuscript.
ACKNOWLEDGEMENTS
We thank Prof. Margot Zoeller from Heidelberg Germany for editing the English grammar of this manuscript, and the efforts and time from anonymous reviewers for reading and commenting on the manuscript. This work was sponsored by grants from the National Nature Science Foundation (82173543 and 81902939), Innovative Research Team of High-level Local Universities in Shanghai (SHSMU-ZLCX20211602), Key laboratory of the Ministry of Education Foundation (2022-MEKLLC-MS-003), Sanming Project of Medicine in Shenzhen (SZSM202311019), Shanghai Key Discipline of Public Health Grants Award (GWVI-11.1-20), and Science and technology commission of Shanghai municipality project (23QA1405700).
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no competing interests.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The animal study was approved by and carried out in compliance with the guidance suggestion of Animal Care Committee of Shanghai Jiao Tong University School of Medicine (permit number: SYXK2018-0027).
Open Research
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.




