Multiomics analysis reveals metabolic subtypes and identifies diacylglycerol kinase α (DGKA) as a potential therapeutic target for intrahepatic cholangiocarcinoma
Weiren Liu, Huqiang Wang, Qianfu Zhao and Chenyang Tao authors contributed equally.
Abstract
Background
Intrahepatic cholangiocarcinoma (iCCA) is a highly heterogeneous and lethal hepatobiliary tumor with few therapeutic strategies. The metabolic reprogramming of tumor cells plays an essential role in the development of tumors, while the metabolic molecular classification of iCCA is largely unknown. Here, we performed an integrated multiomics analysis and metabolic classification to depict differences in metabolic characteristics of iCCA patients, hoping to provide a novel perspective to understand and treat iCCA.
Methods
We performed integrated multiomics analysis in 116 iCCA samples, including whole-exome sequencing, bulk RNA-sequencing and proteome analysis. Based on the non-negative matrix factorization method and the protein abundance of metabolic genes in human genome-scale metabolic models, the metabolic subtype of iCCA was determined. Survival and prognostic gene analyses were used to compare overall survival (OS) differences between metabolic subtypes. Cell proliferation analysis, 5-ethynyl-2'-deoxyuridine (EdU) assay, colony formation assay, RNA-sequencing and Western blotting were performed to investigate the molecular mechanisms of diacylglycerol kinase α (DGKA) in iCCA cells.
Results
Three metabolic subtypes (S1-S3) with subtype-specific biomarkers of iCCA were identified. These metabolic subtypes presented with distinct prognoses, metabolic features, immune microenvironments, and genetic alterations. The S2 subtype with the worst survival showed the activation of some special metabolic processes, immune-suppressed microenvironment and Kirsten rat sarcoma viral oncogene homolog (KRAS)/AT-rich interactive domain 1A (ARID1A) mutations. Among the S2 subtype-specific upregulated proteins, DGKA was further identified as a potential drug target for iCCA, which promoted cell proliferation by enhancing phosphatidic acid (PA) metabolism and activating mitogen-activated protein kinase (MAPK) signaling.
Conclusion
Via multiomics analyses, we identified three metabolic subtypes of iCCA, revealing that the S2 subtype exhibited the poorest survival outcomes. We further identified DGKA as a potential target for the S2 subtype.
Abbreviations
-
- DGKA
-
- diacylglycerol kinase α
-
- iCCA
-
- intrahepatic cholangiocarcinoma
-
- WES
-
- whole-exome sequencing
-
- Human-GEMs
-
- human genome-scale metabolic models
-
- EdU
-
- 5-Ethynyl-2’-deoxyuridine
-
- KRAS
-
- Kirsten rat sarcoma viral oncogene homolog
-
- ARID1A
-
- AT-rich interactive domain 1A
-
- PA
-
- phosphatidic acid
-
- MAPK
-
- mitogen-activated protein kinase
-
- TP53
-
- tumor protein p53
-
- FGFR2
-
- fibroblast growth factor receptor 2
-
- IDH1/2
-
- isocitrate dehydrogenase 1/2
-
- BAP1
-
- BRCA1-associated protein-1
-
- OS
-
- overall survival
-
- QC
-
- quality control
-
- BWT
-
- Burrows-Wheeler transform
-
- GATK
-
- Genome Analysis Toolkit
-
- SNV
-
- single nucleotide variant
-
- Vcf
-
- Variant call format
-
- ANNOVAR
-
- annotate variation
-
- VAF
-
- variant allele frequency
-
- MS
-
- mass spectrum
-
- ITMS
-
- Ion trap mass spectrometry
-
- iBAQ
-
- intensity-based absolute quantification
-
- CV
-
- coefficient of variation
-
- NMF
-
- non-negative matrix factorization
-
- KEGG
-
- Kyoto Encyclopedia of Genes and Genomes
-
- GSVA
-
- gene set variation analysis
-
- DFS
-
- disease-free survival
-
- HR
-
- hazard ratio
-
- BH
-
- Benjamini-Hochberg
-
- PermFIT
-
- permutation-based feature importance test
-
- SVM
-
- support vector machines
-
- DNN
-
- deep neural networks
-
- RF
-
- random forest
-
- AUC
-
- area under the curve
-
- TCA
-
- tricarboxylic acid cycle
-
- tINIT
-
- task-driven integrative network inference for tissues
-
- ACHR
-
- artificially centered hit-and-run
-
- t-SNE
-
- t-distributed stochastic neighbor embedding
-
- scRNA-seq
-
- single-cell RNA sequencing
-
- scCATCH
-
- single-cell cluster-based automatic annotation toolkit for cellular heterogeneity
-
- TPM
-
- transcripts per million
-
- CNV
-
- copy number variation
-
- shRNAs
-
- short hairpin RNAs
-
- CCK8
-
- cell counting kit-8
-
- DMSO
-
- dimethyl sulfoxide
-
- TMA
-
- Tissue microarray
-
- IHC
-
- immunocytochemistry
-
- DAB
-
- 3,3 N-diaminobenzidine tetrahydrochloride
-
- RIPA
-
- radioimmunoprecipitation assay buffer
-
- PVDF
-
- polyvinylidene fluoride
-
- BSA
-
- bovine serum albumin
-
- ERK1/2
-
- extracellular regulated protein kinases 1/2
-
- MEK1/2
-
- mitogen-activated protein kinase kinase1/2
-
- GAPDH
-
- glyceraldehyde-3-phosphate dehydrogenase
-
- HRP
-
- horseradish peroxidase
-
- ECL
-
- enhanced chemiluminescence
-
- GSEA
-
- gene set enrichment analysis
-
- HK1
-
- Hexokinase 1
-
- PFKM
-
- muscle phosphofructokinase
-
- NT5E
-
- ecto-5’-nucleotidase
-
- ENTPD1
-
- ectonucleoside triphosphate diphosphohydrolase 1
-
- ATP
-
- adenosine triphosphate
-
- AMP
-
- adenosine monophosphate
-
- PRODH
-
- proline dehydrogenase
-
- GLS2
-
- glutaminase 2
-
- HEPH
-
- hephaestin
-
- ALOX5
-
- arachidonate 5-Lipoxygenase
-
- PLA2G4A
-
- phospholipase A2, group IVA
-
- CNDP1
-
- cytosolic non-specific dipeptidase 1
-
- GFPT2
-
- glutamine–fructose-6-phosphate aminotransferase 2
-
- HIF-1
-
- hypoxia-inducible factor-1
-
- FcεRI
-
- Fc epsilon Receptor I
-
- LCAT
-
- lecithin-cholesterol acyltransferase
-
- SLC3A2
-
- solute carrier family 3 member 2
-
- GPD2
-
- glycerol-3-phosphate dehydrogenase 2
-
- ALDH1L2
-
- aldehyde dehydrogenase 1 family member L2
-
- DC
-
- dendritic cell
-
- Treg
-
- regulatory T
-
- PLOD1
-
- 2-oxoglutarate 5-dioxygenase 1
-
- ASPH
-
- aspartate beta-hydroxylase
-
- TTN
-
- titin
-
- MUC16
-
- mucin 16
-
- EPHA2
-
- ephrin type-A receptor 2
-
- DG
-
- phosphorylate diacylglycerol
-
- DEGs
-
- differentially expressed genes
-
- TNBCs
-
- triple-negative breast cancer
-
- MPSs
-
- metabolic pathway-based subtypes
-
- PD-1
-
- programmed cell death protein 1
-
- TGF-β-Smad4
-
- transforming growth factor beta-SMAD family member 4
1 BACKGROUND
Intrahepatic cholangiocarcinoma (iCCA), a highly lethal cancer, is the second most prevalent primary liver malignancy, comprising approximately 20% of all hepatic tumors [1]. The incidence of iCCA has increased by more than 140% over the past decades [2] and was estimated to rise by up to 10 folds worldwide in the following decades [3]. Due to silent clinical symptoms at an early stage, most patients are diagnosed at advanced stages. With limited treatment strategies, the 5-year survival rate of iCCA patients remains at approximately 9% [4]. However, the mechanism underlying the pathogenesis of iCCA remains unclear.
Recent studies using high-throughput sequencing technology have revealed the genetic landscape of iCCA, indicating that tumor protein p53 (TP53), KRAS, fibroblast growth factor receptor 2 (FGFR2), isocitrate dehydrogenase 1 (IDH1)/IDH2, and BRCA1-associated protein 1 (BAP1) mutations (fusion) are the primary driver gene variations [5]. Based on these findings, precise treatments targeting FGFR2 and IDH1/IDH2 have been developed and have shown preliminary clinical efficacy [6, 7]. Therefore, multiomics technology may help build more profound insight into the mechanisms underlying iCCA and provide a novel therapeutic strategy. Through integrated multiomics analysis, the results of molecular studies have led to the development of diverse classifications of iCCA. Dong et al. [5] identified four distinct proteomic subgroups with different biological and clinical features, including inflammatory (S1), mesenchymal (S2), metabolic (S3) and differentiated (S4) features. Among these subgroups, patients with S4 showed the best overall survival (OS). Deng et al. [8] classified cholangiocarcinoma, including iCCA and extrahepatic cholangiocarcinoma, into the following three proteomic subtypes: S-I (metabolism), S-II (proliferation), and S-III (stromal), and patients with S-I showed the best OS. Lin et al. [9] described the immunogenomic traits of iCCA, and Song et al. [10] depicted the molecularly distinct features of two histological subtypes of iCCA. In addition, Cho et al. [11] identified three clinically supported iCCA subtypes (stem-like, poorly immunogenic, and metabolism) and found that NCT-501 (an IDH1 family member inhibitor) exhibited synergism with nanoparticle albumin-bound paclitaxel for the stem-like subtype in an iCCA organoid model. These classifications provide us with molecular phenotypes to understand prognostic differences among iCCA patients. However, these studies provide little information on metabolic dysregulation and heterogeneity in iCCA patients.
Metabolic reprogramming is recognized as a hallmark of malignancy [12]. On the one hand, metabolic reprogramming promotes tumorigenesis by promoting the rapid proliferation, survival, invasion, metastasis, and therapeutic resistance of cancer cells. On the other hand, with the development of tumors, cancer cells acquire more mutations and changes, further enhancing metabolic reprogramming, which in turn accelerates tumor growth and development [13, 14]. In addition, accumulating evidence demonstrates that metabolic reprogramming also has a crucial role in regulating the proliferation, differentiation, and function of immune cells, which eventually leads to tumor immune escape [15] and is related to patient prognosis [16]. Nevertheless, little is known about the metabolic differences among iCCA patients with different prognoses. Thus, we performed an integrated multiomics analysis of 116 iCCA and paired adjacent normal tissues and performed metabolic classifications to depict differences in iCCA patient survival, hoping to provide a novel perspective to understand and treat iCCA.
2 MATERIALS AND METHODS
2.1 Clinical summary
We collected iCCA samples and the paired adjacent normal tissues from 116 patients who received liver resection without any anticancer treatments prior to surgery and were pathologically diagnosed with iCCA at Zhongshan Hospital (Shanghai, China), and the follow-up ended in December 2022. We obtained tissue samples and promptly stored them in liquid nitrogen, within 30 min of resection. All collected samples were intratumoral specimens and confirmed by pathologists from Zhongshan Hospital. Additionally, there were two interruptions in perfusion, each lasting for 15 min during the surgical procedure. The sample collection procedure was consistent across all cases. Patients enrolled in this study all signed the informed consent form to allow the use of their data and samples.
The medical ethics committee of Zhongshan Hospital reviewed and approved the use of all the human tissues involved in this research (B2021-611). The baseline clinicopathological characteristics of the 116 patients with iCCA are shown in Supplementary Tables S1-S2.
2.2 Sequencing
-
Genomic analysis (for 116 iCCA samples and the paired adjacent normal tissues), wherein whole-exome sequencing (WES) was performed using the NovaSeq 6000 platform (Illumina, San Diego, CA, USA) with a single sample size of more than 10 G and a sequencing depth >100×.
-
Transcriptomic analysis (for 116 iCCA samples), wherein RNA was extracted for sample quality control, and quality control-compliant samples were subjected to bulk RNA-sequencing using the NovaSeq 6000 platform (Illumina), yielding 10 G raw data per sample.
-
Proteomic analysis (for 116 iCCA samples), wherein approximately 0.1 g issue was lysed in urea solution (8 mol/L urea in 0.1 mol/L Tris-HCl, pH 8.5, containing 1× protease and phosphatase inhibitor), and the whole proteins were extracted by 1 min of sonication (3 s on and 3 s off, amplitude 25%). The peptide segments were isolated by filter-aided sample preparation enzymes and fractionated by C18 reversed-phase liquid chromatography (each sample was divided into 3 fractions). Five hundred nanograms of peptides from each fraction were subjected to mass spectrometry (MS) identification using an Orbitrap Fusion Lumos Tribrid mass spectrum spectrometer (Thermo Fisher Scientific, Carlsbad, CA, USA) coupled online to a nanoflow LC system (EASY-nLC 1200, Thermo Fisher Scientific). The polypeptide samples were dissolved in 10 μL of solvent A (0.1% formic acid in high-performance liquid chromatography grade water). Then, 5 μL of dissolved polypeptide samples were loaded onto the precolumn at a flow rate of 3 μL/min on the EASY-nano-LC chromatography system, and separated on the column at a flow rate of 600 nL/min. The gradient was as follows: 0-10 min, 5%–11% solvent B; 10-110 min,11%–25% solvent B; 110-140 min, 25%–40% solvent B; 140-141 min, 40%–95% solvent B; and 141-150 min, 95% solvent B.
2.3 Quality control and quantification of sequencing data
Genomic and transcriptomic data were sequenced for fragment quality using FastQC software (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to filter out low-quality reads using the FASTX-Toolkit (v 0.0.13, http://hannonlab.cshl.edu/fastx_toolkit/). WES data were analyzed as follows: the Burrows-Wheeler Alignment Tool [17] (v0.7.17, https://bio-bwa.sourceforge.net/) was used to map all sequenced read segments to the hg38 reference genome, with the main algorithm implemented using either spaced-seed indexing or Burrows-Wheeler transform (BWT) techniques. Data pre-processing, including the removal of repetitive sequences, was performed according to the Best Practices Workflows recommended by the Genome Analysis Toolkit (GATK) [18]. We used GATK (v4.1.5.0, https://gatk.broadinstitute.org/hc/en-us) to identify single nucleotide variants (SNVs) in tumor tissues and adjacent normal tissues, using the GATK-recommended af-only gnomad.hg38.vcf with somatic-hg38_1000g_pon.hg38.vcf as a reference. Variant call format (Vcf) was further performed to screen for normal population germline variants and to annotate them with annotate variation (ANNOVAR) (v2019-10-24, http://www.openbioinformatics.org/annovar). Finally, 18,593 SNVs were obtained by screening variant allele frequency (VAF) > 5% and supporting mutations of reads > 10 in tumor tissues, VAF < 1% and variant reads < 5 in adjacent normal tissues. Tumor cellularity and tumor ploidy were estimated by sequenza-utils and sequenza, as well as copy number detection (allele specificity and total copy number) and quantification using the adjacent normal tissues as references.
We chose Salmon [19] (v1.5.2, https://combine-lab.github.io/salmon/) and the reference genome described above for transcriptomic quantitative analysis. The Salmon tool quantifies reads by matching directly to the transcript (cDNA) sequences through a fast and lightweight quasi-mapping program, with the following two main steps to run: build index and sample quantification. Subsequent gene expression analysis was performed using the R package tximport (v1.22.0, https://bioconductor.org/packages/release/bioc/html/tximport.html) [20].
Raw proteomic data were searched using MaxQuant [21] (v1.6.1.0, https://www.maxquant.org/) against UniProt database reference files (Version: 202002; 20,367 sequences, https://www.uniprot.org/). The relevant parameters were set as follows: carbamidomethyl (C) was set as a fixed modification while oxidations (M) and acetyl (protein N-term) were set as variable modifications. MS/MS tolerance (Ion trap mass spectrometry, ITMS) was set to 0.5 Da. Searches allowed for up to 2 missed lysine/arginine cleavages by trypsin (full specificity). The false discovery rate (FDR) for peptides and proteins was set to less than 0.01. Pollution and reverse library results were excluded.
Further proteomic analysis was performed using intensity-based absolute quantification (iBAQ) values, normalized by quantile and treated with log2. We selected proteins expressed in at least 25% of samples while removing liver-specific genes [22, 23] and imputing missing values using MissForest (v1.5, https://cran.r-project.org/web/packages/missForest/index.html) [24].
2.4 General proteomic and metabolic subtyping
Proteins used for general proteomic molecular subtyping were selected based on their coefficient of variation (CV) among samples. The abundance matrix of the top 1,500 proteins with the highest CV values was used as input features after performing min-max normalization across the samples, and the non-negative matrix factorization (NMF) algorithm (https://cran.r-project.org/web/packages/NMF/index.html) was used for subtyping. When k = 3, cophenetic values reached their maximum and began to decline, while silhouette values ranged between 0.64 and 1.00 (Supplementary Figure S1A); thus, we selected three clusters as candidate proteomic clusters.
At the same time, we also used the protein abundance matrix of the top 1,500 proteins with the highest CV, which was identified in at least 50% of the samples and normalized by rowwise min-max scaling, as the input for NMF. When k = 4, cophenetic values reached their maximum and began to decline, while silhouette values ranged between 0.60 and 1.00 (Supplementary Figure S1B); then, we also chose four clusters as candidate general proteomic clusters. Thus, for the proteomics clusters, with the different thresholds of missing values, we obtained two sets of clusters for iCCA: one had three clusters, and the other had four clusters.
For metabolic subtypes, we mainly tried two strategies. For the protein abundance-based strategy, three metabolic protein abundance matrices were selected as input. We input the metabolic proteins in the Kyoto Encyclopedia of Genes and Genomes (KEGG) database [25], the top 1,500 variant metabolic proteins in the Human-GEMs [26], and the 412 variant proteins overlapping between the top 1,500 variant proteins used in the Human-GEMs and top 1,500 proteins used in the three proteomic clusters. For the gene set variation analysis (GSVA) (v1.38.2, https://bioconductor.org/packages/release/bioc/html/GSVA.html) [27] pathway enrichment score-based strategy, 69 metabolic pathways in the c2.cp.kegg.v7.4.symbols.gmt gene set in MsigDB (https://www.gsea-msigdb.org/gsea/msigdb) were selected as the metabolic gene set. We compared all these input silhouette values with silhouette values ranging from 0.77 to 1.00 (Supplementary Figure S1C) when using the top 1,500 variant metabolic proteins in the human genome-wide model as NMF input. Three metabolic subtypes were defined from this input.
2.5 OS, disease-free survival (DFS), and prognostic gene analysis
To compare survival differences in proteomics clusters and metabolic subtypes, Kaplan-Meier curves and log-rank tests were used. The endpoint of the study was OS. OS was defined as the interval between the date of diagnosis and the date of patient death or the last follow-up. DFS was defined as the interval between the date of surgery and the date of disease recurrence or death for any reason or the last follow-up (whichever occurs first). We calculated the hazard ratio (HR) for each gene and corrected the log-rank P value with the Benjamini-Hochberg (BH) algorithm using geneSA (v0.1.1, https://github.com/huynguyen250896/geneSA) [28].
2.6 Metabolic pathway variation analysis of the three metabolic subtypes
To characterize the unique metabolic function of each metabolic subtype, we performed GSVA and selected c2.cp.kegg.v7.4.Symbols.gmt in MsigDB as the gene set together with the gene set composed of metabolic genes in Human-GEMs. To determine the pathway differences between subtypes, we used the R package Limma (v3.46.0, https://bioconductor.org/packages/release/bioc/html/limma.html) [29] to analyze the upregulated and downregulated pathways of each subtype [P < 0.001, corrected with BH algorithm; log2fold change (log2FC) > 0, upregulated; log2FC < 0, downregulated] compared to the other two subtypes.
We also analyzed the differentially expressed proteins of each subtype, which we identified using DeqMS (v1.8.0, https://www.bioconductor.org/packages/release/bioc/html/DEqMS.html) [30] by comparison with the other two subtypes. Proteins with log2FC > 0.5 and P < 0.05 were considered upregulated, and those with log2FC > 1 and P < 0.05 were considered highly upregulated in the subtype.
2.7 Construction of metabolic subtype predictors and metabolic protein signatures of the metabolic subtypes
The proteomic data of this study were used as the training set, and supplemental data from Dong et al. [31] were used as the validation set. We first classified the validation set into three metabolic subtypes using the same metabolic stratification method and aligned the subtype label based on the number of overlapping upregulated proteins with our cohort data. To determine the correspondence between subtypes in the validation set and those in the training set, we compared the number of overlapping differential proteins. In the validation set, we observed that 317 out of the 372 upregulated proteins (log2FC > 1, P < 0.05) identified in the S1 training set exhibited upregulation in one of the subtypes (log2FC > 0, P < 0.05). Consequently, we designated this subtype as the “S1 validation set”. The remaining two subtypes were assigned in the same way, with the S2 validation set overlapping 231 of the 295 upregulated proteins in the S2 training set and the S3 validation set overlapping 547 of the 1,148 upregulated proteins in the S3 training set. The identification of differentially expressed proteins in the validation subtypes was performed using limma, with the same comparison method being used in the training set.
We then constructed metabolic subtype predictors and performed molecular screening of the metabolic signature proteins for each metabolic subtype based on the permutation-based feature importance test (PermFIT) method (https://github.com/SkadiEye/deepTL) [32]. Before predictor construction, we performed feature selection for each subtype, screening for highly upregulated metabolic genes in each subtype in our cohort. Here, metabolic genes were genes in c2.cp.kegg.v7.4.symbols.gmt and genes in the Human-GEMs (version 1.8.0). For each subtype, we screened subtype-specific metabolic signatures using PermFIT-support vector machines (SVMs), PermFIT-deep neural networks (DNNs), and PermFIT-random forests (RFs) and validated the predictive ability of signatures in the validation set. To improve the predictive ability, we replaced one signature based on permutation screening with one prognosis-related metabolic protein for each subtype. For the S2 and S3 training sets with multiple highly upregulated proteins with prognostic implications, the metabolic protein with the highest hazard ratio (HR) was selected for S2, and the metabolic protein with the lowest HR was selected for S3. We finally used the set of metabolic protein signatures with the best accuracy, as well as the area under the curve (AUC) evaluation in the validation set for each subtype.
2.8 Rate-limiting enzyme annotation
A union set of 3,745 metabolic genes was extracted from the Human-GEMs and the KEGG database. Then, a total of 2,728 metabolic genes identified in our cohort remained for further analysis. Based on the gene name and the keyword “rate-limiting”, the abstracts and titles of relevant literature were retrieved and collated from PubMed. Here, we only considered metabolic proteins upregulated in each metabolic subtype such that a total of 293 proteins had retrieval results. After the manual check, we eventually screened a total of 112 upregulated metabolic rate-limiting enzymes in the three subtypes (log2FC > 1, P < 0.05). We classified them according to the KEGG pathway annotations. For glycolysis and the tricarboxylic acid cycle (TCA) pathway, we only focused on the three commonly considered essential rate-limiting enzymes and relaxed log2FC to > 0.5.
2.9 The prognostic gene-related pathway
Metabolic genes (HR > 1, log-rank P < 0.01) annotated in the Human-GEMs or KEGG database were considered high-risk metabolic genes significantly associated with prognosis; 62 of these genes were identified, and 53 low-risk metabolic genes (HR < 1, log-rank P < 0.001) were significantly associated with prognosis. The genes were submitted as a gene list to Metascape (https://metascape.org/gp/index.html) [33] for pathway enrichment analysis. The ten KEGG pathways with the lowest q value are shown.
2.10 Metabolic network construction and analysis
We used Human-GEMs [26] as a reference metabolic network model consisting of 142 metabolic subsystems, 3,626 genes, 8,379 metabolites of different cellular sublocalizations, and 13,084 metabolic reactions. Proteomic and transcriptomic individual metabolic networks were extracted using the task-driven integrative network inference for tissues algorithm (tINIT) [26, 34]. For transcriptomic data, we followed the tutorial at the website https://sysbiochalmers.github.io/Human-GEM-guide/gem_extraction_old_tINIT/. For proteomic metabolic networks, we used the iBAQ values for proteins identified in at least 25% of samples and set discrete values for the protein abundance of each sample according to the following rule: if there was a missing value, we defined it as “none”; if its rounded percentile was in 0-24%, 25%-74%, or 75%-100%, we defined its abundance grade as “low”, “medium”, or “high”, respectively.
We assessed metabolic reactions across all patient subsystems using the Kruskal-Wallis H-test to compare overall differences and the Nemenyi test to compare differences between every two subtypes. BH significance was defined as a corrected P < 0.05. For metabolic task analysis of metabolic networks, we used the CompareMultiModels function in Raven (v2.6.0, https://github.com/SysBioChalmers/RAVEN) [35] and included metabolic tasks to assess whether the task could be passed in an individually specific metabolic network. The enriched tasks in each metabolic subtype were calculated using Fisher's exact test [26, 36].
We converted the proteomic metabolic network from the raven model format to the Cobra (v3.1, http://opencobra.github.io/) model format [37]. We used the sample CbModel function and the artificially centered hit-and-run (ACHR) sampling method to obtain the sampling metabolic fluxes and then took the average value for each reaction. We retained reactions with a non-zero flux value in at least 25% of samples, filled the missing reaction flux value with 0, merged the reactions whose flux similarity was above 0.95 (Spearman correlation coefficients), and merged the reaction sets with the overlapping reactions. The flux values of the merged reaction sets were averaged.
If the d value was less than 0, it indicated the opposite direction to that defined by the model. When screening for upregulated proteins that also had high flux, we replaced condition (iv) with a ratio of the mean metabolic flux of the S2 subtypes to the mean of non-S2 subtypes that was greater than 1, and the corresponding protein was expressed at log2FC > 0.5 and P < 0.05.
2.11 Cell annotation and metabolic heterogeneity analysis of single-cell RNA-sequencing (scRNA-seq) datasets
We used HRA00863 [10] and GSE151530 [38], two scRNA-seq datasets of iCCA. The annotated cells in the HRA000863 dataset were kindly provided by the authors, and GSE151530 was reannotated using SingleR (v1.4.1, https://bioconductor.org/packages/release/bioc/html/SingleR.html) [39]. To obtain an accurate classification of CD8+ T cells in iCCA, we used previously reported markers of CD8+ T cell subtypes [40, 41] and the single-cell cluster-based auto annotation toolkit (scCATCH, v3.2.1, https://github.com/ZJUFanLab/scCATCH) [42] for annotation. We classified CD8+ T cells into exhausted (including precursor exhausted) CD8+ T cells, naive-like CD8+ T cells, early activated CD8+ T cells, and effector memory CD8+ T cells.
Based on a previous study of single-cell pathway activity [43], we performed pathway activity calculations for CD8+ T-cell subtypes using the Pagoda2 (v1.0.10, https://github.com/kharchenkolab/pagoda2) algorithm [44], with a gene set that was previously selected from a metabolic subset of 69 metabolic pathways from c2.cp.kegg.v7.4.symbols.gmt in MsigDB. To compare the differences between subtypes, we used the Kruskal-Wallis H-test to compare the overall differences and the Nemenyi test to compare the differences between every two groups with a significance level of P < 0.05.
2.12 Tumor microenvironment analysis
We performed microenvironment composition analysis using transcriptomic transcripts per million (TPM) data from 116 patients with iCCA and data from Dong's cohort [31]. A web version of xCell (https://comphealth.ucsf.edu/app/xcell) was used in this analysis [45].
In parallel, we used the deconvolutional R package BayesPrism (v2.0, https://github.com/Danko-Lab/BayesPrism) [46] and HRA000863 scRNA-seq datasets to deconvolute the transcriptomic count data of our cohort and transcriptomic TPM data of Dong's cohort [31]. Thirteen cell fractions were deconvoluted: tumor cells, B cells, plasma cells, natural killer cells, muco-associated constant T cells, T regulatory cells, CD4+ T cells, CD8+ T cells, dendritic cells, monocytes, macrophages, endothelial cells, and fibroblasts. Cell fractions for four subtypes of CD8+ T cells were also deconvoluted: exhausted CD8+ T cells, naive-like CD8+ T cells, early activated CD8+ T cells, and effector memory CD8+ T cells. Tumor transcriptome expression profiles were obtained after deconvolution.
We calculated overall significant differences using the Kruskal-Wallis H-test, and we assessed the differences between every two groups using the Nemenyi test with a significance level of P < 0.05. We normalized the cell fraction with the sum of the four CD8+ cell subtypes when analyzing the composition differences of the four CD8+ T-cell subtypes in the metabolic subtypes.
The TME-based subtypes [47] were also generated for the comparison with the results of BayesPrism and xCell. Briefly, 500 differential genes of five TME-based subtypes [47] (100 for each) and NTP algorithm [48] were used to decide the TME-based subtypes for our data.
2.13 Correlation between metabolic pathway activity, metabolic genes, and cell composition in the microenvironment
We used 69 metabolic pathways from the c2.cp.kegg.v7.4.symbols.gmt gene set in MSigDB and used GSVA pathway enrichment scores as pathway activity. We calculated three levels of metabolic pathway activity: the proteomic and transcriptomic levels, and the transcriptomic expression level of tumor cells after BayesPrism deconvolution.
We also analyzed correlations at the metabolic gene level, selecting metabolic immune checkpoints, as well as metabolic genes with consistent prognosis in our dataset and in Dong's cohort data, and again, we calculated three levels of correlation.
To obtain robust correlation results, we referred to previous research methods [49] and set the following criteria for the robust correlation: (1) P < 0.05 for 2,000 permutation tests; (2) P < 0.05 for 2,000 permutation tests after removing any one sample.
We assessed the consistency of metabolic pathway correlations with the cell composition between our dataset and Dong's dataset: 170, 377, and 566 correlations in our cohort were robust using the proteomics, transcriptomics and deconvoluted tumor transriptomic data, respectively, with 74 (43.5%), 219 (58.1%), and 348 (61.5%) also being robust in Dong's cohort.
At the levels of metabolic immune checkpoints, for the proteome, the transcriptome, and the deconvoluted tumor transcriptome, 51, 98, and 105 correlations were robust in our cohort, respectively, of which 33 (64.7%), 63 (64.2%), and 73 (69.5%) were also robust in Dong's cohort.
At the level of metabolic prognosis-related genes, for the proteome, the transcriptome, and the deconvoluted tumor transcriptome, 117, 121 and 165 correlations were robust in our cohort, respectively, of which 58 (49.6%), 66 (54.5%), and 108 (65.5%) were also robust in Dong's cohort.
2.14 Analysis of driver genes and potential functional mechanisms
We used sysSVM2 (https://github.com/ciccalab/sysSVM2) [50] to analyze driver mutations at the individual level in iCCA, selecting 10 candidate driver genes per patient for driver mutation enrichment analysis of metabolic subtypes. In S3, the numbers of patients with BAP1 mutations and IDH1 mutations were 16 and 12, respectively, and a total of 24 patients out of 49 patients. Differentially expressed proteins were identified in patients of S3 with BAP1 mutations (S3_BAP1mut), IDH1 mutations (S3_IDH1mut), and no BAP1 or IDH1 mutations (S3_other) by comparing the protein abundance profiles of these patients to those of patients in non-S3 subtypes (log2FC > 1, P < 0.05 after BH correction). Metascape was used to assess the functions of S3_BAP1mut, S3_IDH1mut, and S3_other.
2.15 Analysis of cis-regulatory effects
We identified significantly amplified or deleted genes using GISTIC2 (2.0.23) with parameters conf set at 0.99, ta and td set at 0.4, and brlen set at 0.7 [51]. We used iProfun (v0.2.0, https://github.com/songxiaoyu/iProFun) [52] to screen potential cis-regulations, with age, sex and hepatitis B surface antigen as covariates. The copy number variation (CNV), proteomic and transcriptomic expression matrix after z score normalization were used as inputs, the permutation number was set to 1,000, and the FDR was set to 0.1. We then selected proteins in significant deletion and amplification regions with cis-regulation of CNV-mRNA-protein. We performed functional enrichment analysis using Metascape for proteins in the two regions.
2.16 Cell lines and cell culture
Human iCCA cell lines RBE and HuCCT1 were maintained in RPMI-1640 (BasalMedia, Shanghai, China) supplemented with 10% fetal bovine serum (FBS, 10099141C, Gibco, Waltham, MA, USA), 100 U/mL penicillin and streptomycin (100×, 15140122, Gibco) at 37°C with 5% CO2. All cells were used within ten passages after thawing. The cell lines were authenticated by short tandem repeat profiling before the experiments started and showed no mycoplasma contamination.
2.17 Lentiviral transfection
Specific diacylglycerol kinase α (DGKA) lentiviral short hairpin RNAs (shRNAs) were synthesized by OBiO Technology (Shanghai, China). The shRNAs were cloned and inserted into the GL401 (pCLenti-U6 -CMV-Puro-WPRE) vector, and an empty vector was used as the negative control. RBE and HuCCT1 cells were transfected following the manufacturer's instructions to knock down the expression of DGKA.
2.18 5-Ethynyl-2’-deoxyuridine (EdU) assay
After transfection, iCCA cells were seeded on 12-well plates at a density of 3 × 105 cells/well. The EdU-594 Kit (C0078S, Beyotime, Shanghai, China) was used to detect the proliferation of the indicated iCCA cells. Cells were treated with EdU and Hoechst dyes. A fluorescence microscope (Olympus, Tokyo, Japan) was utilized to observe and record the experimental results.
2.19 Cell proliferation analysis
For cell viability assays, iCCA cells were digested and seeded in 96-well plates (1,000 cells/well) in 5 replicates. The next day, 30 μmol/L ritanserin (HY-10791, MedChemExpress, Monmouth Junction, NJ, USA), 100 μmol/L phosphatidic acid (PA, 840857, Merck, Darmstadt, Germany), or both were added to the culture medium of each treatment group. Then, the cells were cultured in the respective medium for another 96 h. Cell survival was calculated using cell counting kit-8 (CCK8, CK04, Donjindo, Kumamoto, Japan) by measuring the absorbance at 450 nm using FlexStation 3 (Molecular Devices, San Jose, CA, USA).
2.20 Colony formation assay
For colony formation assays, cells were seeded in 6-well plates (1,000 cells/well) in triplicate and incubated for approximately 2 weeks. In the ritanserin group, cells were treated with 30 μmol/L ritanserin, and an equal volume of dimethyl sulfoxide (DMSO, 41640, Sigma-Aldrich, St Louis, MO, USA) was added to the control group. Then, the cells were fixed with 4% paraformaldehyde and dyed with Giemsa stain. The number of colonies in each well was counted by ImageJ software (National Institutes of Health, Bethesda, MD, USA).
2.21 Tissue microarray (TMA) experiment
A TMA containing 90 iCCA samples was established. After ruling out 2 samples with missing data, 88 samples were included in the present study (Supplementary Table S3). Immunohistochemical (IHC) staining was conducted with IHC reagents from Fuzhou Maixin Biotech Co., Ltd. (Fuzhou, Fujian, China). Briefly, TMA was deparaffinized and rehydrated before antigen retrieval, followed by blocking with 3% hydrogen peroxide and 1× Animal-Free Blocking Solution. Then, the sections were incubated with primary antibodies (DGKA, 11547-1-AP, Proteintech Group, Rosemont, IL, USA) overnight at 4°C. The next day, the sections were incubated with the secondary antibody at 37°C for 1 h, and 3,3 N-diaminobenzidine tetrahydrochloride (DAB) staining was conducted. Finally, they were counterstained with hematoxylin for 2 min. Using the PANNORAMIC 1000 slice scanner (3DHISTECH Ltd., Budapest, Hungary), the tissue sections were scanned and imaged. Quant Center 2.1 analysis software (3DHISTECH Ltd.) was used to calculate the H-score of the target area. H-score = ∑(PI × I) = (percentage of cells of weak intensity × 1) + (percentage of cells of moderate intensity × 2) + (percentage of cells of strong intensity × 3), where PI represents the positive area ratio and I represents the staining intensity. The median H-score was defined as the threshold value to distinguish the low and high DGKA expression groups.
2.22 Western blotting
Using radioimmunoprecipitation assay buffer (RIPA) lysis buffer solution (89900, Thermo Fisher Scientific) containing phosphatase and protease inhibitor cocktail, cell lines were dissolved to extract protein. Protein samples were separated by polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride (PVDF) membranes (IPVH00010, Sigma-Aldrich). After blocking with 5% bovine serum albumin (BSA) solution, the PVDF membrane was incubated with primary antibodies, including DGKA, extracellular regulated protein kinases 1/2 (ERK1/2, T40071, Abmart, Shanghai, China), phospho-ERK1 (T202/Y204) + ERK2 (T185/Y187) (T40072, Abmart), mitogen-activated protein kinase 1/2 (MEK1/2, 11049-1-AP, Proteintech Group), phospho-MEK1/2 (Ser217/221, 9154, Cell Signaling Technology, Danvers, MA, USA), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, 51332, Cell Signaling Technology), followed by incubation with secondary antibodies (7074, 7076, Cell Signaling Technology) conjugated with horseradish peroxidase (HRP). Enhanced chemiluminescence (ECL) Western blotting substrate (WBKLS0500, Sigma-Aldrich) was used to visualize the signal.
2.23 Bulk RNA-seq analysis for cell lines
TRIzol reagent (15596026, Thermo Fisher Scientific) was used to extract total RNA from DGKA-knockdown iCCA cells and control cells. After ensuring the purity and integrity of all RNA samples, the RNA library was then constructed. Sequencing was performed on the Illumina PE150. We defined significance for downstream analysis as fold change > 1 and P < 0.05. Functional enrichment analysis of KEGG was conducted using ClusterProfiler (v3.4.4, https://github.com/YuLab-SMU/clusterProfiler). Gene set enrichment analysis (GSEA) was carried out with GSEA software (v4.0.3, Broad Institute, Cambridge, MA, USA).
2.24 Statistical methods
The chi-square test and Fisher's exact test were used to analyze categorical variables. Kruskal-Wallis's H-test and Student's t-test were utilized to calculate statistical differences for continuous variables, and Spearman and Pearson's correlations were used for continuous variables versus continuous variables. Survival analysis was performed using the Kaplan-Meier method, the log-rank test, and Cox proportional hazards regression analysis. All statistical tests were two-sided, and a P value < 0.05 was considered statistically significant.
3 RESULTS
3.1 Metabolic subtypes and protein signatures of iCCA
In our study, we enrolled 116 patients with iCCA and performed WES and quantification of the transcriptome and proteome (Figure 1A). We attempted several approaches to identify the metabolic subtypes in our cohort of iCCA patients. Based on the NMF method and the protein abundance of metabolic genes in Human-GEMs, three metabolic subtypes (S1, S2, and S3) were identified (Figure 1B, Supplementary Table S1). We further explored the relationship between these three subtypes and potential clinical influencing factors. Notably, HBV infection and diabetes were not significantly associated with any subtype, while obese patients tended to be subtype S1 (Supplementary Table S2).
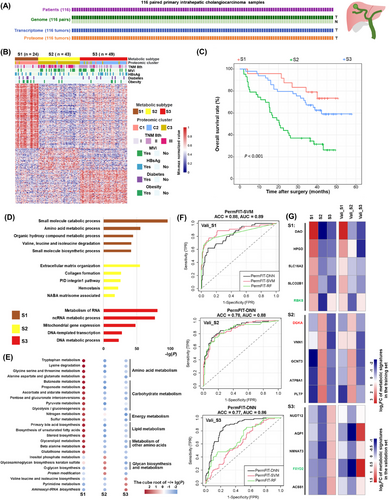
Metabolic subtyping of iCCA. (A) Multiomics sequencing for the enrolled cohort of the iCCA. (B) Metabolic subtypes of iCCA. The heatmap showed the value of subtype-specific metabolic protein abundance, normalized by row min-max. The abbreviations of each metabolic subtype and the number of patients for each metabolic subtype were shown above the pheatmap. (C) Kaplan-Meier plot of OS for the patients in three metabolic subtypes. (D) Pathway and process enrichment analysis using Metascape for each subtype. The top five pathways or GO Biological Processes ordered by the -lg(P) were selected. (E) Gene Set Variation Analysis of KEGG metabolic pathways or Human-GEMs subsystems for three metabolic subtypes. The enrichment degree of each pathway is represented by the cube root of the lg(P). When log2FC < 0, the enrichment degree is represented by the negative cube root of the lg(P). The italicized terms are the subsystems in the human-GEMs, while the normal terms are the metabolic pathways in KEGG. (F) ROC curves of the predictors of metabolic subtypes on the validation set. The black curve represents PermFIT-DNN, the green curve represents PermFIT-RF, and the red curve represents PermFIT-SVM. (G) The log2FC value of metabolic protein signatures screened by predictors for each metabolic subtype. The gene name in red color means this gene is high-risk, the gene name in green color means this gene is associated with a good prognosis.
Abbreviations: iCCA, intrahepatic cholangiocarcinoma; T, tumor; N, adjacent liver tissues; C1, C2 and C3, proteomic cluster 1,2,3; S1, S2 and S3, metabolic subtype 1,2,3 in our training set; Vali_S1, Vali_S2, Vali_S3, the metabolic subtypes in the validation set that are similar to the metabolic phenotypes of the S1, S2, S3 subtypes in the training set, respectively; OS, overall survival; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; GEMs, genome-scale metabolic models; FC, fold change; ACC, accuracy; AUC, area under the curve; ROC, receiver operating characteristic curve; Perm-FIT, Permutation-FIT; DNN, deep neural networks; RF, random forests; SVM, support vector machines.
Out of the three metabolic subtypes, individuals belonging to subtype S1 exhibited the highest survival rate (median OS > 60 months), whereas those in subtype S2 suffered from the shortest duration of survival (median OS = 20.5 months) (Figure 1C). General proteomic clusters were also identified for comparison with the metabolic subtypes (Supplementary Table S1). Interestingly, there was a strong concordance between them, indicating a good correspondence between individual changes in the metabolic protein expression profile and the overall protein expression profile (Supplementary Figure S2A). We compared the differences in proteins used in general proteomic subtyping and metabolic subtyping, both of which resulted in three subtypes. A total of 412 proteins were shared. We used these 412 proteins alone for metabolic subtyping again. Its consistency with the Human-GEMs gene-based method was weaker than that with the KEGG metabolic gene-based method but stronger than that with the GSVA-based method (Supplementary Figure S2B). Other metabolic subtyping results also showed significant differences in OS (Supplementary Figure S2C).
To further explore the molecule-level functional characteristics of each subtype, we submitted the highly upregulated genes in each subtype to Metascape [33] for pathway and biological process analysis (Supplementary Table S4). The top 5 most enriched pathways for each subtype are shown in Figure 1D. The results revealed that metabolism-related pathways, such as small molecule biosynthesis and valine, leucine and isoleucine degradation, were enriched in S1, whereas S2 showed pathways predominantly related to metastasis, and S3-enriched pathways were predominantly related to proliferation. This finding was consistent with the different prognostic characteristics of each subtype.
We also used GSVA to perform a preliminary characterization of the metabolic functional differences among the three metabolic subtypes S1, S2 and S3. Consistent with the above results, the subtype S1 exhibited a wider range of metabolic activities compared to the other two subtypes. The proteins in many metabolic pathways and subsystems of amino acid metabolism, carbohydrate metabolism, energy metabolism, lipid metabolism, and other classes of amino acid metabolism were enriched in S1 (Figure 1E). The pathways of glycosaminoglycan biosynthesis-keratan sulfate, O-glycan biosynthesis, protein modification subsystem, and inositol phosphate metabolism were enriched in S2. Glycosaminoglycan biosynthesis-keratan sulfate [53], O-glycan biosynthesis [54] and inositol phosphate metabolism [55] were reported to be associated with tumor metastasis, which led to the worst prognostic phenotype of S2. The biosynthesis pathways of valine, leucine, isoleucine, and aminoacyl-tRNA and the pyrimidine metabolism pathway were enriched in S3 (Figure 1E), consistent with the vigorous proliferation phenotype mentioned above (Figure 1D).
For potential applications in the future, we constructed a metabolic subtype prediction model using the Permutation-FIT method [32] to screen for the most representative metabolic signature proteins for each metabolic subtype. We used our cohort data as the training set and the proteomic data from the Dong cohort [31] as the validation set (Supplementary Figure S2D-E). The AUC of all three subtypes in our validation set reached above 0.85, indicating the reliability of the model and the representativeness of the selected signatures for metabolic subtypes. The accuracy of the predictor reached 0.88 for S1 and 0.77 for S3 (Figure 1F). The metabolic protein signatures in S1 and S2 were involved in a variety of metabolic functions, and NUDT12, ACSS1, and NMNAT3 signatures in S3 were all associated with the process of nucleotide biosynthesis (Figure 1G).
3.2 Significant and wide metabolic heterogeneity among different metabolic subtypes
To explore the heterogeneity of metabolic characteristics in different metabolic subtypes, three aspects, including “rate-limiting enzymes”, “prognosis-related metabolic genes” and “metabolic network model”, were involved in further analysis. Although the correlations between proteomic and transcriptomic expression of metabolic genes were higher than those of non-metabolic genes, the median Spearman coefficient was only 0.49, indicating that it is of unique value to use proteomic data to characterize metabolic subtypes and their heterogeneity since there was a closer relationship between proteome and phenotype (Supplementary Figure S3A).
First, we annotated metabolic rate-limiting enzymes using differential protein expression in metabolic subtypes by collecting and reviewing the related literature (Supplementary Table S5). We found that the rate-limiting enzymes involved in the fatty acid degradation pathway in S1 were upregulated, while the rate-limiting enzymes upregulated in S2 and S3 were proteins involved in the glycolysis/gluconeogenesis pathway. Hexokinase 1(HK1), HK2, and HK3, the enzymes for converting glucose to 6-phosphate fructose in the first rate-limiting glycolysis step, were upregulated in S2, while muscle phosphofructokinase (PFKM) and PFKP, the enzymes for fructose 1,6-diphosphate production in the rate-limiting step of glycolysis, were upregulated in S3. Subtype-specific variation in rate-limiting enzymes suggests the key heterogeneity in energy utilization for iCCA.
All rate-limiting enzymes in the pyrimidine metabolism pathway were upregulated in S3, except for ecto-5’-nucleotidase (NT5E) and ectonucleoside triphosphate diphosphohydrolase 1(ENTPD1), which were upregulated in S2 (Figure 2A). CD39 (ENTPD1) binds extracellular adenosine triphosphate (ATP) and hydrolyses it to adenosine monophosphate (AMP) [56]; another extracellular nuclease, CD73 (NT5E), hydrolyses AMP to adenosine, and the abnormally high concentration of extracellular adenosine promotes tumor proliferation through various immunosuppressive mechanisms; furthermore, inhibition of CD73 and CD39 can enhance antitumor immunity [56]. Since the S2 subtype had the worst prognosis, we further focused on other specific rate-limiting enzymes of this subtype. The rate-limiting enzymes highly upregulated in S2, such as proline dehydrogenase (PRODH), glutaminase 2 (GLS2), and hephaestin (HEPH), were involved in oxidative stress, while arachidonate 5-Lipoxygenase (ALOX5), phospholipase A2, group IVA (PLA2G4A), and cytosolic non-specific dipeptidase 1 (CNDP1) were involved in inflammatory responses. Additionally, CNDP1 and glutamine-fructose-6-phosphate aminotransferase 2 (GFPT2) were involved in the regulation of protein glycosylation (Supplementary Figure S3B).
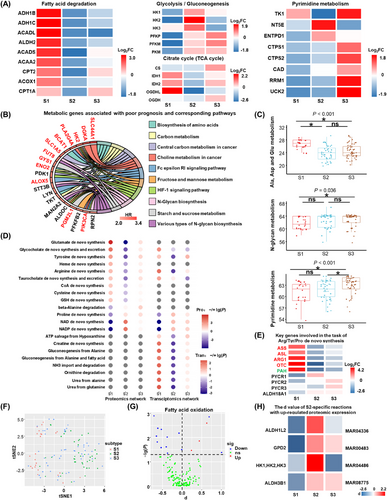
Metabolic feature of metabolic subtypes in iCCA. (A) The log2FC value of key metabolic rate-limiting enzymes in iCCA. (B) Metabolic genes associated with the poor prognosis and the corresponding pathways in intrahepatic cholangiocarcinoma. The proteins in red color were up-regulated in S2. (C) The number of reactions in subsystems for each subtype. The Kruskal-Wallis H-test was used to compare overall differences. Statistical significance between every two groups was calculated using the Nemenyi test. P values shown at the top of each panel were calculated by the Kruskal-Wallis H-test. For the Nemenyi test results, a ‘*’ represents P < 0.05, while ‘ns’ means P > 0.05. (D) The enrichment analysis of metabolic tasks derived from proteomics network and the transcriptomics network for subtypes. Over_representation and under_representation are represented by -lg(P) and lg(P), respectively. (E) The log2FC value for the key gene expression at the proteomics level for Arginine/Tyrosine/Proline de novo synthesis metabolic task. Genes colored in red, green and black represent the genes involved in the Arginine/Tyrosine/Proline de novo synthesis, respectively. (F) The dimensionality reduction map of the sampled flux for the reactions of 116 personal GEMs using tSNE. (G) The d value of reactions in Fatty acid oxidation in S2. d value is the normalized difference of fluxes between subtypes. (H) Reactions in S2 with the up-regulated both in metabolic flux and the corresponding genes at the proteomics level. MAR04336: 10-formyl-THF[c] + H2O[c] + NADP+[c] = > CO2[c] + H+[c] + NADPH[c] + THF[c], MAR00483: sn-glycerol-3-phosphate[c] + ubiquinone[m] = > DHAP[c] + ubiquinol[m], MAR04486: glucose[c] + ITP[c] = > glucose-6-phosphate[c] + H+[c] + IDP[c], MAR08775: glycolaldehyde[c] + H2O[c] + NAD+[c] = > glycolate[c] + 2H+[c] + NADH[c]. MAR00483, MAR04486 and MAR08775 belong to folate metabolism, glycerophospholipid metabolism, purine metabolism, purine metabolism and Tricarboxylic acid cycle and glyoxylate/dicarboxylate metabolism, respectively.
Abbreviations: iCCA, intrahepatic cholangiocarcinoma; S1, S2 and S3, metabolic subtype 1, 2 and 3; FC, fold change; TCA, tricarboxylic acid; HR, hazard ratio; Ala, alanine; Asp, aspartic acid; Glu, glutamic acid; Pro, proteomics network; Tran, Transcriptomics network; Arg/Tyr/Pro, Arginine/Tyrosine/Proline; GEMs, genome-scale metabolic models; tSNE, t distributed stochastic neighbor embedding; THF, tetrahydrofolate; NADP, nicotinamide adenine dinucleotide phosphate; ns, no significant; *, P < 0.05; sig, significant.
Second, we analyzed the metabolic genes associated with different prognostic phenotypes (Figure 2B and Supplementary Figure S3C). Pathway enrichment results showed that the metabolic pathways associated with poor prognosis in iCCA were central carbon metabolism in cancer, choline metabolism in cancer, various types of N-glycan biosynthesis, etc. These metabolic genes were also widely involved in important signaling pathways, including the hypoxia-inducible factor-1 (HIF-1) signaling pathway and the Fc epsilon receptor I (FcεRI) signaling pathway. Notably, among the 20 metabolic genes associated with unfavorable prognosis, 12 genes were upregulated in S2 (Figure 2B).
Third, to further analyze the metabolic heterogeneity of iCCA from a global view, we constructed an individual-level tissue-specific metabolic network based on the tINIT algorithm. We compared the differences in the number of reactions in the metabolic subsystems within S1, S2, and S3. There were 136 subsystems in the individual-specific proteomics metabolic network of iCCA, and 48 metabolic subsystems had subtype differences (Supplementary Table S6). Subsystems generally had a higher number of metabolic reactions in S1 and a lower number of metabolic reactions in S2. The heterogeneous subsystems among subtypes revealed by the reaction numbers, such as alanine, aspartate, glutamate, and pyrimidine metabolism, were consistent with previous GSVA results, but there were also inconsistencies in that the number of N-glycoside synthesis metabolic reactions in S2 was not significantly higher than that in other subtypes (Figure 2C).
The metabolic task is a set of reactions needed to convert a specific metabolite into a specific product. The differences in metabolic tasks between patients at the proteomic level were greater than those based on the transcriptome. A total of 256 metabolic tasks were involved, of which 67 were different across patients at the proteome level, 27 were different at the transcriptome level, and 25 were identified as different metabolic tasks in both the transcriptome and proteome (Supplementary Table S7). A total of 21 metabolic tasks were found to have metabolic intersubtype differences at two omics levels, and these tasks were widely related to the metabolism processes of amino acids, sugars, energy lipids, coenzymes and vitamins, and nucleic acids (Figure 2D).
Multiple metabolic tasks in S1 showed significant enrichment, while most of the tasks were extensively depleted in S2. The proteomics and transcriptomics together showed that glycocholate and taurocholic acid de novo synthesis and secretion were significantly enriched in S1; conversely, proline de novo synthesis, NAD(+) de novo synthesis, and NADP(+) de novo synthesis were significantly deficient in S1. Arginine and tyrosine de novo metabolic tasks were absent in S2 (Figure 2D). Proteomic expression evidence verified that arginine and tyrosine de novo synthesis were significantly depleted in S2, as well as proline de novo synthesis in S1 (Figure 2E); in addition, some genes shared by NAD(+) de novo synthesis reactions had lower expression in S1 than in other subtypes (Supplementary Figure S3D). Additionally, the proteomic data showed that the NAD(+) and NADP(+) de novo synthesis tasks were enriched in S2, while the alanine degradation task was enriched in S3 and absent in S1 (Figure 2D). Metabolic tasks enriched or depleted in different subtypes indicated metabolic reprogramming in the formation of subtypes of iCCA. Depleted metabolic tasks in S1, as well as enriched metabolic tasks in S2 or S3, may be associated with poor prognosis and may be potential metabolic targets.
Metabolic fluxes are the immediate rates at which chemical reactions occur, and metabolic fluxes reflect the characteristics of individual reactions from a global metabolic perspective. We simulated the fluxes of metabolic reactions in the metabolic network based on the ACHR method. The simulated metabolic flux data were dimensionally reduced and were found to differ between S1 and S2 (Figure 2F). S2 generally had smaller metabolic fluxes, such as fatty acid oxidation, than other subtypes (Figure 2G). We found that some reactions in prostaglandin metabolism and synthesis and interleukin metabolism in S2 had higher fluxes than other subtypes (Supplementary Figure S3E). Proteins involved in S2 with significantly high fluxes were also upregulated in S2, such as lecithin-cholesterol acyltransferase (LCAT) and solute carrier family 3 member 2 (SLC3A2) (Supplementary Figure S3E). HK1, HK2, HK3, glycerol-3-phosphate dehydrogenase 2 (GPD2), aldehyde dehydrogenase 1 family member L2 (ALDH1L2), and ALDH3B1 were also found to be upregulated and were found to be involved in the high flux reaction (Figure 2H), but most of the upregulation of the metabolic proteins in S2 was not reflected in the simulated fluxes (125 upregulated metabolic proteins in S2). Thus, the sampling fluxes based on the individual metabolic model indicated that from a global metabolic perspective, except for a small portion of upregulated proteins involved in highly active metabolic reactions, the activity of the entire S2 metabolic reaction was significantly lower than the other two subtypes.
3.3 Analysis of microenvironmental composition and metabolic associations in iCCA
The analysis of the microenvironmental composition of different subtypes showed obvious heterogeneity in cell type fractions. Based on the Kruskal-Wallis H-test, we found that the immune and stromal scores (xCell) of the three metabolic subtypes in S3 were significantly lower than those of the other two subtypes (Supplementary Table S8). BayesPrism results supported the findings based on the xCell method as follows: BayesPrism analysis showed that the tumor cell fraction of S3 was significantly higher than that of the other two subtypes (Nemenyi test, P < 0.05); the results of both methods consistently showed that monocytes and macrophages made up significantly smaller proportions in S3 than in the other subtypes (Nemenyi test, P < 0.05); BayesPrism results showed that the proportions of CD8+ T cells, dendritic cell (DC), and regulatory T (Treg) cells were significantly higher in S1 than in the other subtypes (Nemenyi test P < 0.05). The characteristics of TME-based subtypes [47] were consistent with our findings: the “immune classical” subtype was enriched in S1 (P = 0.003) and the non-inflamed TME-based “Hepatic stem-like” subtype was enriched in S3 (P < 0.001) (Supplementary Table S9).
Due to the importance of CD8+ T cells for iCCA [57, 58], we analyzed the heterogeneity of CD8+ T cell subtypes among different patients using the public iCCA single-cell dataset, as well as the BayesPrism algorithm. We first classified CD8+ T cells from the public single-cell dataset into the following four subtypes: exhausted CD8+ T cells, naive-like CD8+ T cells, early activated CD8+ T cells, and effector memory CD8+ T cells (Supplementary Figure S4A). Both scRNA-seq datasets consistently showed metabolic heterogeneity in the pathways of glycolysis/gluconeogenesis, pyrimidine metabolism, one carbon pool by folate, cysteine and methionine metabolism, and oxidative phosphorylation in the CD8+ T-cell subtypes. We found significant differences in all of these pathways between effector memory CD8+ T cells and exhausted CD8+ T cells. Except for the higher level of oxidative phosphorylation in the effector memory CD8+ T cells, the activity of all other metabolic pathways in effector memory CD8+ T cells was lower than that in exhausted CD8+ T cells (Figure 3A and Supplementary Figure S4B).
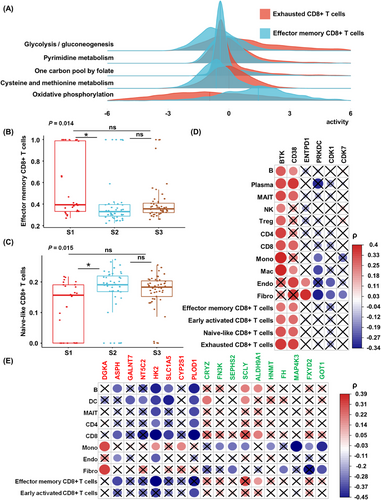
Basic immune characteristics of iCCA. (A) Pathway activities of four CD8+ T cells subtypes in HRA000863 datasets. (B-C) the comparison of cell fractions of effector memory CD8+ T cells and naive-like CD8+ T cells among three metabolic subtypes. Kruskal-Wallis H-test was used to compare the overall significant difference and the Nemenyi test was used to compare the significant difference between every two groups. P values are shown for the Kruskal-Wallis H-test. For the Nemenyi test, ‘*’ and ‘ns’ represent P < 0.05 and P > 0.05, respectively. (D-E) The Spearman correlation between metabolic checkpoints (D) or prognostic metabolic genes (E) and the cell composition in the microenvironment. The gene name in red color means this gene is high-risk, the gene name in green color means this gene is associated with a good prognosis. The crosses indicate that the correlation is not robust in our and Dong's dataset or the correlations in the two datasets are inverse.
Abbreviations: iCCA, intrahepatic cholangiocarcinoma; ns, no significant; B, B cells; Plasma, plasma cells; MAIT, Mucosal-associated invariant T cells; NK, natural killer cells; Treg, T regulatory cells; CD4, CD4+ T cells; CD8, CD8+ T cells; Mono, Monocytes; Mac, Macrophages; DC, dendritic cells; Endo, Endothelial cells; Fibro, Fibroblasts; ρ, the spearman coefficient.
Then, we used BayesPrism to deconvolute the cell fractions of CD8+ T cell subtypes in each patient in the cohort, and the Kruskal-Wallis H-test revealed that the proportion of CD8+ T cells of different subtypes did not differ significantly in patients of different subtypes, but after normalization within CD8+ T cells, effector memory CD8+ T cells made up a significantly higher proportion in S1 than in S2 (Figure 3B) and naive-like CD8+ T cells made up a significantly higher proportion in S2 than in S1 (Figure 3C), indicating the potential trend of internal changes in CD8+ T cell status in the microenvironment of iCCA cells with different metabolic states.
To investigate the association between the microenvironment and metabolism in iCCA, we analyzed the correlation of metabolic pathway activity, represented by metabolic gene expression level, with the non-tumor cell fraction. A total of 49 robust correlations were identified between metabolic pathway activities and non-tumor cell fractions, with endothelial cells associated with the activities of the most metabolic pathways at the proteomic level, followed by CD4+ T cells, CD8+ T cells, and mucosa-associated constant T cells. In contrast, the arachidonic acid metabolic pathway was associated with the greatest proportion of non-tumor cells, followed by the glycosylphosphatidylinositol (GPI)-anchored biosynthetic pathway (Supplementary Figure S4C).
From the single-gene viewpoint, we first analyzed the correlation of 12 metabolic immune checkpoints with the proportion of microenvironmental cells, 6 of which had robust correlations (Figure 3D and Supplementary Table S10). Then, more generally, we analyzed correlations of 24 prognostic metabolic proteins with the fraction of cells in the tumor microenvironment (TME). We identified 42 robust correlations, among which HK2 was associated with the largest proportion of non-tumor cells in 8 cell types, followed by procollagen-lysine, 2-oxoglutarate 5-dioxygenase 1 (PLOD1). Notably, these two proteins were highly upregulated in S2. Effector memory CD8+ T cells were negatively associated with aspartate beta-hydroxylase (ASPH) and HK2 and positively associated with ALDH9A1, and early activated CD8+ T cells were negatively associated with PLOD1. In addition, DGKA was positively correlated with fibroblasts and monocytes (Figure 3E and Supplementary Table S11). We compared the overlap of the transcriptome and the tumor transcriptome after deconvolution with the proteome, and we found that the correlation results of metabolic immune checkpoint and prognostic genes in different omics were more consistent with metabolic pathways (Supplementary Figure S4D-F).
3.4 The potential genetic drivers of metabolic and immune features in iCCA
After profiling the metabolic and immune features of different iCCA subtypes, we sought to explore the potential genetic drivers of the metabolic and immune phenotypic features. To this end, we analyzed the role of driver genes and CNV cis-regulation for each metabolic subtype.
In our cohort, a total of 18,593 somatic mutations were identified, among which ARID1A, titin (TTN), BAP1, TP53, and IDH1 had the highest mutation frequency. The region with significant copy number amplification was 19p13.2, and the region with significant deletion was 1p36.11 (Figure 4A and Supplementary Table S12). The median mutation burden was 0.61/MB, and some mutations were mutually exclusive and cooccurring, e.g., ARID1A mutations co-occurring with mucin 16 (MUC16) and ephrin type-A receptor 2 (EPHA2) mutations (Supplementary Figure S5A-B).
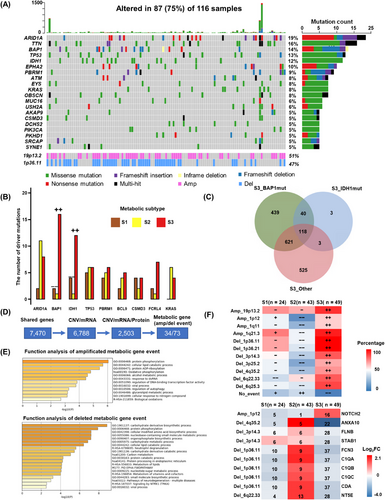
The formation mechanism of metabolic and immune subtype regulation in iCCA. (A) The top 20 high-frequency SNVs and the most amplification and deletion CNV events identified in our cohort. (B) Driver mutation subtype enrichment analysis. (C) The number of up-regulated genes compared between S3_BAP1mut or S3_IDH1mut or S3_Other. (D) CNV in cis-regulatory regions of metabolic genes within significantly deleted and amplified regions. (E) Pathway and process enrichment analysis for amplified or deleted metabolic genes. (F) The percentage of amplified or deleted events in each subtype (upper), The log2FC of genes affected by amplified or deleted events. The number of amplified or deleted events in each subtype was shown (down). ++: significant over-representation, P < 0.05. −: significant under-representation, P < 0.05. +: not significant over-representation, P > 0.05. −: not significant under-representation, P > 0.05. The enrichment analysis was using Fisher's exact test.
Abbreviations: iCCA, intrahepatic cholangiocarcinoma; SNV, single nucleotide variant; CNV, copy number variation; Amp, Amplification; Del, Deletion; S1, S2 and S3, metabolic subtype 1, 2 and 3; S3_BAP1mut, patients in S3 with BAP1 mutations; S3_IDH1mut, patients in S3 with IDH1 mutations; S3_Other, patients in other subtypes with neither BAP1 mutation nor IDH1 mutation; GO, gene ontology; FC, fold change.
Genes with driver mutations in at least 10 individuals were screened for subtype enrichment analysis. The results showed that BAP1 and IDH1 mutations were significantly enriched in S3 (P < 0.001, P = 0.017) and absent in S2 (P = 0.001, P = 0.007), while the remaining driver mutations showed no trend of subtype enrichment (Figure 4B). We divided S3 into S3_BAP1mut, S3_IDH1mut, and S3_Other and compared them with each other to identify differential proteins. In terms of the overlapping upregulated proteins, S3_BAP1mut was more unique than S3_IDH1mut (Figure 4C).
Shared upregulated proteins in S3 exhibited the basic features of S3 subtypes, including histone modification and DNA damage repair. In contrast, the shared proteins in S3_BAP1mut and S3_IDH1mut were involved in biological functions, including bile acid synthesis and stromal adhesion-dependent cell spreading (Supplementary Figure S5C-D).
Next, we analyzed the role of CNVs for each metabolic subtype. A total of 2,503 genes were found to be cis-regulated by iProFun, and 348 genes were mutated at the chromosome arm level; of these genes, 116 were amplified genes, 232 were deleted genes, and 107 were metabolism-related genes, accounting for 30% of the total. Among metabolism-related genes, 34 were amplified, and 73 were deleted (Figure 4D).
The amplification events mainly affected protein phosphorylation, intracellular lipid catabolism, and adenosine diphosphate ribosylation; the deletion events mainly affected carbohydrate derivative biosynthesis processes, protein phosphorylation, cellular modification of amino acid biosynthetic processes, and nucleobase-containing small molecule metabolic processes (Figure 4E).
We found a significant enrichment of chromatin deletion and amplification events in S3 and fewer chromatin deletion and amplification events in S2 (Figure 4F). We screened for genes that were significantly amplified or deleted in S3 with corresponding differentially up- and downregulated proteins. Our analysis revealed that most of these proteins were involved in inflammatory response-related processes; these proteins typically exhibited opposite expression trends in S2 (Figure 4F).
3.5 DGKA inhibition prevented iCCA cell proliferation in vitro
Multiomics analysis revealed that patients with the S2 metabolic subtype had the shortest survival, and DGKA, the protein encoded by DGKA, was the signature protein in the S2 subtype. Through IHC analysis, we found that patients with high DGKA expression had shorter OS (median OS = 33.7 months) and DFS (median DFS = 19.6 months) (Figure 5A-C). Multivariate survival analysis indicated that DGKA was an independent prognostic factor for the OS and DFS of patients with iCCA (Supplementary Figure S6A). To determine the role of DGKA in iCCA progression, we knocked down the expression of DGKA in the iCCA cell lines RBE and HuCCT1 by two different shRNAs (shDGKA #1 and shDGKA #2) (Figure 5D and Supplementary Figure S6B). CCK8 assays showed reduced proliferation of iCCA cells with DGKA knockdown, and colony formation assays and EdU staining revealed similar findings (Figure 5E-G). Meanwhile, we utilized ritanserin, a selective DGKA inhibitor, to inhibit the expression of DGKA in iCCA cells. Likewise, colony formation assays and EdU staining indicated that inhibition of DGKA reduced the proliferation of iCCA cells (Supplementary Figure S6C-D). Therefore, these results showed that DGKA inhibition prevented iCCA cell proliferation in vitro.
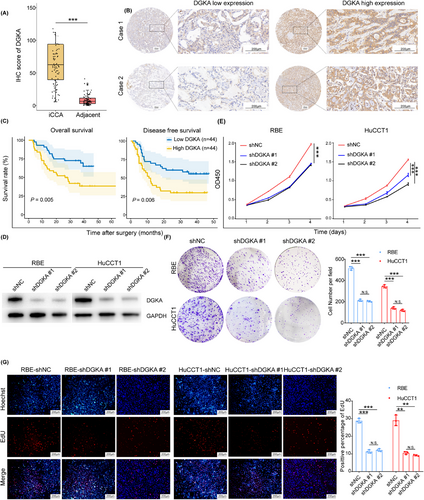
DGKA inhibition prevented iCCA cell proliferation in vitro. (A) The expression of DGKA in iCCA and adjacent normal liver tissues (n = 88). (B) Representative IHC images of DGKA staining. (C) Survival Kaplan-Meier curves of iCCA patients of OS (left) or DFS (right) with low DGKA expression (blue, n = 44) or high DGKA expression (yellow, n = 44), Log-Rank test was used to calculate P values. (D) Protein expression levels of DGKA in iCCA cells. (E-G) Knockdown of DGKA repressed the proliferation of iCCA cell lines (RBE, HuCCT1) through CCK8 assays (E), colony formation assays (F) and EdU assays (G). P values in E-G were calculated by two-tailed Student's t-tests, **, P < 0.01, ***, P < 0.001, and N.S. means P > 0.05. Error bars in A and E-G indicate the mean ± s.d. The experiments in E-G were repeated three times with similar results.
Abbreviations: DGKA, diacylglycerol kinase α; iCCA, intrahepatic cholangiocarcinoma; IHC, immunocytochemistry; OS, overall survival
DFS, disease-free survival; CCK8, cell counting kit-8; OD, optical density; EdU, 5-Ethynyl-2’-deoxyuridine; N.S., not significant; s.d., standard deviation.
3.6 DGKA regulated iCCA cell proliferation via the PA-MEK/ERK pathway
Previous studies reported that DGKA could phosphorylate diacylglycerol (DG) to generate PA [59, 60]. PA, a lipid secondary messenger, plays a role in several critical biological events by regulating downstream signaling pathways [61, 62]. Therefore, we speculated that DGKA regulates iCCA proliferation via the metabolism of PA and the regulation of signaling pathways. Due to the lack of significant differences in the effects of different shRNAs on iCCA cell proliferation, we proceeded with further research using shDGKA #1. Next, we added exogenous PA to the medium of DGKA-knockdown cells, and an increase in the proliferation of iCCA cells was observed by CCK8 assays (Figure 6A). Furthermore, cell viability assays indicated that exogenous PA rescued the viability of ritanserin-treated iCCA cells (Figure 6B). To further explore the mechanism of DGKA in iCCA, RNA-seq was performed in DGKA-knockdown cells. A total of 4,870 differentially expressed genes (DEGs) were found (Supplementary Figure S7A).
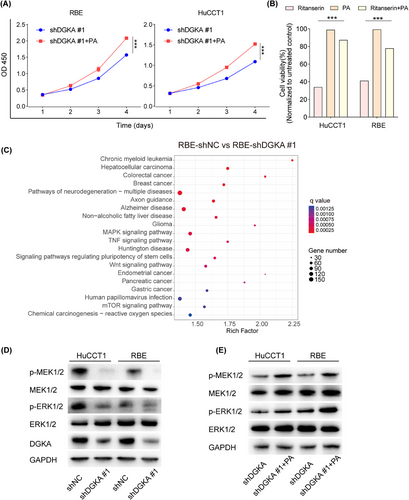
DGKA regulated iCCA cell proliferation via the PA-MEK/ERK pathway. (A) Exogenous PA was added to the medium of DGKA knockdown cells, and an increase in the proliferation of iCCA cells was observed by CCK8 assays, indicating that exogenous PA enhanced the proliferation of DGKA knockdown iCCA cells. (B) Exogenous PA was added to the medium of ritanserin-treated cells, and an increase in the proliferation of iCCA cells was observed by CCK8 assays, indicating that exogenous PA rescued the cell viability of ritanserin-treated iCCA cells (RBE, HuCCT1). (C) KEEG enrichment analysis showed the top 20 enriched signaling pathways based on all the differential expressed genes, n = 3 for each cell line. (D) Knockdown of DGKA inhibited the phosphorylation of MEK1/2 and ERK1/2. (E) Exogenous PA increased the phosphorylation of MEK1/2 and ERK1/2 in DGKA knockdown iCCA cells. P values in A-B were calculated by two-tailed Student's t-tests, ***, P < 0.001. Error bars in A-B indicate the mean ± s.d. The experiments in A-B and D-E were repeated three times with similar results.
Abbreviations: DGKA, diacylglycerol kinase α; iCCA, intrahepatic cholangiocarcinoma; PA, phosphatidic acid; OD, optical density; CCK8, cell counting kit-8; ERK, extracellular regulated protein kinases; MEK, mitogen-activated protein kinase; KEGG, Kyoto Encyclopedia of Genes and Genomes; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; s.d., standard deviation.
Consistent with the results of functional assays, GSEA showed that negative regulation of cell proliferation was enriched in the DGKA-knockdown group (Supplementary Figure S7B-D). Furthermore, KEGG enrichment analysis revealed that the MAPK signaling pathway was significantly enriched based on all DEGs (Figure 6C). Thus, we assessed the expression of p-MEK1/2, MEK1/2, p-ERK1/2, and ERK1/2 in DGKA-knockdown iCCA cells. The results showed that DGKA knockdown reduced the expression of MEK1/2 and ERK1/2 phosphorylation in iCCA cells (Figure 6D). In contrast, exogenous PA rescued phosphorylation of MEK1/2 and ERK1/2 in DGKA-knockdown iCCA cells (Figure 6E). These results suggested that DGKA regulated iCCA cell proliferation via the PA-MEK/ERK pathway.
4 DISCUSSION
In this study, we identified three metabolic subtypes (S1-S3) with subtype-specific biomarkers of iCCA by integrating WES, bulk RNA-seq and proteomics analyses. The S2 subtype with the worst survival showed the activation of some special metabolic processes, immune-suppressed microenvironment and KRAS/ARID1A mutations. Among the S2 subtype-specific upregulated proteins, DGKA was further identified as a potential drug target for iCCA.
Understanding the metabolic profiles is important in heterogeneous diseases. Gong et al. [63] classified triple-negative breast cancer (TNBC) into three heterogeneous metabolic pathway-based subtypes (MPSs) with distinct metabolic features and found that inhibition of LDH could enhance tumor response to anti-programmed cell death protein 1 (PD-1) immunotherapy in the MPS2 subgroup. In our study, we identified three metabolic subtypes (S1, S2, and S3) with distinct clinical outcomes and metabolic features. In addition, a strong concordance was found between metabolic subtypes and proteomic molecular subtypes (using not only metabolic proteins), which suggested that studying metabolic differences provided a novel perspective to understand the heterogeneity of iCCA. Patients in S2 had downregulated expression of key rate-limiting enzymes in fatty acid degradation and upregulation of key rate-limiting enzymes involved in glycolysis, which is consistent with common patterns of metabolic adaptation among various cancer types [64], while patients in S1 subtypes showed almost opposite expression. This metabolic heterogeneity might help explain the survival difference.
Over the past few years, immunotherapy has shown robust therapeutic activity in treating cancers clinically, including iCCA [65]. Through BayesPrism, we deconvoluted the proportion of naive-like CD8+ T cells and effector memory CD8+ T cells in each patient. We found that patients in the S2 subtype had a higher proportion of naive-like CD8+ T cells and a lower proportion of effector memory CD8+ T cells. Furthermore, higher expression of CD39 and CD73 was found in the S2 subtype. CD39, together with CD73, can ultimately convert ATP to adenosine, which binds the adenosine receptors of immune cells and plays an immunosuppressive role [56]. We speculated that the immunosuppressive microenvironment in S2 subtype might be due to the high expression levels of CD39 and CD73, which could be potential targets. Martin-Serrano et al. [47] described a comprehensive TME-based stratification of iCCA, which was composed of five subtypes, encompassing “Immune Classical”, “Inflammatory Stroma”, “Hepatic Stem-like”, “Tumor Classical” and “Desert-like”.Then, they used 500 DEGs of five TME-based subtypes (100 for each) of iCCAs from the training set and the NTP algorithm [48] to decide the TME-based subtypes for other datasets of iCCAs. We also used this approach to carry out a TME-based subtyping for the iCCA patients in this study (Supplementary Table S9). The characteristics of these TME-based subtypes were consistent with our findings. For example, S3 had fewer proportions of immune cells and stroma-related cells (Supplementary Tables S8-S9).
To identify the molecular drivers of distinct metabolic subtypes in iCCA, we found that S2 tended to have a higher mutation frequency of KRAS and ARID1A. Consistent with a previous report [5], KRAS mutations were mutually exclusive with IDH1 and BAP1 alterations. In addition, it was reported that KRAS/ARID1A mutations could drive cholangiocarcinoma synergistically from a biliary origin via failed engagement of the transforming growth factor beta-SMAD family member 4 (TGF-β-Smad4) tumor suppressor pathway [66]. Furthermore, KRAS mutation could upregulate the expression and/or activity of key glycolytic enzymes to promote glycolysis [67, 68] and utilize fatty acid synthesis to regenerate NAD(P)+ and synthesize lipids or fatty acid oxidation [69], which is consistent with what we found in the S2 subtype. Additionally, S2 had more stable genomes than S3, which allowed some of the inflammation-related proteins in S2 to have fewer copy number deletion events and may therefore lead to the formation of an immunosuppressive microenvironment, resulting in poorer survival.
We identified DGKA in the S2 subtype and found that it might be a putative biomarker for predicting the clinical outcomes of iCCA patients. A tumor-promoting role of DGKA in iCCA, consistent with previous studies in other cancer types [70, 71], was observed. Functionally, DGKA could phosphorylate diacylglycerol to generate PA, which may activate downstream signaling pathways in cancer cells and regulate the malignant phenotype of cancer cells. Consistent with these findings, we found that downregulation of DGKA suppressed iCCA cell proliferation, and exogenous PA rescued the level of proliferation, which activated the MEK-ERK pathway. Therefore, inhibiting the expression of DGKA might suppress the proliferation of iCCA cells. Thus, DGKA may be a target in the treatment of iCCA. In addition, we found a positive correlation between DGKA expression and fibroblast fractions, and a previous report indicated that DGKA was involved in fibroblast activation after irradiation [72]. The relationship between DGKA and fibroblasts in iCCA needs further investigation.
Our research had some limitations. First, bioinformatic analysis of genetic drivers could not determine the causal effects, and driver events that metabolically specify the three subtypes require further functional investigation. Second, in vivo experiments should be carried out to determine the potential value of DGKA as a therapeutic target.
5 CONCLUSIONS
In summary, our study provided a metabolic classification of iCCA and deciphered the metabolic features for different prognostic subtypes through the integration of multiomics analysis. More importantly, we proposed and validated that DGKA might be a potential target for the worst subtype. These new findings have significant implications for understanding the formation mechanism of iCCA heterogeneity and improving precise treatment strategies.
DECLARATIONS
AUTHOR CONTRIBUTIONS
Yinghong Shi, Dong Yang, Zheng Tang, Weiren Liu, Huqiang Wang, Qianfu Zhao, Jian Zhou and Jia Fan conceived the study and designed the experiments. Dong Yang, Huqiang Wang, Yushan Hou, Zhixiang Yang and Rongyu Ping performed all computational experiments and analyses. Dong Yang, Zimei Sun and Yushan Hou prepared the samples for multiomics experiments. Weiren Liu, Qianfu Zhao, Chenyang Tao, Weifeng Qu and Rui Yang performed and analyzed all validation experiments. Weifeng Qu, Run Huang, Guiqi Zhu, Xifei Jiang, Chenyang Tao, Yuan Fang, Jun Gao, Xiaoling Wu, Jiafeng Chen and Tianhao Chu collected and prepared patient samples. Yinghong Shi, Dong Yang, Tang Zheng, Weiren Liu, Huqiang Wang and Qianfu Zhao reviewed the results, interpreted the data and wrote the manuscript. All authors reviewed and approved the final manuscript for submission.
ACKNOWLEDGMENTS
We thank the proteomics and bioinformatics platforms in the National Center for Protein Sciences (Beijing) for support in proteomics data generation and large-scale data processing. This project was supported by grants from the National Natural Science Foundation of China (82273387, 82273386, 82073217, 32270711, 82073218 and 82003084), the National Key Research and Development Program of China (2018YFC1312100), Beijing Nova Program (20220484230), Shanghai Municipal Science and Technology Major Project (2018SHZDZX05), Shanghai Municipal Key Clinical Specialty, CAMS Innovation Fund for Medical Sciences (CIFMS) (2019-I2M-5-058) and the State Key Laboratory of Proteomics (SKLP-K202004).
CONFLICT OF INTEREST STATEMENT
The authors declare no competing interests.
CONSENT FOR PUBLICATION
Not applicable.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the medical ethics committee of Zhongshan Hospital of Fudan University (Y2020-622). The tissue samples were obtained with written informed consent from each patient.
Open Research
DATA AVAILABILITY STATEMENTS
RNA-sequencing data that support the findings of this study have been deposited in the Genome Sequence Archive in the National Genomics Data Center (https://ngdc.cncb.ac.cn/), China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (GSA-Human: HRA004766), which are publicly accessible at https://ngdc.cncb.ac.cn/gsa-human. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD042525. Researchers who wish to gain more information are needed to send emails to the corresponding author.




