Blockage of EGFR/AKT and mevalonate pathways synergize the antitumor effect of temozolomide by reprogramming energy metabolism in glioblastoma
Xiaoteng Cui, Jixing Zhao and Guanzhang Li authors contributed equally to this work.
Abstract
Background
Metabolism reprogramming plays a vital role in glioblastoma (GBM) progression and recurrence by producing enough energy for highly proliferating tumor cells. In addition, metabolic reprogramming is crucial for tumor growth and immune-escape mechanisms. Epidermal growth factor receptor (EGFR) amplification and EGFR-vIII mutation are often detected in GBM cells, contributing to the malignant behavior. This study aimed to investigate the functional role of the EGFR pathway on fatty acid metabolism remodeling and energy generation.
Methods
Clinical GBM specimens were selected for single-cell RNA sequencing and untargeted metabolomics analysis. A metabolism-associated RTK-fatty acid-gene signature was constructed and verified. MK-2206 and MK-803 were utilized to block the RTK pathway and mevalonate pathway induced abnormal metabolism. Energy metabolism in GBM with activated EGFR pathway was monitored. The antitumor effect of Osimertinib and Atorvastatin assisted by temozolomide (TMZ) was analyzed by an intracranial tumor model in vivo.
Results
GBM with high EGFR expression had characteristics of lipid remodeling and maintaining high cholesterol levels, supported by the single-cell RNA sequencing and metabolomics of clinical GBM samples. Inhibition of the EGFR/AKT and mevalonate pathways could remodel energy metabolism by repressing the tricarboxylic acid cycle and modulating ATP production. Mechanistically, the EGFR/AKT pathway upregulated the expressions of acyl-CoA synthetase short-chain family member 3 (ACSS3), acyl-CoA synthetase long-chain family member 3 (ACSL3), and long-chain fatty acid elongation-related gene ELOVL fatty acid elongase 2 (ELOVL2) in an NF-κB-dependent manner. Moreover, inhibition of the mevalonate pathway reduced the EGFR level on the cell membranes, thereby affecting the signal transduction of the EGFR/AKT pathway. Therefore, targeting the EGFR/AKT and mevalonate pathways enhanced the antitumor effect of TMZ in GBM cells and animal models.
Conclusions
Our findings not only uncovered the mechanism of metabolic reprogramming in EGFR-activated GBM but also provided a combinatorial therapeutic strategy for clinical GBM management.
List of abbreviations
-
- GBM
-
- glioblastoma
-
- CNS
-
- central nervous system
-
- RTKs
-
- receptor tyrosine kinases
-
- EGFR
-
- epidermal growth factor receptor
-
- ACSS3
-
- acyl-CoA synthetase short-chain family member 3
-
- ACSL3
-
- acyl-CoA synthetase long-chain family member 3
-
- ELOVL2
-
- long-chain fatty acid elongation-related gene ELOVL fatty acid elongase 2
-
- GFAP
-
- glial fibrillary acidic protein
-
- CHI3L1
-
- chitinase 3 like 1
-
- TCA
-
- citric acid cycle
-
- OLIG2
-
- oligodendrocyte transcription factor 2
-
- SOX6
-
- SRY-box transcription factor 6
-
- RFA
-
- RTK-fatty acid-gene signature
-
- PDGFA
-
- platelet-derived growth factor subunit A
-
- DMEM
-
- Dulbecco's modified Eagle's medium
-
- siRNAs
-
- small interfering RNAs
-
- ChIP
-
- chromatin immunoprecipitation
-
- LC‒MS/MS
-
- liquid chromatography-tandem mass spectrometry
-
- CNV
-
- copy number variation
-
- CGGA
-
- Chinese glioma genome atlas
-
- TCGA
-
- The Cancer Genome Atlas Program
-
- Rembrandt
-
- repository of molecular brain neoplasia data
-
- ROC
-
- receiver operating characteristic
-
- ANOVA
-
- analyses of variance
-
- IDH
-
- isocitrate dehydrogenase
-
- OCR
-
- oxygen consumption rate
-
- ECAR
-
- extracellular acidification rate
-
- PCs
-
- phosphatidylcholines
-
- LysoPCs
-
- lysophosphatidylcholines
-
- PE
-
- phosphatidylethanolamine
-
- PG
-
- phosphatidyl glycerol
-
- PI
-
- phosphatidylinositol
-
- PS
-
- phosphatidylserine
-
- VLCFA
-
- very long-chain fatty acid
-
- HMG-CoA
-
- hydroxymethylglutaryl-CoA
-
- TMZ
-
- temozolomide
-
- BBB
-
- blood-brain-barrier
-
- ssGSEA
-
- single-sample gene set enrichment analysis
-
- TKI
-
- tyrosine kinase inhibitor
-
- NSCLC
-
- non-small cell lung carcinoma
-
- DMSO
-
- dimethyl sulfoxide
-
- OSI
-
- Osimertinib
-
- ATO
-
- atorvastatin
-
- HFD
-
- high-fat diet
1 BACKGROUND
Glioblastoma (GBM) is one of the most malignant primary neoplasms of the central nervous system (CNS) in clinics. GBM is characterized by high intra- and inter-tumor heterogeneities, insensitivity to conventional radiotherapy and chemotherapy, and frequent recurrence [1-4]. In general, GBM patients exhibit rapid progression and poor prognosis, with a median survival time of 16-18 months, despite surgical resection combined with radio- and temozolomide-based chemotherapy since the first diagnosis [5]. Therefore, there is an urgent need for better combinational therapeutic strategies for GBM management.
Receptor tyrosine kinases (RTKs) are a family of cell membrane surface proteins [6] that participate in the regulation of important cellular functions, including cell growth and protein synthesis, through signal transduction [7]. Hyperactivation of the RTK/PI3K/AKT pathway is the typical molecular signature of GBM pathomechanism [8]. Epidermal growth factor receptor (EGFR) amplification, overexpression, and pathogenic mutations are commonly identified in GBM [9]. Notably, the alternatively spliced oncogenic EGFR-vIII variant lacks exons 2-7, resulting in the constitutive tyrosine kinase activity without any ligand stimulations [10]. GBM with EGFR amplification and EGFR-vIII mutation often exhibits severe malignancies [11]. Although the hyperactivated EGFR pathway plays roles in lipogenesis and cholesterol uptake [12, 13], whether the EGFR pathway contributes to fatty acid metabolism and energy generation remains unclear.
Reprogramming of cellular metabolism is another pathological hallmark in tumorigenesis, supporting the high demand for energy of tumor cells and promoting the establishment of an immunosuppressive microenvironment [14, 15]. Abnormal metabolisms of lipids, amino acids, and nucleotides, as well as enhanced glycolysis, are commonly observed in GBM cells [16]. Interestingly, a high level of cellular cholesterol plays a vital role in tumor cell survival and disease progression [17]. However, the actual effects of inhibiting lipid metabolism and/or reducing cholesterol levels on the metabolic remodeling of GBM cells need to be further characterized.
In this study, we explored the regulatory correlation between EGFR expression and fatty acid metabolism-associated genes for establishing a signature in prognostic prediction, investigated the functional role of EGFR/AKT and mevalonate pathway in energy metabolic reprogramming, and developed a combinational therapeutic strategy against GBM.
2 MATERIALS AND METHODS
2.1 Clinical GBM sample acquisition
In total, 14 tumor tissue samples, namely TBD528-T1, TBD528-T2, TBD629-T1, TBD629-T2, TBD629-T3, TBD706-T1, TBD706-T2, TBD706-T3, TBD717-T1, TBD717-T2, TBD717-T3, TBD717-T4, TBD720-T1, and TBD720-T2, from 5 patients who were diagnosed with the primary GBM based on their head magnetic resonance imaging (MRI) examinations, were obtained by multipoint sampling for single-cell RNA sequencing. Another 66 GBM samples were freshly harvested for RNA sequencing and untargeted metabolomic analyses. Clinical information, MRI images, and tumor specimens of typical GBM cases were collected from Beijing Tiantan Hospital and Affiliated Hospital of Hebei University. Collection and analysis of all clinical GBM samples were approved by the medical ethics committee of Beijing Tiantan Hospital (Approval No. KY 2020-093-02) and Hebei University affiliated Hospital (Approval No. HDFY-LL-2020-017). Written informed consent was acquired from the patients or immediate family members of the patients.
2.2 Single-cell RNA sequencing and data processing
Fresh GBM samples were collected directly from the sampling room immediately after the surgery and immersed in a tissue storage solution (#130-100-008, Miltenyi, Biotec, Bergisch Gladbach, Germany). Samples were mechanically and enzymatically dissociated using a tumor dissociation kit (#130-095-929, Miltenyi), according to the manufacturer's instructions. Briefly, GBM tissues were cut into small pieces and digested into single-cell suspensions. For single-cell RNA sequencing, barcode sequences were labeled on the single cells and respective single-cell libraries were constructed using the BD Rhapsody system (San Jose, CA, USA). Libraries were finally sequenced on the Illumina NovaSeq sequencing platform (San Diego, CA, USA).
Raw data were aligned to the human reference genome (GRCh38). A sparse matrix containing gene expressions and barcode information was constructed by UMI tools (https://github.com/CGATOxford/UMI-tools) [18], and imported into the Seurat R package [19]. The sequencing data were processed as previously described [20]. Briefly, cells with a less than 200-library size or a higher than 0.3% mitochondrial transcript ratio were excluded. The filtered data were analyzed for batch effect removal, dimensionality reduction, and unsupervised clustering by the Seurat package in the default parameters.
The human primary cell atlas (HPCA) was used to identify the cell types described in our previous study [20]. Briefly, the correlation coefficients between the transcriptomes of each cell and the expression profiles of each cell type-specific genes in HPCA were analyzed by using SingleR package. The CellMarker database (ttp://xteam.xbio.top/CellMarker/) was employed to determine the cell type based on their marker gene expressions [21]. Marker genes, such as CD3e for T-cells, CD14 for macrophages, FCGR3B for oligodendrocytes, MAG for neutrophils, CLDN5 for endothelial cells, MYL9 for smooth muscle cells and fibroblast (SMC&Fibro), and PTPRZ1 for tumor cells were visualized as violin plots using the ggplot2 R package. Marker genes of each cell cluster were identified by the “FindAllMarkers” function of the Seurat package and visualized as violin plots.
The CNV for each sample was predicted by analyzing the expression levels of selected gene markers located on each chromosome using the InferCNV package. The CNV in tumor cells was determined by comparison with nonmalignant cells, and visualized as a heatmap.
2.3 GBM public data collection and correlation analysis
Data from the Chinese Glioma Genome Atlas (CGGA), The Cancer Genome Atlas (TCGA), and Repository for Molecular Brain Neoplasia Data (Rembrandt) databases were obtained for analysis. Clinical information and RNA sequencing data of glioma patients in the TCGA database were obtained from the University of California Santa Cruz (UCSC) Xena Functional Genomics Explorer (https://xenabrowser.net/). RNA sequencing data of CGGA, microarray data of Rembrandt, and corresponding clinical data were downloaded from the CGGA website (http://www.cgga.org.cn/). The RNA data processing was performed as previously described [20]. Briefly, count data were transformed to fragments per kilobase per million reads (FPKM).
Correlation analysis between the expression levels of 2 genes was measured by the ggcorrplot R package. The gene expression distributions and respective correlation coefficients were visualized by the Performance Analytics package. Survival curves were analyzed using the Survival R package. All the analyses were performed with default parameters.
2.4 GBM patient grouping and RTK-fatty acid-gene signature (RFA) construction
The expression levels of glial fibrillary acidic protein (GFAP), chitinase 3 like 1 (CHI3L1), oligodendrocyte transcription factor 2 (OLIG2), SRY-box transcription factor 6 (SOX6), EGFR, and platelet-derived growth factor subunit A (PDGFA) in GBM patients from the CGGA, TCGA, and Rembrandt databases were selected. For grouping, patients were first separated into the G1/G2 versus G3 groups based on the value of . Next, the upper median value of was employed for the definition of G1 versus G2. For RFA signature construction, The RFA score was calculated according to this formula: , where βi was the coefficient calculated by univariate COX regression analysis of gene , and Ei refers to the expression of gene . Gene included GFAP, CHI3L1, EGFR, PDGFA, acyl-CoA synthetase short chain family member 3 (ACSS3), acyl-CoA synthetase long chain family member 3 (ACSL3), and ELOVL fatty acid elongase 2 (ELOVL2) in the TCGA, CGGA, and Rembrandt cohorts. Patients with RFA scores above the median were considered high-score individuals.
2.5 Correlation between RFA score and immune microenvironment of GBM
Single sample gene set enrichment analysis (ssGSEA) and CIBERSORT tools were performed for evaluate the correlation between RFA scores and infiltrated immune cells in the microenvironment of GBMs. Briefly, for ssGSEA analysis, the immune metagene set was obtained [22] and the scores of each immune cells in GBM patients from the CGGA, TCGA, and Rembrandt databases were analyzed by using GSVA v1.46.0 R package (https://github.com/rcastelo/GSVA). The results were visualized as heatmaps. For CIBERSORT analysis, the expression data of GBMs from the CGGA, TCGA, and Rembrandt databases were uploaded to the online tool (https://cibersortx.stanford.edu/) [23] and LM22 containing 22 immune cell types was chosen as a signature matrix. The results were downloaded and the correlation between RFA scores and macrophage M2 was visualized as a scatter plot.
2.6 RNA sequencing analysis
Total RNA was isolated from fresh GBM samples using TRIzol reagent (#15596-026, Thermo Fisher, Waltham, MA, USA), according to the manufacturer's protocol. Then rRNA was removed and RNA was fragmentated for reverse transcription. The cDNA libraries were generated and sequenced using the Illumina HiSeq4000 platform (San Diego, CA, USA). The sequencing data were aligned to the human reference genome (hg19) using the Hisat2 tool (http://daehwankimlab.github.io/hisat2/main/) [24] and analyzed by the DESeq2 R package.
2.7 Cell culture, lentivirus, and chemicals
The human GBM cell line U-87 MG was purchased from the American Type Culture Collection (#HTB-14), human GBM primary cell TBD0220 was constructed by our laboratory, and mouse GBM cell line CT2A were purchased from BLUEFBIO Co. Ltd. (#BFN60810497, Shanghai, China). All the cells were maintained in our laboratory. Lentivirus vectors encoding the EGFR-vIII protein and luciferase reporter were constructed by IBS Biotech. Co., Ltd (Shanghai, China). The U-87 MG, and CT2A cells were cultured in Dulbecco's modified Eagle medium (DMEM; #PM150210, Procell Life Science&Technology Co.,Ltd., Wuhan, China) supplemented with 10% fetal bovine serum (FBS; #HN-FBS-500, Shanghai Chuan Qiu Biotechnology Co.,Ltd., Shanghai, China). TBD0220 cells were cultured in DMEM/F12 (#PM150312, Procell Life Science&Technology Co.,Ltd., Wuhan, China) with 10% FBS. All cells were grown at 37°C in a cell incubator with 5% CO2. U-87 MG cells were transduced with EGFR-vIII lentiviral particles and screened with 2 μg/mL of puromycin (#P8230, SolarBio Science & Technology Co., Ltd., Beijing, China) to generate the U-87 MG-EGFR-vIII cell line.
Temozolomide (TMZ; #HY-17364), MK-2206 (#HY-10358), MK-803 (#HY-N0504), Osimertinib (OSI) (#HY-15772), and Atorvastatin (ATO) (#HY-B0589) chemicals were purchased from MedChemExpress (Shanghai, China). For treating GBM cells, TMZ, MK-2206, and MK-803 were dissolved in DMSO (#D8371, SolarBio Science & Technology Co., Ltd., Beijing, China) to the storage concentrations of 100 mmol/L, 5 mmol/L, and 5 mmol/L, respectively. For animal experiments, drugs were dissolved in DMSO and diluted in the following solvents to the final concentrations of 10% DMSO, 40% PEG300 (#HY-Y0873, MedChemExpress), 5% Tween-80 (#HY-Y1891, MedChemExpress), and 45% saline. Small interfering RNAs (siRNAs) were synthesized by IBS Biotech. Co., Ltd. siRNA sequences are summarized in Supplementary Table S1.
2.8 Untargeted metabolomic analysis
TBD0220 cells were treated with DMSO, 5 μmol/L of MK-2206, or 5 μmol/L of MK-803. After treatment, cells were washed once using prechilled PBS and rapidly frozen in liquid nitrogen after the treatment. Then, cells were scrape-harvested using collection buffer (prechilled HPLC grade methanol: Milli-Q water = 4: 1, v/v) into 1.5 mL Eppendorf tubes. Fresh GBM tissues were harvested after surgery and transferred into collection buffer. Samples were dehydrated and redissolved in detection buffer (prechilled HPLC grade methanol: Milli-Q water = 1: 4, v/v) at -20°C condition after sonication. The metabolites were analyzed using ultra-performance liquid chromatography combined with a QE high-resolution mass spectrometer (BRE0032850, Thermo Fisher, Waltham, MA, USA). Data were preprocessed under the criteria of RSD < 0.3 in the QC sample, normalized using Progenesis QI software (v2.3, Nonlinear Dynamics, Newcastle, UK) with the parameters of appropriate precursor tolerance (5 ppm/10 ppm), product tolerance (10 ppm/20 ppm), and production threshold (5%), and excluded by any peak with missing values of > 50%. Different metabolites were identified using the OPLS-DA method with default parameters.
2.9 Conjoint analysis of RNA sequencing and metabolomics
RNA sequencing data and metabolomics data of 66 fresh GBM tissues were integrated. The correlations between gene expression profile and metabolites were analyzed.
2.10 Chromatin immunoprecipitation (ChIP)
TBD0220, U-87 MG, and U-87 MG-EGFR-vIII cells were cross-linked by formaldehyde (#F8775, Sigma Aldrich, St. Louis, MO, USA) at a final concentration of 1% for 10min, followed by quenching of cross-links using glycine solution. Nuclear lysates were prepared and sonicated for chromatin fragmentation. Adequate lysates containing 100 μg of total protein per reaction were incubated with 5 μL p-NF-κB antibody (#3033S, Cell Signaling Technology), and ChIP grade magnetic beads (#16-663, Millipore, Billerica, MA, USA) at 4°C overnight. The immunoprecipitated and purified DNA samples were obtained using a QIAquick PCR Purification kit (#28104, Qiagen, Duesseldorf, Germany) after reversing the cross-link, and measured by the quantitative real-time polymerase chain reaction (qPCR) method with specific primers. qPCR reactions were performed using SYBR Green Mix (#Q711, Vazyme Biotech Co., Nanjing, China) in a thermocycler instrument (ABI QuantStudio 3 Real-time PCR System). All primers were synthesized by GENEWIZ (Suzhou, China). The ChIP primer sequences are summarized in Supplementary Table S2.
2.11 Colony formation assay and cell growth assay
Colony formation and cell growth assays were performed as described elsewhere [25]. Briefly, for colony formation assay, a total of 500 cells were plated into 6-well plates and treated with DMSO, 1 μmol/L of MK-2206, or 1 μmol/L of MK-803 for 14 days. Then cells were prefixed by 4% paraformaldehyde for 10 min at room temperature and stained by crystal violet staining solution (#C0121, Beyotime Biotechnology, Shanghai, China) for 30 min at room temperature. After washing by distilled water, the samples were captured using a bright-field microscope (#CX41, Olympus Corporation, Tokyo, Japan). For cell growth assay, 5 × 103 cells per well were seeded into 96-well plates and treated with DMSO, 5 μmol/L of MK-2206, or 5 μmol/L of MK-803 for 5 days. Cell counting kit-8 (#HY-K0301, MedChemExpress, Shanghai, China) was used to measure the proliferation of cells.
2.12 Seahorse XF Cell Mito Stress and Glycolysis Stress assays
TBD0220, U-87 MG, and U-87 MG-EGFR-vIII cells were used for the Seahorse XF Cell Mito Stress assay. First, cells were seeded on the Seahorse XF24 cell culture microplates (#102342-100, Agilent Technologies, Inc., Santa Clara, CA, USA) at a density of 1 × 104 cells per well. To measure the effects of inhibitors, DMSO, 5 μmol/L MK-2206, 5 μmol/L MK-803, or 5 μmol/L OSI was added to the culture medium. To detect the mechanistic effects of ACSS3, ACSL3, and ELOVL2, respective siRNAs were transfected using Lipofectamine-3000 reagent (#L3000150, Thermo Fisher). After 24 h of transfection, the mitochondrial stress and glycolytic function were measured. The Seahorse XF Cell Mito Stress Test kit (#103015-100, Agilent, Santa Clara, CA, USA) and Seahorse XF Glycolysis Stress Test Kit (#103020-100, Agilent) were used following the manufacturer's protocol. Briefly, a sensor cartridge (#102342-100, Agilent Technologies, Inc., Santa Clara, CA, USA) was hydrated using Seahorse XF Calibrant at 37°C in a non-CO2 incubator overnight. The medium were prepared by supplementing Seahorse XF DMEM medium (#103575-100, Agilent Technologies, Inc., Santa Clara, CA, USA) with essential additives (1 mmol/L pyruvate, 2 mmol/L glutamine, and 10 mmol/L glucose for MitoStress; 2 mmol/L glutamine for Glycolysis Stress) and warmed to 37°C. The compounds included in the kit were solubilized by the medium and loaded into the ports on the sensor cartridge. After calibrating the prepared sensor cartridge and placing the cell culture microplate at 37°C in a non-CO2 incubator for 45 min to 1 h, the oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) were measured on a Seahorse XFe24 Analyzer (Agilent Technologies, Inc., Santa Clara, CA, USA).
2.13 ATP detection assay
ATP detection assays were performed using an ATP detection kit (#S0026, Beyotime Biotechnology, Shanghai, China). Briefly, a total of 1 × 104 cells per well were seeded into 96-well plates and treated with DMSO, 5 μmol/L MK-2206, or 5 μmol/L MK-803 for 24 h. Cells were lysed and incubated with ATP detection buffer for 3-5min. Then, relative luminescence units were measured on a BioTek Gen5 Microplate Reader (BioTek Instruments, Vermont, USA), and ATP concentrations were calculated from the standard curve.
2.14 Protein preparation, western blotting, and antibodies
Total cell lysates were prepared from GBM cells using RIPA buffer (150 mmol/L NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mmol/L Tris-HCl pH = 7.4) supplied with Protease Inhibitor Cocktail (#HY-K0010, MedChemExpress), and PMSF (P0100, SolarBio Science & Technology Co., Ltd., Beijing, China). Total protein samples were analyzed by 8% or 10% sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE). Antibodies against ACSS3 (1:500; #16204-1-AP, Proteintech, Wuhan, China), ACSL3 (1:1000; #20710-1-AP, Proteintech), ELOVL2 (1:1000; #ab176327, abcam, Cambridge, MA, USA), p-EGFR (1:1000; #3777S, Cell Signaling Technology), EGFR (1:1000; #18986-1-AP, Proteintech), p-AKT (1:1000; #4060S, Cell Signaling Technology), AKT (1:1000; #9272S, Cell Signaling Technology), p-NF-κB (1:1000; #3033S, Cell Signaling Technology), NF-κB (1:1000; #8242S, Cell Signaling Technology), Na-K-ATPase (1:1000; #3010S, Cell Signaling Technology), CDK2 (1:1000; #AB40719, Absci, Vancouver, WA, USA), CDK4 (1:1000; #DF6102, Affinity Biosciences, Jiangsu, China), CDK6 (1:1000; #ab124821, abcam), Cyclin D (1:1000; #AF0931, Affinity Biosciences), p-Rb (1:1000; #AF3103, Affinity Biosciences), Rb (1:500; #DF6840, Affinity Biosciences), LKB1 (1:500; #YT2572, Immunoway, Plano, TX, USA), p-LKB1 (1:500; #YP0900, Immunoway), AMPKα1/2 (1:1000; #YT0216, Immunoway), p-AMPKα1/2 (1:500; #YP0575, Immunoway), β-actin (1:5000; #AF7018, Affinity Biosciences), β-Tubulin (1:5000; #66240-1-Ig, Proteintech), and GAPDH (1:5000; #60004-1-Ig, Proteintech) were used for western blotting, according to the manufacturer's instructions. Briefly, proteins separated in gel were transferred to polyvinylidene fluoride (PVDF) membrane (#IPVH00010, Millipore, Billerica, MA, USA). Then the membrane was incubated with blocking buffer [5% skim milk [#8340, SolarBio Science & Technology Co., Ltd., Beijing, China] and 0.1% Tween-20 [#T8220, SolarBio Science & Technology Co., Ltd., Beijing, China] in PBS) for 2 h at room temperature, followed by incubated with the above specific antibodies at 4°C overnight. After washed and incubated with HRP-conjugated secondary antibodies (1:10,000; #RS0001 and #RS0002, ImmunoWay Biotechnology Company, Plano, TX, USA), immunoblots were performed using a FluoChem instrument (ProteinSimple, San Jose, CA, USA).
2.15 Cell cycle analysis
Cell cycle status was detected using a Cell Cycle Assay Kit (#C543, Dojindo, Kumamoto, Japan), according to the manufacturer's protocol. Briefly, cells were digested by trypsin and fixed in 70% of ethanol overnight at 4°C. The cells were then centrifuged to remove excess ethanol and resuspended in a working solution with propidium iodide and RNase at 37°C for 30 min and at 4°C for 30 min, respectively. Cell cycle distribution was detected by BD FACSVerse instruments (BD Biosciences, San Jose, CA, USA). Flow cytometry results were analyzed by FlowJo v10.6.2 software (FlowJo, LLC, Ashland, OR, USA).
2.16 In vivo orthotopic xenograft mouse model and treatments
Female BALB/C nude mice aged 4 weeks were used to construct the GBM orthotopic model. TBD0220 cells (1 × 105 cells in 3 μL PBS) infected with luciferase lentiviruses were intracranially injected using the guidance of a stereotactic instrument. TMZ (5 mg/kg), MK-2206 (100 mg/kg), and MK-803 (50 mg/kg) were administered by oral gavage every day after 7 days of injections. Tumor growth was monitored by in vivo Imaging System (IVIS; PerkinElmer Inc, Waltham, Massachusetts, USA) spectrum on Day 7, Day 14, and Day 21. Kaplan-Meier curves were used to analyze survival. Four weeks after modeling, the brain tissues were carefully harvested, fixed by formalin, embedded in paraffin, and used for immunohistochemistry (IHC) analysis.
Female C57BL/6J mice aged 4 weeks were used to establish the intracranial tumor model. A total of 2 × 104 CT2A cells in 3 μL PBS were injected into mouse brains. After 7 days of tumor transplantation, mice received daily oral gavage of TMZ (5 mg/kg), OSI (25 mg/kg), and/or ATO (10 mg/kg). Bioluminescence imaging using IVIS was employed on Days 3, 7, and 14 to detect tumor growth. On Day 21, the mice were sacrificed and the tumors were carefully extracted for flow cytometry.
The BALB/C nude mice (#401) and C57BL/6J mice (#219) were obtained from the Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). All mice were housed in a specific pathogen free (SPF) breeding barrier with individual ventilated cages. Tribromethyl alcohol (#HY-B1372, MedChemExpress, Shanghai, China) was used for anesthetize mice. Mice should be sacrificed by cervical dislocation after anesthesia in the following conditions: near death or immobile, have significant weight loss, or unable to feed or drink. All animal experimentations were approved by the Animal Ethical and Welfare Committee of Hebei University (Approval No. IACUC-2020XS001).
2.17 Intertumoral distributions of drugs in vivo
Drug distribution analysis was performed as previously described [26]. Briefly, GBM orthotopic models were constructed using TBD0220 cells in a total of 1 × 105 cells per mouse. After 7 days, MK-2206, MK-803, OSI or ATO was administered by oral gavage. The brain tissues were isolated and homogenized by a freezing grinder. The drug concentrations were analyzed using a triple quadrupole liquid chromatography-tandem mass spectrometry (LC-MS; Waters Corporation, Milford, MA, USA). Briefly, chromatographic separation was performed on a Waters BEH C18 column (2.1 × 100 mm, 1.7 μm). Mobile phase A constitution contained 0.1% formic acid solution, and mobile phase B was acetonitrile at a flow rate of 0.3 mL/min. The operation parameters of MS were as follows: capillary voltage (3.0 kV), desolvation temperature (500°C), cone gas flow (150 L/Hr), and desolvation gas flow (800 L/Hr).
2.18 Hematoxylin and eosin (H&E) staining and IHC assay
Paraffin-embedded tissues were stained with H&E, and IHC assays were performed according to the protocol described in our previous study [25]. Briefly, tissues were cut into sections, followed by dewaxed, hydrated, and performed for antigen retrieval. For H&E, hematoxylin (#G1080, SolarBio Science & Technology Co., Ltd., Beijing, China) and eosin (#G1100, SolarBio Science & Technology Co., Ltd., Beijing, China) were sequentially stained. For IHC, after blocked by PBS containing 10% FBS for 1 h at room temperature, samples were incubated with the following antibodies at 4°C overnight: Ki67 (1:200; #ZM-0167, ZSGB-BIO, Beijing, China), p-AKT (1:200; #4060S, Cell Signaling Technology), ACSS3 (1:100; #16204-1-AP, Proteintech), ACSL3 (1:100; #20710-1-AP, Proteintech), ELOVL2 (1:200; #ab176327, abcam), MHC-II (1:1000; #68258S, Cell Signaling Technology), CD206 (1:400; #91992S, Cell Signaling Technology), and CD8a (1:400; #98941S, Cell Signaling Technology). Then samples were washed twice by PBS, incubated with horse radish peroxidase-conjugated secondary antibody, and colorized by diaminobenzidine (DAB). The stained slices were mounted by neutral resin and imaged using a bright-field microscope (#CX41, Olympus Corporation, Tokyo, Japan).
2.19 Detection of tumor infiltrated CD8+ T-cells and macrophages
Tumors were digested into single-cell suspensions, and the red blood cells were removed using red blood cell lysis buffer (#R1010, SolarBio Science & Technology Co., Ltd., Beijing, China). The cells were centrifuged and resuspended into MACS buffer (0.5% bovine serum albumin, 2 mmol/L EDTA in PBS). The anti-CD16/CD32 antibody (1:50; #101319, BioLegend, San Diego, CA, USA) was utilized to block the nonspecific binding sites. Tumor-infiltrating cytotoxic T lymphocytes were stained using anti-CD45.2 (1:200; #109806, BioLegend), anti-CD3 (1:40; #100236, BioLegend), and anti-CD8a (1:20; #100734, BioLegend); tumor-infiltrating macrophages were stained by antibodies of anti-CD45.2 (1:200; #109806, BioLegend), anti-F4/80 (1:20; #123110, BioLegend), anti-CD11b (1:80; #101226, BioLegend), anti-CD11c (1:80; #117318, BioLegend), and anti-CD206 (1:40; #141708, BioLegend) antibodies, according to the manufacturer's protocol. Briefly, the cells were blocked for 10 min on ice, followed by stained with specific antibodies for 30 min on ice away from the light. Then the samples were detected on a BD FACSVerse instrument (BD Biosciences). Flow cytometry results were analyzed by FlowJo v10.6.2 software.
2.20 Statistical analysis
All statistical analyses were performed using GraphPad Prism 8 software and R language (v3.6.2). Independent-sample Student's t-test was used for comparison between two experimental groups. One-way and two-way analyses of variance (ANOVA) were used to compare at least three experimental groups. The error bars in the figures represent the mean ± standard deviation (SD). A P value of less than 0.05 was considered statistically significant, defined as ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
3 RESULTS
3.1 Single-cell RNA sequencing depicts a unique transcriptional landscape of GBM cells
High intra- and inter-tumor heterogeneities of GBM cells, characterized by diverse cell populations and cell cycle states, led to insensitivity to conventional treatments and poor prognosis. To better understand the complexity of the GBM microenvironment, a total of 14 tumor tissue samples were collected by multipoint sampling from 5 GBM patients for single-cell RNA sequencing library construction using the BD Rhapsody system. After a series of stringent cell quality control analyses, the cells were annotated into 7 subtypes, of which the vast majority of cells were tumorigenic, accounting for 69.22% of the cell population (Figure 1A). The other cell types in that population were macrophages (10.76%), oligodendrocytes (6.17%), neutrophils (5.60%), T cells (4.32%), smooth muscle cells and fibroblasts (SMC&Fibro; 3.37%), and endothelial cells (0.56%) (Figure 1A, Supplementary Figure S1A-C). Copy number variation (CNV) is associated with tumorigenesis, leading to increased genomic instability and abnormal protein expression in tumor cells [27, 28]. The combination of chromosome 7 amplification and chromosome 10 deletion is the pathological hallmark feature of GBM [29]. Hence, we determined the CNVs of tumor cells compared with nonmalignant cells using the InferCNV R package [30, 31], and the chromosomal changes were observed in all samples but to different extents (Supplementary Figure S1D). Moreover, TBD717-T1-T4 and TBD528-T1 samples displayed chromosome 3 amplification. TBD717-T1-T4 samples also exhibited changes in chromosome 12 amplification and chromosome 11 deletion. The TBD629-T3, TBD706-T1, TBD706-T2, TBD706-T3, TBD720-T1, and TBD720-T2 samples indicated abnormal amplification of chromosome 20. Only the TBD706-T3 sample showed an amplification change for chromosome 19. These results not only confirmed that all samples were GBM but also reflected high intra- and inter-tumor heterogeneities.
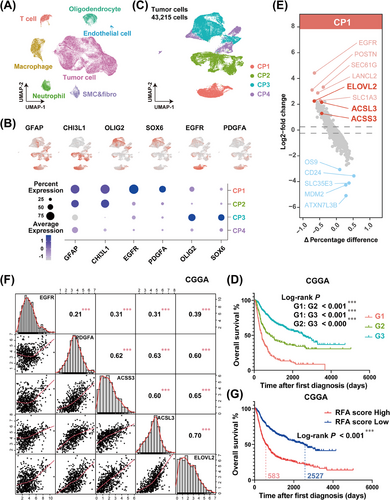
GBM patients with high levels of EGFR accompanied by ACSS3, ACSL3, and ELOVL2 expressions show worse prognoses. (A) Cell types were annotated and visualized as a UMAP plot in GBM single-cell RNA sequencing data. (B) Expressions of GFAP, CHI3L1, OLIG2, SOX6, EGFR, and PDGFA in subcellular populations of tumor cells from GBM single-cell data were stained. (C) A total of 43,215 tumor cells were grouped into 4 cell populations according to the expression features of key genes. (D) Kaplan-Meier curve was generated to evaluate the overall survival time of GBM patients in distinct groups from the CGGA cohort, based on the expressions of GFAP, CHI3L1, OLIG2, SOX6, EGFR, and PDGFA. G1 (red line) represented patients with GFAPhigh, CHI3L1high, OLIG2low, SOX6low, EGFRhigh, and PDGFAhigh; G2 (green line) represented patients with GFAPhigh, CHI3L1high, OLIG2low, SOX6low, EGFRlow, and PDGFAlow; and G3 (cyan line) represented patients with GFAPlow, CHI3L1low, OLIG2high, SOX6high. ∗∗∗P < 0.001 (Log-rank test). (E) Differentially expressed genes in CP1 were identified and visualized as a volcano plot. The dotted lines represent 0.25 and -0.25 of log2-fold change. (F) The correlation analysis among the expressions of EGFR, PDGFA, ACSS3, ACSL3, and ELOVL2 in the CGGA cohort. ∗∗∗P < 0.001 (Pearson). (G) Kaplan-Meier curve analysis showed that patients with high RFA scores in the CGGA cohort had a poor prognosis. ∗∗∗P < 0.0001 (Log-rank test). Abbreviations: GBM, glioblastoma; UMAP, uniform manifold approximation and projection; GFAP, glial fibrillary acidic protein; CHI3L1, chitinase 3 like 1; OLIG2, oligodendrocyte transcription factor 2; SOX6, SRY-box transcription factor 6; EGFR, epidermal growth factor receptor; PDGFA, platelet-derived growth factor subunit A; ACSS3, acyl-CoA synthetase short-chain family member 3; ACSL3, acyl-CoA synthetase long-chain family member 3; ELOVL2, long-chain fatty acid elongation-related gene ELOVL fatty acid elongase 2; CGGA, Chinese glioma genome atlas.
3.2 High levels of EGFR, ACSS3, ACSL3, and ELOVL2 confer a poor prognosis for GBM patients
Next, we focused on the tumor subtypes, which accounted for the largest proportion. We picked a tumor cell subset (a total of 43,215 cells) for reanalysis and visualization using UMAP (Supplementary Figure S2A). By identifying the marker genes in clusters, we traced the gene expression characteristics of different tumor cell types. Based on the expression features of the astrocyte marker gene GFAP, mesenchymal GBM marker gene CHI3L1, proneural GBM marker gene OLIG2 and SOX6 [32], tumor cells could easily be distinguished. Furthermore, we identified a mass of cells with GFAP and CHI3L1 positivity that showed high expression levels of EGFR and platelet-derived growth factor subunit A (PDGFA). Therefore, according to the gene expression profiles and distribution patterns of those genes, tumor cells were grouped into 4 different cell populations: CP1 (GFAP+CHI3L1+EGFR+PDGFA+OLIG2−SOX6−), CP2 (GFAP+CHI3L1+EGFR−PDGFA−OLIG2−SOX6−), CP3 (GFAP−CHI3L1−EGFR−PDGFA−OLIG2+SOX6+), and CP4 (GFAP−CHI3L1−EGFR−PDGFA−OLIG2−SOX6−) (Figure 1B-C, Supplementary Figure S2B). Based on the expression profiles of GFAP, CHI3L1, OLIG2, SOX6, EGFR, and PDGFA, we divided the glioma patients from the CGGA [33], TCGA, and Rembrandt cohorts into 3 groups: Group G1 presented high expression of GFAP and CHI3L1, along with RTK pathway activation (high expression levels of EGFR and PDGFA genes); Group G2 had a similar gene expression profile to G1, except for the inactive RTK pathway (low levels of EGFR and PDGFA gene activations); Group G3 was characterized by high expression levels of OLIG2 and SOX6 (Supplementary Figure S3). Kaplan-Meier survival analysis revealed that patients with G1 characteristics had the shortest overall survival time, whereas patients with G3 features had the longest lifetime and the patients with G2 features exhibited a moderate survival time (Figure 1D, Supplementary Figure S4). Given that O-6-Methylguanine-DNA Methyltransferase (MGMT) plays a vital role in GBM chemoresistance via removing the TMZ-induced O6-methylguanine (O6-MeG) in DNA and has been recognized as an important predictor of the responsiveness to TMZ [34, 35], we performed subgroup analysis of the G1, G2, and G3 groups based on the expression level of MGMT and analyzed the overall survival curves. The results in Supplementary Figure S5A-C showed that there was no statistical difference between MGMTlow and MGMThigh subgroups in each group. It is well-known that the promoter methylation status is the primary reason to determine the expression level of MGMT in GBM [36]. Therefore, we resorted each group into 2 subgroups based on the promoter methylation status of MGMT (MGMTp) in TCGA and CGGA cohorts (There is no MGMTp data in Rembrandt cohort), and performed the Kaplan-Meier analysis. The results in Supplementary Figure S5D-E revealed that the overall survival time of MGMTpmethy and MGMTpunmethy in each group exihibited no statistical differences, although G2 + MGMTpunmethy subgroup had a shorter survival time compared to G2 + MGMTpmethy subgroup in the TCGA cohort. Taken together, these findings demonstrated that the above grouping strategy contributes to prognostic prediction of glioma patients through an MGMT-independent mechanism.
By identifying differentially expressed genes of each population (Figure 1E, Supplementary Figure S6, Supplementary Table S3), we noticed that acyl-CoA synthesis-associated genes (ACSS3, ACSL3, and ELOVL2) were enriched in the CP1 subgroup (Figure 1E, Supplementary Table S3), implying that RTK pathway activation in GBM could be accompanied by vigorous fatty acid biosynthesis and metabolism. The positive correlations among these genes were further verified using glioma cohorts in the CGGA, TCGA, and Rembrandt databases (Figure 1F, Supplementary Figure S7, Supplementary Table S4-S6). Because of the positive expression correlations and the great prognostic values of the aforementioned genes, we attempted to integrate their expression profiles to construct an RFA network for GBM patients for clustering and prognosis prediction. As shown in Supplementary Figure S8A, a higher RFA score was significantly associated with a higher tumor grade or malignancy. Kaplan-Meier curve showed a shortened overall survival time for patients with high RFA scores (Figure 1G, Supplementary Figure S8B). Through the univariate and multivariate Cox analyses, we found that the efficacy of the RFA score in prognosis prediction was similar to that of the conventional evaluation criteria, such as the WHO classification, isocitrate dehydrogenase (IDH) gene mutation status-based classification, or 1p/19q co-deletion classification (Supplementary Table S7-S9). In addition, the receiver operating characteristic (ROC) curve for RFA scores that stratified prognosis exhibited higher sensitivities (Supplementary Figure S8C).
3.3 The EGFR/AKT pathway upregulates the expression of ACSS3, ACSL3, and ELOVL2 by promoting the NF-κB phosphorylation
Given the positive correlation between EGFR and fatty acid metabolism-associated genes (Figure 1F, Supplementary Figure S7), we hypothesized that the expression of ACSS3, ACSL3, and ELOVL2 might be transcriptionally regulated by the EGFR/AKT pathway. To prove this hypothesis, we employed the GBM cell lines U-87 MG and primary GBM cell TBD0220 with an endogenous EGFR-vIII heterozygous mutation to interrogate the expression patterns of ACSS3, ACSL3, and ELOVL2 upon activation or inhibition of the EGFR/AKT pathway. As shown in Figure 2A, the expression levels of ACSS3, ACSL3, and ELOVL2 were significantly increased after the EGF stimulation or EGFR-vIII mutant overexpression in U-87 MG cells. Additionally, the AKT inhibitor MK-2206 also decreased the expression of these genes in TBD0220 cells. Studies have shown that EGFR/AKT pathway-driven NF-κB signaling activation plays an important role in the tumorigenesis and metastasis of GBM [37-39]. We found that NF-κB could be phosphorylated by the overexpressing EGFR-vIII mutant and dephosphorylated by MK-2206, indicating the regulatory associations among these genes (Figure 2B). The NF-κB inhibitor JSH-23 suppressed the expression of ACSS3, ACSL3, and ELOVL2 by decreasing the levels of phosphorylated NF-κB (p-NF-κB) in EGFR-vIII-overexpressing U-87 MG (U-87 MG-EGFR-vIII), and TBD0220 cells (Figure 2C). ChIP assays further demonstrated that the enrichment of p-NF-κB at these gene promoters was significantly increased upon activation of the EGFR/AKT pathway and decreased after MK-2206 treatment (Figure 2D-F). Collectively, these results suggested that the hyperactivated EGFR/AKT pathway can enhance the expression of the ACSS3, ACSL3, and ELOVL2 genes via NF-κB-dependent transcriptional activation.
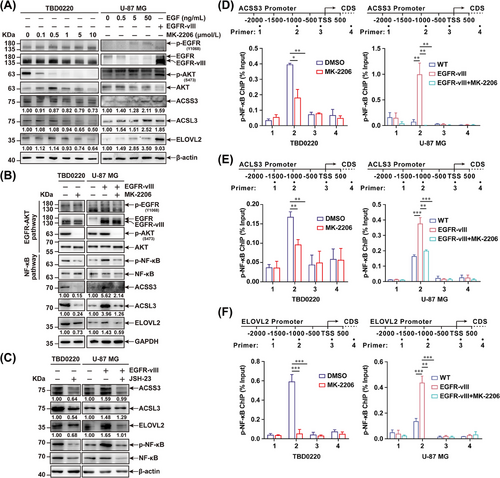
EGFR/AKT pathway activation enhances the expression of ACSS3, ACSL3, and ELOVL2 via NF-κB activation. (A) TBD0220 cells were treated with 0, 0.1, 0.5, 1, 5, and 10 μmol/L MK-2206 for 24 h. U-87 MG cells were stimulated with 0, 0.5, 5, and 50 ng/mL of EGF for 24 h, or overexpressed with EGFR-vIII mutant. The expression levels of EGFR, p-EGFR, AKT, p-AKT, ACSS3, ACSL3, ELOVL2, and β-actin were tested by western blotting. Protein was normalized to their respective β-actin loading control and expressions were quantified by ImageJ software. (B) Western blotting analysis of EGFR, p-EGFR, AKT, p-AKT, NF-κB, p-NF-κB, ACSS3, ACSL3, ELOVL2, and GAPDH expressions in TBD0220, U-87 MG and U-87 MG-EGFR-vIII cells treated with DMSO or 5 μmol/L of MK-2206 for 24 h. (C) Western blotting to check the expression levels of ACSS3, ACSL3, ELOVL2, NF-κB, p-NF-Κb, and β-actin in TBD0220, U-87 MG, and U-87 MG-EGFR-vIII cells treated with DMSO or 100 μmol/L of JSH-23 treatment. Protein was normalized to their respective β-actin loading control and expressions were quantified by ImageJ software. (D-F) TBD0220, and U-87 MG-EGFR-vIII cells were treated with DMSO or 5 μmol/L MK-2206 for 24 h. ChIP analysis of the regulatory regions of p-NF-κB binding to the promoters of ACSS3 (D), ACSL3 (E), and ELOVL2 (F). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 (independent-sample Student's t-test for TBD0220; one-way ANOVA for U-87 MG). Abbreviations: EGFR, epidermal growth factor receptor; p-EGFR, phosphorylation of epidermal growth factor receptor; EGF, epidermal growth factor; AKT, AKT serine/threonine kinase 1; GBM, glioblastoma; ANOVA, analysis of variance; ACSS3, acyl-CoA synthetase short-chain family member 3; ACSL3, acyl-CoA synthetase long-chain family member 3; ELOVL2, long-chain fatty acid elongation-related gene ELOVL fatty acid elongase 2.
3.4 Inhibition of the EGFR/AKT pathway represses ATP production, cell proliferation, and cell cycle progression of GBM cells in vitro
To further dissect the mechanism of energy metabolism reprogramming under the condition of EGFR/AKT pathway inhibition, we performed Seahorse analysis to evaluate the metabolic change after stimulation with MK-2206 in TBD0220 and U-87 MG cells. The results showed that the oxygen consumption rate (OCR) was increased in U-87 MG-EGFR-vIII cells compared with that of WT cells but decreased in both TBD0220 and U-87 MG-EGFR-vIII cells when treated with MK-2206, when compared with the control cell status (Figure 3A-B). The changes in the basal respiration, proton leak, and ATP production rates were in agreement with OCRs for those cell lines (Figure 3A-B). Subsequently, we tried to investigate whether the EGFR/AKT pathway induced energy metabolism remodeling via regulating ACSS3, ACSL3, and ELOVL2 expressions. Depleting ACSS3, ACSL3, and ELOVL2 expression respectively or together decreased OCR, basal respiration, proton leak, and ATP production rates in TBD0220 and U-87 MG-EGFR-vIII cells, compared with cells transfected siNC (Supplementary Figure S9). Overexpression of ACSS3, ACSL3, and ELOVL2 alleviated the inhibitory energy metabolism and ATP production by MK-2206 in TBD0220 and U-87 MG-EGFR-vIII cells (Supplementary Figure S10). Moreover, the results of the ATP assay demonstrated an increased ATP production in EGFR-vIII-overexpressing U-87 MG cells, but ATP production was significantly decreased upon MK-2206 treatment in TBD0220 and U-87 MG-EGFR-vIII cells (Figure 3C). Cell growth and colony formation assays showed that EGFR/AKT pathway activation could improve the number of colonies and the rate of GBM cell proliferation, while those effects were reversed in MK-2206 treated cells (Figure 3D-E, Supplementary Figure S11). Supplementing ATP could partially rescue the proliferation inhibition caused by MK-2206 (Figure 3D-E). The cell cycle distributions of GBM cells under different treatment conditions were monitored by flow cytometry analysis, revealed an increasing number of G0/G1 cells transitioning into the S phase when the EGFR/AKT pathway was hyperactivated, and an inverse effect was observed after stimulation with MK-2206 (Figure 3F, Supplementary Figure S12). The expressions of cell cycle-associated proteins, such as CDK2, CDK4, CDK6, and Cyclin D, as well as the phosphorylation of RB, presented a significantly increased change in EGFR/AKT pathway activation, which could be diminished by the MK-2206 (Figure 3G).
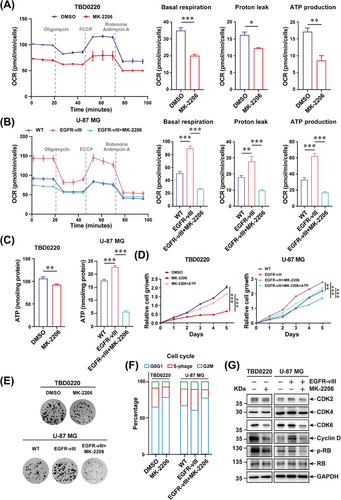
EGFR/AKT pathway regulates mitochondrial respiration and proliferation in GBM cells. (A-B) TBD0220 (A), and U-87 MG-EGFR-vIII cells (B) were treated with DMSO or 5 μmol/L MK-2206 for 24 h. The mitochondrial functions were monitored by Seahorse XF Cell Mito Stress test. The OCR, basal respiration, proton leak, and ATP production rates were measured as illustrated. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 (independent-sample Student's t-test for TBD0220; one-way ANOVA for U-87 MG). (C) ATP levels in TBD0220, U-87 MG, and U-87 MG-EGFR-vIII cells were analyzed after 24 h of treatments with DMSO or 5 μmol/L of MK-2206. ∗∗P < 0.01, ∗∗∗P < 0.001 (independent-sample Student's t-test for TBD0220; one-way ANOVA for U-87 MG). (D) The cell growth assay for TBD0220, U-87 MG, and U-87 MG-EGFR-vIII lines treated with DMSO, 5 μmol/L MK-2206, or 5 μmol/L MK-2206 plus 50 μmol/L ATP were performed. ∗∗∗P < 0.001 (two-way ANOVA). (E) The colony formation assay of TBD0220, U-87 MG, and U-87 MG-EGFR-vIII lines treated with DMSO or 1 μmol/L MK-2206. (F) Cell cycle distributions were analyzed by flow cytometry in TBD0220, U-87 MG, and U-87 MG-EGFR-vIII cells treated with DMSO or 5 μmol/L MK-2206. (G) Western blotting to show changes in expressions of CDK2, CDK4, CDK6, Cyclin D, RB, p-RB, and GAPDH in TBD0220, U-87 MG, and U-87 MG-EGFR-vIII cells treated with DMSO or MK-2206. Abbreviations: EGFR, epidermal growth factor receptor; AKT, AKT serine/threonine kinase 1; GBM, glioblastoma; ANOVA, analysis of variance; DMSO, dimethyl sulfoxide; CDK2, cyclin-dependent kinase 2; CDK4, cyclin-dependent kinase 4; CDK6, cyclin-dependent kinase 6; OCR, oxygen consumption rate.
In summary, these results suggested that the EGFR/AKT pathway may contribute to the energy metabolism reprogramming, cell proliferation, and cell cycle progression in GBM cells, which could be blocked by treating these cells with the AKT inhibitor MK-2206.
3.5 Hyperactivation of the EGFR/AKT pathway promotes energy metabolism by elevating fatty acid metabolism and cholesterol levels in GBM
The RTK signaling pathway, especially the EGFR/PI3K/AKT axis, promotes fatty acid biosynthesis and lipogenesis in GBM [12]. Considering the effect of transcriptional regulation of the EGFR/AKT pathway on the gene expression profiles of ACSS3, ACSL3, and ELOVL2, we noticed that EGFR/AKT pathway activation in GBM cells could induce the fatty acid metabolism-associated metabolic reprogramming. To investigate the metabolic characteristics of GBM cells with hyperactivated EGFR/AKT pathway, we obtained 66 fresh samples from GBM patients to interrogate the metabolites and transcriptome features. The conjoint analysis illustrated that most of the detected phosphatidylcholines (PCs) were positively correlated with increased expressions of EGFR, ACSS3, ACSL3, and ELOVL2, whereas lysophosphatidylcholines (LysoPCs) were negatively related to the activation of these genes. Other metabolites, such as phosphatidylethanolamine (PE), phosphatidyl glycerol (PG), phosphatidylinositol (PI), and phosphatidylserine (PS), had no significant changes in their levels in samples with differentially expressed fatty acid metabolism-associated genes (Figure 4A-B). Furthermore, we showed that the levels of medium and long-chain fatty acids increased proportionally with the upregulation of EGFR expression in GBM samples (Figure 4C).
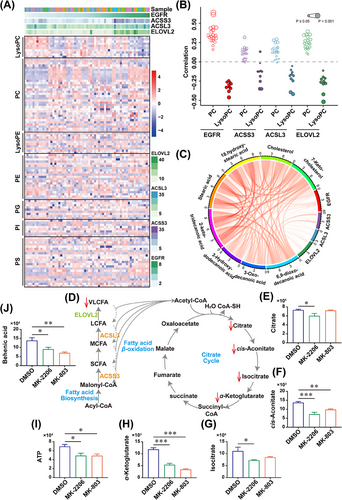
The hyperactivated EGFR/AKT pathway correlates with phospholipid metabolism and accelerated cholesterol biosynthesis in GBM cells. (A) A total of 66 GBM samples were analyzed using RNA sequencing and untargeted metabolomics. The expressions of EGFR, ACSS3, ACSL3, ELOVL2 and corresponding metabolites such as LysoPC, PC, LysoPE, PE, PG, PI, and PS were analyzed. (B) Untargeted metabolomic analysis of PC and LysoPC correlated with the expressions of EGFR, ACSS3, ACSL3, and ELOVL2 in GBM samples. (C) The long-chain fatty acids and cholesterol levels were positively correlated with EGFR, ACSS3, ACSL3, and ELOVL2 expressions in GBM samples. (D) The schematic diagram of fatty acid beta-oxidation and TCA cycle. (E-J) The levels of crucial intermediates in the TCA cycle, such as citrate (E), cis-aconitate (F), isocitrate (G), α-ketoglutarate (H), ATP (I), and VLCFA behenic acid (J) were downregulated in TBD0220 cells by 24 h of treatment with DMSO, 5 μmol/L MK-2206 or 5 μmol/L MK-803. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 (One-way ANOVA). Abbreviations: EGFR, epidermal growth factor receptor; AKT, AKT serine/threonine kinase 1; GBM, glioblastoma; ANOVA, analysis of variance; ACSS3, acyl-CoA synthetase short-chain family member 3; ACSL3, acyl-CoA synthetase long-chain family member 3; ELOVL2, long-chain fatty acid elongation-related gene ELOVL fatty acid elongase 2; TCA, citric acid cycle; VLCFA, very long-chain fatty acid; LysoPC, lysophosphatidylcholine; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidyl glycerol; PI, phosphatidylinositol; PS, phosphatidylserine.
It is well characterized that fatty acids can be catabolized by the beta-oxidation pathway to produce acetyl-CoA, which is an important component of the TCA cycle for ATP production [40] (Figure 4D). Hence, we applied a series of inhibitor assays to explore the effects of restraining the EGFR/AKT pathway on energy metabolism remodeling. Metabolite changes in TBD0220 cells were monitored upon stimulation with MK-2206. The results showed that compared with the DMSO control, multiple intermediates in the TCA cycle were decreased under MK-2206 treatment (Figure 4E-H), leading to reduced ATP production (Figure 4I). The level of behenic acid (C22: 0), a very long-chain fatty acid (VLCFA), was decreased in MK-2206 treated GBM cells (Figure 4J). Metabolism pathway enrichment analysis further revealed that differential metabolites were enriched in pathways associated with energy metabolism, for example, “central carbon metabolism in cancer”, “TCA cycle”, and “glycerophospholipid metabolism” (Supplementary Figure S13A), indicating the effect of the EGFR/AKT pathway modulation on the energy metabolism remodeling.
It was worth noting that elevated cholesterol levels were measured in GBM samples with higher EGFR expression (Figure 4C). Considering that catalytic conversion of acetyl-CoA to hydroxymethylglutaryl-CoA (HMG-CoA) is essential for both the mevalonate pathway and cholesterol biosynthesis, we attempted to explore the metabolic change in TBD0220 cells with mevalonate pathway inhibition by MK-803 treatment. LC-MS analysis showed that changes in the content of TCA cycle intermediates as well as behenic acid levels after MK-803 treatment were similar to those after MK-2206 stimulation (Figure 4E-J). MK-803-induced altered metabolites were enriched in the energy metabolism-associated pathways, such as “oxidative phosphorylation” and “central carbon metabolism in cancer”, which were comparable to the changes upon MK-2206 treatment (Supplementary Figure S13B).
Overall, these results suggested that EGFR/AKT pathway activation may increase the content of various fatty acids and cholesterol levels, conferring sthenic energy metabolism changes to GBM cells.
3.6 Blocking the mevalonate pathway reduces membrane-localized EGFR levels, affecting the EGFR/AKT signal transduction
Next, we attempted to elucidate the mechanistic involvement of the mevalonate pathway in the metabolic remodeling of GBM cells. Given that cholesterol acts as a regulator of cell proliferation and membrane homeostasis [41], we isolated the cytosolic contents and membrane components of TBD0220, U-87 MG, and U-87 MG-EGFR-vIII cells upon MK-803 treatment. Compared to the DMSO control, the population of either EGFR, or EGFR-vIII on the cell membrane was significantly decreased after stimulating GBM cells with MK-803, and cholesterol supplementation rescued the membrane-localized levels of EGFR or EGFR-vIII (Figure 5A). Then, we investigated the effect of MK-803 on the EGFR/AKT pathway by the selective ligand activation method. TBD0220, U-87 MG, and U-87 MG-EGFR-vIII cells were stimulated with different durations of EGF after MK-803 treatment, which showed a significantly weakened response to EGF and EGFR/AKT pathway transduction and a rapidly decreasing AKT phosphorylation level after EGF stimulation (Figure 5B). The expression of ACSS3, ACSL3, and ELOVL2 was suppressed in GBM cells treated with MK-803 treatment (Figure 5C).
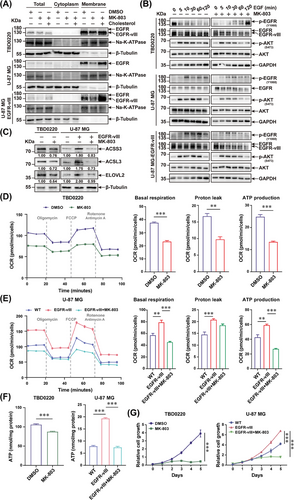
MK-803 inhibits EGFR/AKT pathway transduction via decreasing EGFR levels on the cell membrane. (A) The components of cytosol and membrane in TBD0220, U-87 MG, and U-87 MG-EGFR-vIII cells treated with DMSO, MK-803, or MK-803 plus 50 μmol/L water-soluble cholesterol were fractionated. The distribution levels of EGFR, Na-K-ATPase, and β-Tubulin were measured by western blotting. (B) TBD0220, U-87 MG, and U-87 MG-EGFR-vIII cells were stimulated by 50 ng/mL EGF at different time points after 24 h pre-treatment with DMSO or MK-803. Western blotting analysis was performed to detect the expression and phosphorylation levels of EGFR, and AKT. GAPDH was used as the loading control. (C) Western blotting analysis of ACSS3, ACSL3, ELOVL2, and β-Tubulin expressions in TBD0220, U-87 MG, and U-87 MG-EGFR-vIII cells treated with DMSO or 5 μmol/L MK-803 for 24h. Proteins were normalized to their respective β-Tubulin loading control and expressions were quantified by ImageJ software. (D-E) TBD0220 (D), and U-87 MG-EGFR-vIII cells (E) were treated with 5 μmol/L of MK-803 for 24 h. The mitochondrial functions in these cells were monitored by Seahorse XF Cell Mito Stress assay. The OCR, basal respiration, proton leak, and ATP production rates were measured as illustrated. ∗∗P < 0.01, ∗∗∗P < 0.001 (independent-sample Student's t-test for TBD0220; one-way ANOVA for U-87 MG). (F) ATP levels in TBD0220, U-87 MG, and U-87 MG-EGFR-vIII cells were analyzed after 24 h of treatment with DMSO or 5 μmol/L MK-803. ∗∗∗P < 0.001 (independent-sample Student's t-test for TBD0220; one-way ANOVA for U-87 MG). (G) The proliferation rates of TBD0220, U-87 MG, and U-87 MG-EGFR-vIII cells were determined after treatment with DMSO or 5 μmol/L MK-803. ∗∗∗P < 0.001 (Two-way ANOVA). Abbreviations: EGFR, epidermal growth factor receptor; AKT, AKT serine/threonine kinase 1; ANOVA, analysis of variance; ACSS3, acyl-CoA synthetase short-chain family member 3; ACSL3, acyl-CoA synthetase long-chain family member 3; ELOVL2, long-chain fatty acid elongation-related gene ELOVL fatty acid elongase 2; DMSO, dimethyl sulfoxide; OCR, oxygen consumption rate.
Subsequently, we measured the effect of MK-803 on energy metabolism and cell proliferation in GBM cells. MK-803 treatment significantly decreased the OCR in TBD0220 cells and antagonized the increase in OCR by EGFR-vIII in U-87 MG cells (Figure 5D-E). Basal respiration, proton leak, and ATP production were significantly suppressed in GBM cells treated with MK-803 (Figure 5D-E). Significantly decreased intracellular ATP levels and inhibited cell proliferation rates were also observed in both TBD0220, and U-87 MG-EGFR-vIII cells after MK-803 treatment (Figure 5F-G).
Taken together, these results suggested that mevalonate pathway inhibition by MK-803 can reprogram energy metabolism and repress the proliferation of GBM cells by blocking EGFR/AKT signal transduction and the expression of fatty acid metabolism-associated genes.
3.7 Targeting the EGFR/AKT and mevalonate pathways enhances the antitumor effects of TMZ, contributing to the GBM growth suppression in vivo
TMZ is the first-line chemotherapy drug for the clinical treatment of malignant gliomas [42]. We assessed the TMZ sensitization effects on MK-2206 and MK-803 treatments in vivo. First, we evaluated the ability of MK-2206 and MK-803 to cross the blood-brain-barrier (BBB). TBD0220 cells were intracranially injected into nude mice to establish the GBM orthotopic model. The brain tissues were isolated for drug concentration measurement. As shown in Supplementary Table S10 and Supplementary Figure S14-S15, the tissue content of MK-2206 was 573.03 ± 156.98 ng/g, significantly superior to TMZ (8.84 ± 3.23 ng/g), while the concentration of MK-803 (4.35 ± 0.74 ng/g) was approximately half of that of TMZ. The above results indicated that MK-2206 and MK-803 had good BBB permeability. Subsequently, we administered chemotherapeutics to tumor-bearing mice by oral gavage, according to the experimental design shown in Figure 6A. Bioluminescence imaging data indicated that compared with the sham controls, single drug interruption of TMZ, MK-2206, or MK-803 could reduce the tumor growth in xenograft mice, among which TMZ had the most effective antitumor effect. Moreover, multidrug treatments exhibited a better tumor inhibition effect than single-drug therapy. The combination of TMZ, MK-2206, and MK-803 showed the best therapeutic effect on tumor-bearing mice (Figure 6B-C). Kaplan-Meier survival curve revealed that mice treated with a 3-drug regimen had the best prognosis (Figure 6D). H&E staining confirmed a significant decrease in the tumor burden in the treatment group compared with that of vehicle-treated sham mice, which was consistent with the bioluminescence results. The proliferative abilities of xenografts were suppressed after treatment based on the Ki67 expression levels in IHC assays. The expression of ACSS3, ACSL3, ELOVL2, and AKT phosphorylation were inhibited in GBM mice treated with either MK-2206, MK-803, or different drug combinations (Figure 6E, Supplementary Figure S16).
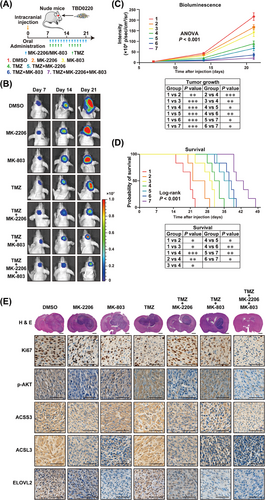
Targeting EGFR/AKT and mevalonate pathways suppresses GBM proliferation and prolongs the survival time of tumor-bearing mice. (A) The flow diagram of nude mice xenograft model. (B) Representative brain bioluminescence images of nude mice on Day 7, Day 14, and Day 21 after implantation. (C) Tumor growth curves were quantitated and illustrated. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 (two-way ANOVA). (D) Kaplan-Meier curves for different experimental and control groups. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 (log-rank test). (E) Representative images of FFPE brain tissues for H&E and IHC staining of Ki67, p-AKT, ACSS3, ACSL3, and ELOVL2. Scale bar = 1 mm for H&E, 50 μm for IHC. Abbreviations: EGFR, epidermal growth factor receptor; AKT, AKT serine/threonine kinase 1; GBM, glioblastoma; ANOVA, analysis of variance; FFPE, formalin-fixed and paraffin-embedded; ACSS3, acyl-CoA synthetase short-chain family member 3; ACSL3, acyl-CoA synthetase long-chain family member 3; ELOVL2, long-chain fatty acid elongation-related gene ELOVL fatty acid elongase 2.
3.8 Inhibition of the EGFR/AKT and mevalonate pathways remodels the immune microenvironment of GBM tumors
Given that enhanced fatty acid metabolism and cholesterol biosynthesis support GBM growth and immunosuppressive microenvironment formation [43-46], we explored the relationship between RFA score and immune cell metabolism by parsing sequencing data of glioma cohorts in the CGGA, TCGA, and Rembrandt databases. We found that the RFA score was positively correlated with immunosuppressors obtained from Wang et al. [47] (Figure 7A, Supplementary Figure S17A, Supplementary Figure S18A, Supplementary Table S11). We overlapped positively correlated genes with RFA from the CGGA, TCGA, and Rembrandt cohorts and visualized the top 10 genes. The results revealed that RFA scores were significantly correlated with Caspase 4 (CASP4), E74 Like ETS Transcription Factor 4 (ELF4), Leukocyte Associated Immunoglobulin Like Receptor 1 (LAIR1), LYN Proto-Oncogene, Src Family Tyrosine Kinase (LYN), Major Histocompatibility Complex, Class I-Related (MR1), Macrophage Scavenger Receptor 1 (MSR1), Nuclear Factor Kappa B Subunit 1 (NFKB1), Ras-Related Protein Rab-27A (RAB27A), Switch-Associated Protein 70 (SWAP70), and Transforming Growth Factor Beta 1 (TGFB1) expressions (Figure 7B, Supplementary Figure S17B, Supplementary Figure S18B). By employing single-sample gene set enrichment analysis (ssGSEA), we observed that patients with high RFA scores had a significantly strong correlation with immune cell lineages (Figure 7C, Supplementary Figure S17C, Supplementary Figure S18C). To further comprehensively investigate the RFA-associated immunological processes in GBM, we calculated the tumor purities of glioma cohorts using the ESTIMATE R package, which indicated negative associations between RFA scores and tumor purities, implying that GBM with high RFA score might have higher intra-tumor heterogeneity and more infiltrated immune cells (Figure 7D, Supplementary Figure S17D, Supplementary Figure S18D). The immune cell compositions of tumors were analyzed by the CIBERSORT algorithm. A high RFA score was correlated with increased M2 macrophage infiltration in CGGA and Rembrandt cohorts, which represented immunosuppression (Figure 7E, Supplementary Figure S17E, Supplementary Figure S18E). Taken together, our results demonstrated that RFA scores might provide valid evidence regarding the immune microenvironment status of GBM tumors.
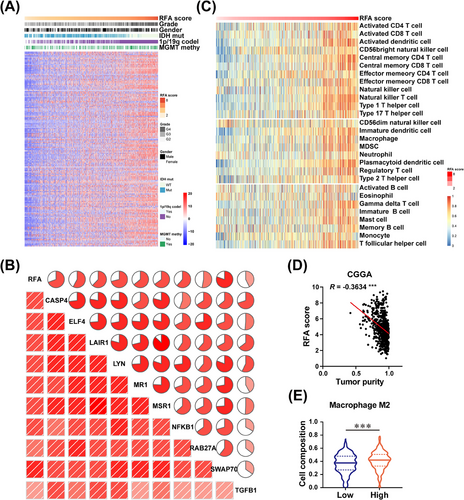
RFA score positively correlates with the immunosuppressive GBM microenvironment. (A) Expressions of immunosuppressors were positively correlated with respective RFA scores in glioma samples of the CGGA cohort. The clinical information of gender, tumor grade, IDH mutation status, 1p/19q co-deletion status, and MGMT promoter methylation status in GBM samples are displayed. (B) Correlations between RFA scores and the expressions of CASP4, ELF4, LAIR1, LYN, MR1, MSR1, NFKB1, RAB27A, SWAP70, and TGFB1 were analyzed in the CGGA cohort. (C) The ssGSEA for correlation of RFA scores with corresponding immune cell lineages in the CGGA cohort. (D) Tumor purity was negatively correlated with RFA scores in GBM samples of the CGGA cohort. ∗∗∗P < 0.001 (Pearson) (E) CIBERSORT analysis of immune cell compositions in GBM samples of the CGGA cohort. The M2 macrophage levels were directly correlated with RFA scores. ∗∗∗P < 0.001 (independent-sample Student's t-test). Abbreviations: GBM, glioblastoma; RFA, RTK-fatty acid-gene signature; CGGA, Chinese glioma genome atlas; MGMT, O6-methylguanine DNA methyltransferase; CASP4, caspase 4; ELF4, E74-like factor 4, LAIR1, leukocyte-associated Ig-like receptor 1; LYN, LYN Proto-Oncogene, Src family tyrosine kinase; MR1, major histocompatibility complex, class I-related; MSR1, macrophage scavenger receptor 1; NFKB1, nuclear factor kappa B subunit 1; RAB27A, RAB27A, member RAS oncogene family; SWAP70, switching B cell complex subunit; TGFB1, transforming growth factor beta 1; ssGSEA, single-sample gene set enrichment analysis.
To investigate the role of EGFR/AKT and mevalonate pathway inhibition in GBM microenvironment modulation in vivo, we utilized a syngeneic CT2A, a mouse-derived GBM cell line with Pten deficiency [48], to establish an intracranial tumor model in immunocompetent C57BL/6J mice. To apply the combinatorial therapeutic strategy to GBM therapy better and faster, the AKT inhibitor MK-2206 was changed to OSI, a BBB-permeable third-generation EGFR tyrosine kinase inhibitor (TKI) authorized for clinical non-small cell lung carcinoma (NSCLC) treatment [49, 50], and MK-803 was replaced by ATO, a commonly used cholesterol-lowering drug that inhibits the mevalonate pathway [51]. In vitro experiments demonstrated that OSI treatment reduced the expressions of ACSS3, ACSL3, and ELOVL2 via EGFR/AKT pathway blockade (Supplementary Figure S19), and decreased energy metabolism (Supplementary Figure S20), resulting in GBM proliferation inhibition and cell cycle arrest (Supplementary Figure S21), similar to MK-2206. Intertumoral distribution assays demonstrated a high level of OSI (1656.19 ± 61.32 ng/g) in the tumor tissues and a comparable drug concentration of ATO (9.22 ± 0.22 ng/g) to TMZ (Supplementary Table S10, Supplementary Figures S22-S23). The mice received different drug combinations via intragastric administration after CT2A intracranial injection (Figure 8A). Monitoring by bioluminescence imaging revealed that tumor growth was significantly decreased in the TMZ, TMZ + OSI, TMZ + ATO, and TMZ + OSI + ATO groups compared with the DMSO control group (Figure 8B-C). Furthermore, combined drug strategies had better tumor inhibition effects than TMZ alone. Notably, the TMZ + OSI + ATO regimen displayed the best antitumor outcome in this animal cohort (Figure 8C). Considering that ATO is mainly used against hyperlipidemia in the clinic, we developed a mouse model of high-fat diet (HFD)-induced hyperlipidemia in addition to these drug treatments. Compared to the chow diet as a control, the HFD showed a slightly increased tumor growth in the TMZ therapy group, whereas there was no significant difference in tumor growths between the HFD and chow diet animals receiving the TMZ + OSI + ATO regimen, suggesting that the effect of HFD could be suppressed by ATO treatment (Figure 8C). In both TMZ + OSI + ATO and TMZ + OSI + ATO + HFD treated tumors, the M1/M2 ratios of macrophages and tumor-infiltrating cytotoxic T lymphocytes were significantly increased (Figure 8D-F, Supplementary Figure S24, Supplementary Figure S25), implying an enhanced antitumor immune response.
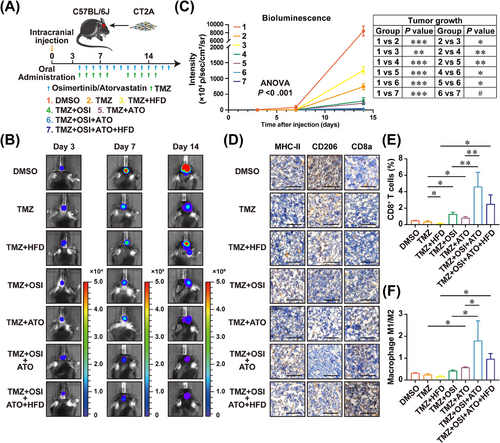
The combination therapeutic strategy of TMZ + OSI + ATO reduces tumor proliferation and improves GBM microenvironment. (A) The flow diagram of the C57BL/6J mice xenograft model is shown. (B) Representative brain bioluminescence images of C57BL/6J mice on Day 3, Day 7, and Day 14 post-implantation. (C) Quantitation of tumor growth curves. #not significant, ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 (two-way ANOVA). (D) IHC staining of MHC-II, CD206, and CD8a in FFPE tumor tissue sections. Scale bar = 100 μm. (E) Flow cytometry analysis to evaluate the infiltration rate of cytotoxic T-lymphocytes under different combination treatments of TMZ, OSI, and ATO. ∗P < 0.05, ∗∗P < 0.01 (one-way ANOVA). (F) Flow cytometry analysis to evaluate M1/M2 ratios of macrophages under different combination treatments of TMZ, OSI, and ATO. ∗P < 0.05 (one-way ANOVA).
Abbreviations: GBM, glioblastoma; TMZ, temozolomide; OSI, Osimertinib; ATO, Atorvastatin; MHC-II, major histocompatibility complex, class II; FFPE, formalin-fixed paraffin-embedded; ANOVA, analysis of variance.
To further verify the clinical effect of ATO on sensitizing GBM to TMZ, we examined GBM patients who had undergone standard treatments and follow-ups in Beijing Tiantan Hospital and Affiliated Hospital of Hebei University (Supplementary Figure S26A). It is worth noting that these patients routinely received ATO orally due to their history of hyperlipidemia. GBM patients who took statins orally every day had significantly prolonged OS and progression-free survival (PFS) compared to those without statins (Figure 9A-B). As the typical case (patient #1) showed, owing to the STUPP regimen and long-term TMZ treatment combined with ATO, PFS extended to 21 months, which was significantly longer than the average of GBM patients without statins (Figure 9B-C). H&E and Ki67 staining identified that EGFR/AKT signal activation represented by phosphorylation of EGFR and AKT was still at a low level, although the EGFR level in primary GBM tissue was significantly high. Moreover, these tumor tissue samples were weakly positive for ACSS3, ACSL3, and ELOVL2, as well as mild signals for CD8+ T cells and M2 macrophages, presumably due to patient #1 receiving oral ATO daily before the first diagnosis of GBM (Figure 9D). For comparison, the GBM tissue of patient #15, who was a GBM patient without hyperlipidemia and enrolled in the “without statins” group (Figure 9A-B), was used for IHC-staining. The results showed that p-EGFR and p-AKT staining displayed a significantly higher level compared to those of patient #1, indicating EGFR/AKT pathway hyperactivation in the GBM tissue of patient #15. The signals of ACSS3, ACSL3, and ELOVL2 were strongly positive in the tumor tissue sample of patient #15, as well as more M2 macrophage infiltration and few signals for CD8+ T cells, compared to those of patient #1 (Supplementary Figure S26B). In addition, transcriptomic analysis of paired samples of primary and recurrent GBM showed that RFA scores of recurrent samples were significantly increased, which supports the importance of statins co-therapy for PFS prolongation (Supplementary Figure S26C-D).
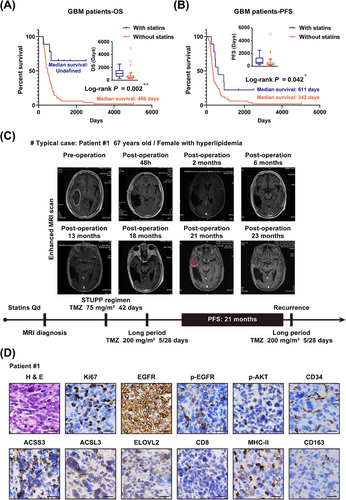
TMZ synergizes ATO's antitumor potency in clinical GBM treatment. (A-B) Kaplan-Meier curve of OS (A) and PFS (B) in GBM patients with statins (n = 9) or without statins (n = 51). ∗P < 0.05, ∗∗P < 0.01 (log-rank test). (C) Intracranial images of the typical GBM case from diagnosis to postoperative follow-ups by MRI and clinical examinations. (D) Representative images of FFPE sections of primary tumor tissues from patient #1 for H&E and IHC staining of Ki67, EGFR, p-EGFR, p-AKT, CD34, ACSS3, ACSL3, ELOVL2, CD8, MHC-II, and CD163. Scale bar = 20 μm.
Abbreviations: TMZ, temozolomide; ATO, Atorvastatin; GBM, glioblastoma; MRI, magnetic resonance imaging; FFPE, formalin-fixed paraffin-embedded; OS, overall survival; PFS, progression-free survival; EGFR, epidermal growth factor receptor; p-EGFR, phosphorylation of epidermal growth factor receptor; p-AKT, phosphorylation of AKT serine/threonine kinase 1; ACSS3, acyl-CoA synthetase short-chain family member 3; ACSL3, acyl-CoA synthetase long-chain family member 3; ELOVL2, long-chain fatty acid elongation-related gene ELOVL fatty acid elongase 2; MHC-II, major histocompatibility complex, class II.
Taken together, these in vivo results provided strong evidence that OSI-mediated EGFR/AKT pathway inhibition could enhance the tumor inhibitory effect of TMZ. Additionally, TMZ could synergize with the effect of OSI + ATO combination therapy, benefitting GBM patients to the greatest extent, especially for patients comorbid with hyperlipidemia.
4 DISCUSSION
GBM is one of the most malignant primary tumors in clinics, featured by high heterogeneity, resistance to conventional therapeutic strategies and easy relapse. Aberrant EGFR amplification and mutation is a typical feature of GBM, contributing to a more vicious phenotype. In this study, we demonstrated augmented EGFR expression accompanied with fatty acid metabolism-associated genes ACSS3, ACSL3, and ELOVL2 in GBM. RTK signaling pathway activated GBM had vigorous fatty acid metabolism and high cholesterol levels. MK-2206 and MK-803 decreased tumor proliferation via remodeling energy metabolism. Mechanically, inhibition of AKT phosphorylation and cholesterol biosynthesis reduced EGFR level on the cell membrane, affecting transduction of the EGFR/AKT signaling pathway. Targeting EGFR/AKT and mevalonate pathway enhanced the antitumor effect of TMZ.
Excessive RTK pathway activation caused by EGFR amplification and mutation is commonly observed in 57.4% of GBM subjects [9]. EGFR is known to facilitate SREBP1 cleavage to promote fatty acid synthesis [12]. In this study, we found that EGFR was co-expressed with ACSS3, ACSL3, and ELOVL2 by single-cell RNA sequencing, which was further verified by bulk sequencing / microarray analysis of the CGGA, TCGA, and Rembrandt databases. Furthermore, using dimensional reduction to improve clustering and marker gene expression characteristics in single-cell transcriptomic analysis and verification in public databases, we established a novel RFA signature with the companion diagnostics that indicated that GBM patients with high RFA scores had poor prognoses and short survival.
GBM tumor progression is always accompanied by metabolic abnormalities, including increased glucose uptake and lipid metabolism. Remodeling metabolic disorders may benefit GBM treatment. It has been demonstrated that blocking glucose-mediated SCAP glycosylation ameliorates EGFR-vIII-driven GBM growth [52]. Our previous research also showed that targeting PTRF/cPLA2 axis-mediated lipid metabolism reprogramming could suppress tumor growth and remodel the immune microenvironment in GBM by reversing abnormal LysoPC generation [53]. Importantly, we found that the EGFR pathway activation promoted vigorous fatty acid metabolism and a higher level of cholesterol, compared to the respective controls, through comprehensive metabolomic and transcriptomic analyses of GBM samples. The hyperactivated EGFR/AKT pathway upregulated ACSS3, ACSL3, and ELOVL2 expression levels via transcriptional regulation of NF-κB phosphorylation. Inhibiting the EGFR/AKT pathway, along with the mevalonate pathway, suppressed energy generation and tumor growth in vitro and enhanced the antitumor effect of TMZ in vivo. Inhibition of the mevalonate pathway could reduce membrane-localized protein levels, including glucose transporters.
EGFR translocation on mitochondria is reported to regulate mitochondria dynamics by interacting with MFN1 and disturbing MFN1 polymerization [54]. In gliomas, inhibition of EGFR translocation on mitochondria by iPA promotes PUMA-induced cell death [55]. These findings implied the correlation between mitochondria-localized EGFR and the balance of mitochondrial fission/fusion, high level of EGFR increased the number of mitochondria in tumor cells. In current study, we demonstrated that hyperactivation of the EGFR/AKT pathway contributed to energy production. Blockade of EGFR/AKT pathway inhibits mitochondrial function and reduces ATP production (Figure 3A-C, Supplementary Figures S9-S10) by decreasing the expression of ACSS3, ACSL3, and ELOVL2. Metabolomic data revealed that multiple intermediates in the TCA cycle and behenic acid were decreased under MK-2206 treatment (Figure 4D-J). Collectively, we speculate that a high level of EGFR/EGFR-vIII in GBMs might not only promote mitochondrial function by elevating the expression of ACSS3, ACSL3, and ELOVL2 but also increase the number of mitochondria by promoting mitochondrial fission, leading to vigorous energy production for tumor growth. Besides, we uncovered that mevalonate pathway inhibition suppressed EGFR/AKT pathway transduction by decreasing the level of membrane-localized EGFR in this study. Some studies have reported that ATO augments the antitumor effect of TMZ by inhibiting the prenylated Ras-mediated signaling pathway [56]. These studies suggest that statins may be involved in multiple crucial regulators of the EGFR signaling pathway. In addition, statins-induced decrease of EGFR on the cell membrane could be alleviated by supplement of cholesterol (Figure 5A), indicating that cholesterol-dependent membrane dynamics plays a vital role in EGFR/AKT pathway transduction. Given that mitochondrial membranes are composed of phospholipid bilayers, proteins, and cholesterol, statins treatment may affect the mitochondrial translocation of EGFR/EGFR-vIII, leading to mitochondrial dysfunction.
It is reported that cancer-derived succinate promotes macrophage polarization and malignant progression of tumors [57]. Our results confirmed that triple drugs treatment increased the rate of M1/M2 macrophages (Figure 8D and F), possibly because of the decrease of succinate levels caused by TCA cycle inhibition resulting from the blockade of the EGFR/AKT pathway. In addition, adenosine is reported as an important regulator produced by tumor cells and immune cells in tumor microenvironments that impair T cell-mediated antitumor response [58, 59]. CD39 and CD73, two ectonucleotidases expressed on the cell surface to catalyze extracellular ATP to adenosine, have been recognized as novel immune checkpoints that are involved in antitumor immune response [60]. The in vivo results found that combination treatment elevated the infiltration of CD8+ T cells in tumor tissues (Figure 8D-E), which might be due to the decreased level of adenosine caused by ATP production inhibition. These interesting hypotheses deserve further exploration and verification.
The antitumor effects of RTK pathway inhibitors have been verified in various cancers. For example, the first-generation TKI Lapatinib was approved by the U.S. Food and Drug Administration (FDA) to treat advanced breast cancer [61]. The first-generation EGFR-TKIs Erlotinib, Gefitinib, and the second-generation inhibitor Afatinib have been approved by FDA to administrate NSCLC patients [62-64]. In view of the vital roles of EGFR in GBM malignancy and metabolic remodeling, EGFR-TKIs should have potential inhibitory effects on tumor growth, but the therapeutic effects of the first-generation TKIs in clinical trials of GBM have not been satisfactory [65, 66]. This may be associated with the poor permeability of these drugs to the CNS [67]. A case report has shown that the third-generation EGFR-TKI OSI has a highly brain-penetrating ability and a good antitumor effect against GBM [68]. Consistently, we found that OSI enhanced the therapeutic effect of TMZ in combination therapy. Therefore, combined therapeutic strategies involving TMZ auxiliary to OSI and ATO could obtain a better treatment outcome.
Our in vitro and in vivo results demonstrated that statins could effectively inhibit energy production in GBM cells, and the inhibitory effect was more potent upon prolonging the treatment time. The morphology of statin-treated GBM cells exhibited stark changes (data not shown), indicating that the mechanism of action of statins was not only limited to modulating energy metabolism and the TCA cycle but also to the regulation of cholesterol-mediated cell membrane homeostasis. Statin-based treatments suppressed membrane-localized EGFR levels and EGFR/AKT pathway transduction in GBM cells. Cholesterol is essential for tumor survival. Recent studies have found that the major portion of cholesterol in GBM cells relies on exogenous uptake, which is different from that in normal brain tissues [13, 17]. Therefore, the ATO treatment also blocked the production and secretion of cholesterol by neurons and astrocytes, thereby inhibiting the uptake by glioma cells and facilitating the antitumor effect of ATO.
In addition, we found that a phase II clinical trial (NCT02029573) of ATO in combination with radiotherapy and TMZ for GBM was completed with negative results [69]. The study showed that the combination of ATO and conventional therapy could not prolong the PFS of GBM patients but found that high levels of LDLs were associated with poor prognosis. Our research has differences from the above. In this study, we first identified a metabolism-associated signature for GBM sorting through single-cell sequencing analysis. The signature contained EGFR and lipid metabolism pathway-associated genes. GBMs with a high signature score had a better effect on the combinational therapeutic strategy. In addition, 9 GBM patients (of 60 patients in total) included in this study had hypercholesterolemia and a high signature score (Figure 9, Supplementary Figure S26). They had to take statin drugs every day, even before the first diagnosis of GBM. We speculate that this would lay the metabolic foundation and provide the sensitization condition for the follow-up radiotherapy and chemotherapy (Figure 10).

Schematic illustration depicting the mechanism of hyperactivation of EGFR/AKT and mevalonate pathway promoting energy metabolism, leading to malignant progression of GBM. Combinational therapeutic strategy of TMZ, TKI, and statins benefited GBM patients.
Abbreviations: EGFR, epidermal growth factor receptor; AKT, AKT serine/threonine kinase 1; GBM, glioblastoma; TMZ, temozolomide; TKI, tyrosine kinase inhibitor.
This study has certain limitations which need to be carefully reviewed. We demonstrated that blocking the EGFR/AKT and mevalonate pathways could remodel tumor metabolic disorder and significantly inhibit GBM growth in both in vitro and in vivo settings. However, due to the clinical usage limitation of TKI, we were unable to collect samples from GBM patients receiving TMZ + OSI + ATO treatment at the same time. Therefore, whether this combinatorial therapeutic strategy can benefit clinical GBM patients, in general, needs further clinical investigations. Moreover, the liver damage caused by statins should also be carefully considered, especially for long-term treatment plans.
5 CONCLUSIONS
In summary, our study uncovered a regulatory mechanism by which EGFR/AKT pathway activation promotes energy metabolism by upregulating ACSS3, ACSL3, and ELOVL2 expressions in GBM cells. Inhibitors against the EGFR/AKT pathway and cholesterol synthesis could reverse disease phenotypes by regulating the expressions of fatty acid metabolism-associated genes and decreasing the level of membrane-localized EGFR. Furthermore, by employing multi-omics approaches to comprehensively analyze and utilize glioma data from multiple databases, we established a metabolism-associated RFA signature based on the degree of malignancy and metabolic characteristics of gliomas, which could predict the prognosis of patients. Furthermore, the combinatorial therapeutic strategy of TMZ, auxiliary EGFR-TKI, and statins can benefit GBM patients with high RFA scores by significantly prolonging their survival time.
DECLARATIONS
AUTHOR CONTRIBUTIONS
Chunsheng Kang, Chuan Fang, and Tao Jiang conceptualized and designed the study. Xiaoteng Cui, Jixing Zhao, Guanzhang Li, Chao Yang, Shixue Yang, Qi Zhan, Junhu Zhou, Yunfei Wang, Menglin Xiao, Biao Hong, Kaikai Yi, Fei Tong, Yanli Tan, and Qixue Wang conducted the experiments. Hu Wang, Tao Jiang, and Chuan Fang provided support with managing GBM clinical samples. Xiaoteng Cui, Jixing Zhao, and Guanzhang Li analyzed the data, generated the figures, and prepared the manuscript. All authors approved the final version of the manuscript.
ACKNOWLEDGMENTS
We are thankful to the patients who donated GBM samples to this study. We acknowledge Dr. Liang Xu from Shanghai IBS Biotech. Co., Ltd for supporting lentiviral reagents, Shanghai Genechem Co., Ltd for their support in the single-cell RNA sequencing and data processing, and Shanghai OE Biotech Co., Ltd for RNA sequencing and metabolomic analysis support.
CONFLICT OF INTERESTS STATEMENT
The authors declare that there is no conflict of interest in this study.
FUNDING INFORMTAION
This work was supported by grants from the National Natural Science Foundation of China (82002657, 82073322, 81761168038), the Hebei Natural Science Foundation Precision Medicine Joint Project (H2020201206), the Tianjin Key R&D Plan of Tianjin Science and Technology Plan Project (20YFZCSY00360), Brain Tumor Precision Diagnosis and Treatment and Translational Medicine Innovation Unit, Chinese Academy of Medical Sciences (2019-I2M-5-021), the Science and Technology Project of Tianjin Municipal Health Commission (TJWJ2021QN003), Key-Area Research and Development Program of Guangdong Province (2023B1111020008), and Multi-input Project by Natural Science Foundation of Tianjin Municipal Science and Technology Commission (21JCQNJC01250).
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Collection and analysis of all clinical GBM samples were approved by the medical ethics committee of Beijing Tiantan Hospital (Approval No. KY 2020-093-02) and Hebei University affiliated Hospital (Approval No. HDFY-LL-2020-017). All participants had signed the informed consents. All animal experimentations were approved by the Animal Ethical and Welfare Committee (Approval No. IACUC-2020XS001).
CONSENT FOR PUBLICATION
Not applicable.
Open Research
DATA AVAILABILITY STATEMENT
All sequencing and metabolomics data in this study are available from the lead contact, Chunsheng Kang ([email protected]), upon reasonable request.




