CD146 promotes malignant progression of breast phyllodes tumor through suppressing DCBLD2 degradation and activating the AKT pathway
Jiewen Chen and Qingji Xu authors contributed equally to this work.
Abstract
Background
As a rapid-progressing tumor, breast malignant phyllodes tumors (PTs) are challenged by the lack of effective therapeutic strategies and suitable prognostic markers. This study aimed to clarify the role and mechanism of CD146 on promoting PTs malignant progression, and to identify a novel prognosis marker and treatment target of breast malignant PTs.
Methods
The expression and prognostic significance of CD146 in PTs was detected through single-cell RNA-sequencing (scRNA-seq), immunostaining, real-time PCR and other methodologies. Functional experiments including proliferation assay, colony formation assay, transwell assay, and collagen contraction assay were conducted to validate the role of CD146 in malignant progression of PTs. The efficacy of anti-CD146 monoclonal antibody AA98 against malignant PTs was corroborated by a malignant PT organoid model and a PT patient-derived xenograft (PDX) model. Transcriptome sequencing, proteomic analysis, co-immunoprecipitation, and pull-down assay was employed to identify the modulating pathway and additional molecular mechanism.
Results
In this study, the scRNA-seq analysis of PTs disclosed a CD146-positive characteristic in the α-SMA+ fibroblast subset. Furthermore, a progressive elevation in the level of CD146 was observed with the malignant progression of PTs. More importantly, CD146 was found to serve as an independent predictor for recurrence in PT patients. Furthermore, CD146 was found to augment the viability and invasion of PTs. Mechanistically, CD146 acted as a protective “shield” to prevent the degradation of Discoidin, CUB, and LCCL domain-containing protein 2 (DCBLD2), thereby activating the phosphoinositide 3-kinase (PI3K)/protein kinase B (AKT) signaling pathway and enhancing malignant behaviors of PT cells. In the malignant PT organoid and PDX model, a significant suppression of malignant PT growth was observed after the application of AA98.
Conclusions
These findings suggested that CD146 served as an efficacious marker for predicting PT malignant progression and showed promise as a prognosis marker and treatment target of breast malignant PTs. The study further unveiled the essential role of the CD146-DCBLD2/PI3K/AKT axis in the malignant progression of PTs.
List of abbreviations
-
- PT
-
- phyllodes tumor
-
- α-SMA
-
- α-smooth muscle actin
-
- scRNA-seq
-
- single-cell RNA sequencing
-
- qRT-PCR
-
- quantitative real-time-polymerase chain reaction
-
- PDX
-
- patient-derived xenograft
-
- Co-IP
-
- Co-immunoprecipitation
-
- DCBLD2
-
- discoidin, CUB, and LCCL domain-containing protein 2
-
- PI3K
-
- phosphoinositide 3-kinase
-
- AKT
-
- protein kinase B
-
- MCAM
-
- melanoma cell adhesion molecule
-
- MUC18
-
- mucin 18
-
- CAM
-
- cell adhesion molecule
-
- PCA analysis
-
- principal component analysis
-
- uMAP
-
- uniform manifold approximation and projection
-
- DMEM/F12
-
- Dulbecco's modified eagle medium: F-12
-
- FBS
-
- fetal bovine serum
-
- EGF
-
- epidermal growth factor
-
- ATCC
-
- American type culture collection
-
- DMEM
-
- Dulbecco's modified Eagle medium
-
- IHC
-
- immunochemistry
-
- IF
-
- immunofluorescence
-
- HRP
-
- horseradish peroxidase
-
- DAB
-
- 3,3’-diaminobenzidine
-
- RIPA
-
- radio immunoprecipitation assay
-
- SDS-PAGE
-
- sodium dodecyl sulfate - polyacrylamide gel electrophoresis
-
- PVDF
-
- polyvinylidene fluoride
-
- MTS
-
- 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4- sulfophenyl)- 2h-tetrazolium
-
- siRNA
-
- small interfering RNA
-
- BSA
-
- bovine serum albumin
-
- mRNA
-
- message RNA
-
- KEGG
-
- Kyoto Encyclopedia of Genes and Genomes
-
- NCBI
-
- National Center for Biotechnology Information
-
- GEO
-
- Gene Expression Omnibus
-
- IgG
-
- immunoglobulin G
-
- ORF
-
- open reading frame
-
- PDGFRβ
-
- platelet derived growth factor receptor beta
-
- sgRNA
-
- single guide RNAs
-
- lipo3000
-
- lipofectamine 3000
-
- WB
-
- western blot
-
- PMSF
-
- phenylmethyl sulfonyl fluoride
-
- MWCO
-
- molecular weight cutoff
-
- AP-MS
-
- affinity purification–mass spectrometry
-
- SEM
-
- standard error of the mean
-
- KM
-
- kaplan-meier
-
- ROC
-
- receiver operator curve
-
- HSCs
-
- hematopoietic stem cells
-
- NK
-
- natural killer
-
- ACTA2
-
- actin alpha 2
-
- COL1A1
-
- collagen type I
-
- COL1A2
-
- collagen type II
-
- S100A4
-
- S100 calcium binding protein A4
-
- FAP
-
- fibroblast activation protein alpha
-
- VIM
-
- vimentin
-
- CAV1
-
- caveolin 1
-
- RGS5
-
- regulator of G protein signaling 5
-
- CCL2
-
- C-C motif chemokine ligand 2
-
- CCL8
-
- C-C motif chemokine ligand 8
-
- OS
-
- overall survival
-
- LRFS
-
- local recurrence-free survival
-
- AUC
-
- area under curve
-
- NC
-
- negative control
-
- CDK2
-
- cyclin-dependent kinase 2
-
- CDK4
-
- cyclin-dependent kinase 4
-
- N-Cad
-
- N-Cadherin
-
- E-Cad
-
- E-Cadherin
-
- GAPDH
-
- glyceraldehyde-3-phosphate dehydrogenase
-
- GSEA
-
- Gene Set Enrichment Analysis
-
- TNFα
-
- tumor necrosis factor-alpha
-
- NFκB
-
- nuclear factor kappa B
-
- mTOR
-
- mammalian target of rapamycin
-
- KRAS
-
- kirsten rat sarcoma viral oncogene homolog
-
- AGE
-
- advanced glycation end products
-
- RAGE
-
- AGEs-receptor
-
- ECM-receptor
-
- extracellular matrix receptor
-
- PRAS40
-
- the proline-rich AKT substrate of 40 KDa
-
- AP2B1
-
- adaptin-β2
-
- ARHGDIA
-
- Rho GDP dissociation inhibitor alpha
-
- ROBO1
-
- roundabout guidance receptor 1
-
- GJA1
-
- gap junction protein, alpha 1
-
- STX4
-
- syntaxin 4
-
- RAC1
-
- rac family small GTPase 1
-
- VDAC3
-
- voltage dependent anion channel 3
-
- MARCKS
-
- myristoylated alanine rich protein kinase C substrate
-
- EGFR
-
- epidermal growth factor receptor
-
- STK24
-
- serine⁄threonine kinase 24
-
- TIMER
-
- Tumor IMmune Estimation Resource
-
- CCLE
-
- Cancer Cell Line Encyclopedia
-
- IP
-
- immunoprecipitation
-
- CHX
-
- cycloheximide
-
- SEMA4B
-
- Semaphoring 4B
-
- CSC
-
- cancer stem cell
-
- NRAS
-
- neuroblastoma-RAS
-
- CCL18
-
- C-C motif chemokine ligand 18
-
- TAMs
-
- tumor-associated macrophages
-
- RTK
-
- receptor tyrosine kinase
-
- VEGFR2
-
- vascular endothelial growth factors receptor 2
1 BACKGROUND
Breast phyllodes tumors (PTs) are pathologically classified as benign, borderline or malignant tumors. Malignant PTs are clinically notorious for their high recurrence and distant metastasis rates. In addition, adjuvant treatments like chemotherapy or radiotherapy yield minimal effectiveness against these malignant PTs [1]. Wide excision remains the primary therapeutic strategy to mitigate recurrence and metastasis. However, even with surgical intervention, local recurrence rates stagger between 18% and 65% [2, 3], the distant recurrence rate of malignant PTs can reach a stark 21%-30% [4]. Patients with distant metastasis suffer from rapid progression and face significantly high mortality rates. Therefore, novel therapeutic strategies against breast PTs, as well as biomarkers for predicting prognosis of breast PTs are warranted.
Unlike breast cancer, PTs are fibroepithelial tumors characterized by a lesser nonmalignant epithelial component and a more abundant spindle stromal cell population [5]. These stromal cells serve as the key determinants for pathologic classification [5]. The features and functions of the spindle stromal cells, however, remain unexplored. Previous research suggested that fibroblasts, especially α-smooth muscle actin-positive (α-SMA+) fibroblasts, are a major component of PTs and contribute significantly to the recurrence or metastatic behavior of PTs [6]. However, how α-SMA+ fibroblasts function on the malignant progression of PTs remains unknown.
CD146, also known as melanoma cell adhesion molecule (MCAM), melanoma-cell adhesion molecule (Mel-CAM), and mucin 18 (MUC18), belongs to the immunoglobulin superfamily and is a cell adhesion molecule (CAM), that found abundantly in malignant tumors [7]. Previous studies have proved CD146 as a stromal cell marker that defines fibroblast subtypes within the hematopoietic stem cell (HSC) niche [8]. It shows a positive correlation with α-SMA [9, 10]. However, the involvement of CD146 in PT development and its potential as a prognostic marker or therapeutic target for PTs remains to be further investigated. This study aimed to explore the clinical significance of CD146 and unveil its regulatory role and underlying molecular mechanism in PT progression.
2 MATERIALS AND METHODS
2.1 Patients and tissue samples
Breast PT tissues were sourced from 298 patients with 146 benign, 90 borderline, and 62 malignant PTs, among which samples consisted of 7 benign, 3 borderline, and 7 malignant PTs were utilized for single-cell RNA-sequencing (scRNA-seq, detailed information of donors seen in Supplementary Table S1). Fibroadenoma tissues were obtained from 4 patients with fibroadenoma. These patients were treated at the Breast Tumor Center, Sun Yat-sen Memorial Hospital, Sun Yat-sen University (Guangzhou, Guangzhou, China) between April, 2006 and February, 2020. Inclusion criteria were as follows: (1) breast PTs (including benign, borderline and malignant cases) and fibroadenoma; (2) treatment with surgical resection; (3) pathologically confirmed breast PTs or fibroadenoma by two independent pathologists; and (4) agreement for the use of surgical specimens for research purposes. Exclusion criteria were as follows: (1) diagnosed with other breast tumor such as primary breast ductal carcinoma; and (2) rejection from patients for the use of their surgical specimens for research purposes. The study was approved by the Ethics Committee of Sun Yat-sen Memorial Hospital Institutional Review Board. Informed consent was obtained from all participants. Overall survival (OS) was defined as the duration from initial diagnosis to the date of death from any cause or the most recent follow-up. Local recurrence-free survival (LRFS) was defined as the timeframe from the initial diagnosis to the date of local disease recurrence or the most recent follow-up.
2.2 scRNA-seq
Following the fresh surgical resections, PT tissues were segmented and subjected to digestion in Dulbecco's Modified Eagle Medium/F-12 (DMEM/F12, Gibco, Waltham, MA, USA) alongside collagenase IV (1 mg/mL, Gibco) for a duration of 60 min at a temperature of 37°C. Post-digestion, samples were filtered using a 100 μm cell strainer before undergoing in red blood cell lysis buffer (Dakewe, Beijing, China) to facilitate erythrocyte removal. A dead cell removal kit (130-090-101, Milteny, Bergisch Gladbach, Germany) was utilized to eliminate dead cells, and remaining cell underwent scRNA-seq analysis.
Gene expression profiles were generated by processing the raw reads using the Cell Ranger toolkit (version 2.1.1; 10× Genomics, Shanghai, China). The raw base call (BCL) files were converted into FASTQ files via the “mkfastq” command. Post-extraction of cell barcodes and unique molecular identifiers (UMIs), adapters and poly A tails were excised. Subsequently, Read 2 alignment was performed with the GRCh38 Ensemble build 92 genome (fastp 2.5.3a and featureCounts 1.6.2). Reads sharing cell barcode, UMIs, and gene were grouped for UMIs per gene per cell using the “count” command. The resulting UMI count tables provide the basis for further analysis.
The raw output data underwent individual processing using the Seurat package (version 3.1.5; http://satijalab.org/seurat/) within the R software (version 3.6.1). Cells expressing less than 200 genes or with over 15% mitochondrial genes were filtered out. A total of 33,682 filtered cells were considered suitable for further bioinformatic analysis.
Post-processing, the Seurat object was subjected to the Read10× () function. For each sample, gene expression was adjusted to mirror the gene fraction, multiplied by 10,000, and then transformed into a natural logarithm after an increment of 1 to circumvent the mathematical error of taking a log of 0. The top 3,000 highly variable genes (HVGs) were identified, centered, and scaled before the principal component analysis (PCA). The Harmony package (version 1.0) was employed to mitigate batch effects, considering the top 50 PCA components.
Differential over-expressed genes in specific cluster were identified by comparing to other clusters using the Wilcoxon Rank-Sum Test with the FindMarkers function in Seurat (adjusted P <0.05, only. pos = T and log fc. threshold = 0.1). The cluster Profiler package (version 3.14.3) was used to calculate the cluster-specific overrepresented GO biological process. Lately, data visualization was achieved using the Uniform Manifold Approximation and Projection (UMAP).
Gene Set Enrichment Analysis (GSEA) was conducted using curated gene sets to discern pathways that were either induced or repressed in the fibroblast's clusters distinguished by low and high CD146 expression levels. Specifically, the average gene expression level was calculated, and the logarithmic 2-fold change (FC) was used as the test statistic, contrasting the specific fibroblasts cluster against other cells. The 50 hallmark gene sets derived from the Molecular Signatures Database (MSigDB) (accessing from https://www.gsea-msigdb.org/gsea/msigdb) for the GSEA analysis.
2.3 Cell culture
Benign PT cell lines (SYSH-BPT-01) and malignant PT cell lines (SYSH-MPT-01, 02, 03, and 04) were established from primary benign and malignant PT cells isolated from PT tissues. Both cells types were maintained in DMEM/F12 (Gibco) supplemented with 15% fetal bovine serum (FBS; Gibco), 10 μg/mL insulin (Sigma, Burlington, MA, USA), 0.5 mg/mL hydrocortisone (Sigma), 20 ng/mL EGF (Peprotech, Cranbury, NJ, USA), and 100 U/mL penicillin-streptomycin (Gibco) at 37°C in 5% CO2 environment [11]. The HEK293T cell line, purchased from American Type Culture Collection (ATCC, Manassas, Virginia, USA), was cultured in Dulbecco's modified Eagle medium (DMEM; Gibco) supplemented with 10% FBS at 37°C in 5% CO2. The HEK293T cell line was utilized to further verify the mechanism being studied.
2.4 Immunohistochemistry (IHC) and immunofluorescence (IF)
PT tissues from patients and mice were fixed in 10% neutral formalin, embedded in paraffin, and subsequently sectioned. For IHC, sections were deparaffinized with xylene, hydrated with a gradient of alcohol, and subjected to heat-induced antigen retrieval with EDTA (pH8.0; Biosharp, Anhui, Hefei, China). These antigen-retrieval sections were blocked with 10% goat serum (Gibco), and incubated sequentially with primary antibody (4°C overnight), biotin-conjugated secondary antibodies (Zhongshanjinqiao, Beijing, China) at room temperature for 20 min, and horseradish peroxidase (HRP)-conjugated streptavidin (Zhongshanjinqiao) at room temperature for 20 min. The staining was visualized using 3,3’-diaminobenzidine (DAB, Zhongshanjinqiao). The staining index (SI), calculated as staining intensity (grade 1-4) × staining density (grade 1-4) for 5 random fields, was assessed (scale bar: 100 μm). For IF, the blocked sections were incubated with mouse monoclonal anti-CD146 (Abcam, Cambridge, MA, USA) and rabbit monoclonal anti-αSMA (Abcam) at 4°C overnight, and visualized using fluorescent dye-conjugated secondary antibodies (Invitrogen, Danvers, MA, USA). After staining nuclei with 4′,6- diamidino-2-phenylindole (DAPI), the positive signal was examined by confocal microscopy. Detailed information on the antibodies used in this study can be found Supplementary Table S2.
2.5 Western blot (WB) analysis
Tissue and cell line protein samples were extracted by Radio Immunoprecipitation Assay (RIPA) buffer (Beyotime, Shanghai, China) incubation with protease inhibitors (CWBIO, Taizhou, Jiangsu, China) and phosphatase inhibitors (CWBIO). Protein samples (30 μg) were separated by 10% sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE), transferred to PVDF membranes (Millipore, Burlington, Massachusetts, USA), blocked with 5% nonfat milk (Biosharp), incubated with primary antibodies, and sequentially incubated with Horseradish Peroxidase (HRP)-conjugated secondary antibodies (Proteintech, Sankt Leon-Rot, Opelstraße 1, Germany). Enhanced Chemiluminescence (ECL) (Thermo Fisher, Waltham, MA, USA) was used for visualization. Information on the antibodies used in this study is provided in Supplementary Table S2.
2.6 Quantitative Real-time PCR (qRT-PCR)
Tissue and cell line RNA was isolated using TRIzol (Invitrogen) [12]. Reverse transcription was performed by using reverse transcription kit (ABM, BC, Canada). SYBR qPCR kit (Vazyme, Nanjing, Jiangsu, China) were used for qRT-PCR. Primer sequences are shown in Supplementary Table S3.
2.7 Proliferation assay
Proliferation of PT cells was measured by a 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2h-tetrazolium (MTS) assay. PT cells (2 × 103/well, 200 μL) were seeded into 96-well plates. 10 μL 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (Promega, Madison, WI, USA) was added to each well and incubated for 2 h at 37°C in the dark according to the manufacturer's instructions. After incubation, the count of proliferating cells was confirmed by measuring the absorbance at 492 nm.
2.8 Transwell assay
Transwell chambers (3422, Corning, Bedford, Massachusetts, USA) were used for transwell experiments. A total of 1 × 104 cells maintained in serum-free medium were seeded into the transwell chambers with (for the invasion assay) or without (for the migration assay) 20% Matrigel (354234, Corning) seeded in advance. Medium supplemented with 10% FBS was added to the lower chambers. Migratory and invasive cells were stained with crystal violet after 24 h of culture and counted in 5 randomly selected fields (scale bar: 100 μm).
2.9 Collagen contraction assay
A sterile solution of acid-soluble collagen type I (Corning) was prepared for the collagen contraction assay [6]. Benign and malignant PT cells were suspended in the collagen solution. The collagen/cell mixture (1:1) was seeded into 24-well plates coated with 1% Bovine Serum Albumin (BSA) and polymerized at 37°C for 30 min. Subsequently, 1 mL of medium was added to each plate and cultured for 36 h. Collagen contraction of PT cells was determined by measuring the area of the gel surface.
2.10 Establishment of PTs organoid
Fresh tumor tissues from malignant PT patients were procured, minced into 1-2 mm pieces, and subjected to enzymatic digestion with 1 mg/mL collagen III (Worthington, Aberdeen, MD, USA) at 37°C for 2 h. Post-digestion, DMEM/F12 containing 10% FBS was added to the mixture which was further centrifuged at 300 × g for 10 min and filtered through a 70 μm sieve. The filtered mixture was centrifuged again, and the pellets were resuspended in PT-specific medium (DMEM/F12 containing 15% FBS, 10 μg/mL insulin, 0.5 mg/mL hydrocortisone, 20 ng/mL EGF, and 100 U/mL penicillin-streptomycin, 2 × 106 cells/mL). The suspension was then embedded in 50 μL Matrigel domes and plated in 96-well plates (174929, Thermo Fisher) and incubated at 37°C for 15 min. After this incubation period, 200 μL of PT-specific medium was added. The medium was replaced every 3-4 days, and the diameter of the organoid was measured weekly.
2.11 Transcriptome sequencing and analysis
Total RNA in cells from different treatment groups (benign PT cells with or without CD146 overexpression; malignant PT cells with or without CD146 silencing) was obtained using Trizol (Invitrogen). Transcriptome sequencing was applied to analyze the expression profiling of human transcripts in the samples and performed on BGISEQ machines (MGI, Shenzhen, Guangdong, China) using the standard RNA sequencing protocol with a read length of 50 bases. The raw data was filtered with SOAPnuke (v1.5.2) followed by quantifying by Salmon (v1.90) in mapping-based mode according to human reference genome GRCh38 and the Ensembl release version 74 gene annotation. To compare expression between genes within samples, the data was imported to tximport (v1.24.0) and summarized abundances, counts and lengths to gene-level. Finally, the gene counts were normalized and differentially expressed genes were identified by EdgeR (v3.84.4). mRNA expression analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG; https://www.genome.jp/kegg/) pathway enrichment analysis were further performed to screen the differential genes and putative pathways. The criteria for screening of differentially expressed mRNA were FC >1.41 in the benign PT cell line overexpressing CD146 and FC <0.71 in the malignant PT cell line silencing expression of CD146 while an adjusted P < 0.05. Raw sequences were deposited in the NCBI GEO database (https://www.ncbi.nlm.nih.gov/geo) under the accession number GSE227380.
2.12 Gene expression correlation analysis
The gene expression data of all types of cancers were explored in the Tumor IMmune Estimation Resource (TIMER; https://cistrome.shinyapps.io/timer) for The Cancer Genome Atlas (TCGA; https://www.cancer.gov/ccg/research/genome-sequencing/tcga) or Dependency Map (depmap portal; https://depmap.org/portal/) for the Cancer Cell Lines encyclopedia (CCLE; https://sites.broadinstitute.org/ccle/). The relationship between CD146 and DCBLD2 was calculated using website's default parameter.
2.13 In vitro therapeutic anti-CD146 antibody application
SYSH-MPT-02 cells were treated with 150 μg/mL IgG or therapeutic anti-CD146 antibody (AA98; in-house) for 16 h. Post-treatment, SYSH-MPT-02 cells were harvested for protein extraction or functional assays.
2.14 PT patient-derived xenograft (PDX) model and in vivo therapeutic anti-CD146 antibody application
Six-week-old female NOD/SCID mice were procured from Shanghai Model Organisms Center, lnc. (Shanghai, China) and housed at the Guangdong Laboratory Animals Monitoring Institute (Guangzhou, Guangdong, China). PT tissue was procured from 3 patients with malignant PT (information seen in Supplementary Table S4) and minced into 1 mm pieces. These minced tumor tissues were seeded into the fat pad (left flank) of NOD/SCID mice. One week later, mice with similar tumor size were divided into 4 groups of 6 mice each. The first group was treated with PBS by intraperitoneal injection for 24 days (twice a week). The second group was treated with IgG (Sigma) by intraperitoneal injection for 24 days (0.25 mg/mice, twice a week). The third group was treated with therapeutic anti-CD146 antibody (AA98) by intraperitoneal injection for 24 days (0.25 mg/mice, twice a week). The tumor size was measured every 4 days. Tumor volumes were calculated in accordance with the equation Volume = (length × width2)/2. At the endpoint, mice were euthanized by CO2 inhalation and PDX tissues were collected for weighing and further analysis. All animal experiments were approved by the Animal Care and Use Committee of Sun Yat-sen University.
2.15 Plasmids, small interfering RNA (siRNA) construction and transfection
The pcDNA3-CD146 plasmid (in-house) was utilized to overexpress CD146 in benign PT cell lines. Briefly, cells (1 × 105 cells/well) were incubated with Viafect (Promega)/plasmid complex for 12 h. After this period, cells were washed with PBS and cultured in a medium with FBS for another 48 h.
For CD146-TurboID or MEM-TurboID plasmid constructs, the CD146 encoding cDNA or PDGFRβ transmembrane domain which C-terminal tagged with linker/BirA*/Flag was synthesized and cloned into the pCDH-CMV-MCS-Puro (provided by Dr. Pengyuan Yang) by seamless assembly strategy.
For single guide RNA (sgRNA) plasmid constructs, sgRNA targeting CD146 were meticulously designed and curated using the Benchling platform (https://benchling.com/). These sgRNA were then assembled and integrated into the LentiCRISPR v2 plasmid (Addgene, Watertown, MA, USA), adhering to protocols established by the Feng Zhang laboratory [13].
In order to transiently suppress the expression of CD146 in malignant PT cell lines, siRNA was employed. Briefly, a concise procedure involved incubating the cells with complexes of lipo3000/siCD146-1, lipo3000/siCD146-2 or lipo3000/siCD146-3 complex (Invitrogen) for 12 h. Following this incubation, the cells were rinsed with PBS and subsequently cultured in medium fortified with FBS for an additional 48 h. The sequences of the RNAs are provided in Supplementary Table S3 for further reference.
2.16 Viral transduction and CD146 knock out stable cell line generation
Lentiviral vector encoding CD146 sgRNA, together with psPAX2 and pMD2.G packaging plasmids (Addgene), were transfected into HEK293T cells at ∼60%-70% confluence using Lipofectamine 2000 (Invitrogen) in serum-free medium. After a 6-h incubation, the cells were cultured in fresh medium supplemented with FBS. After 48 h, the cell medium containing the lentiviral particles was collected and filtered through a 0.45 μm filter (Corning). To generate stable cell lines, cells were treated with puromycin (2 μg/mL; Beyotime) for a minimum of 7 days. The viability of these cells was then determined using WB and qRT-PCR.
2.17 Co-immunoprecipitation (Co-IP)
Co-IP was performed by lysing the cells in cell lysis buffer (Beyotime) supplemented with protease inhibitor cocktails (CWBIO) at 4°C. The lysate was then centrifuged at 12,000 × g for 10 min at the same temperature. The supernatants were precleared with Protein G PLUS agarose (Santa cruz, Dallas, TX, USA) and then immunoprecipitated with the appropriate antibodies or anti-FLAG M2 agarose beads (Millipore) at 4°C overnight with rotation. After three washes, the immunoprecipitants were detected by WB.
2.18 Protein pull-down assay
His-DCBLD2 and CD146-Fc or human Fc protein (200 ng/mL; Sino Biological, Beijing, China) were incubated in PBS at 4°C for 1 h. After complete binding, the proteins were immunoprecipitated with Protein G PLUS-agarose beads (Santa cruz) and the bound proteins were analyzed by WB. Mouse IgG, anti-CD146 AA1, or AA98 antibodies were then incubated with CD146 and Protein G PLUS-agarose beads prior to incubation with DCBLD2. After three washes in PBS with 0.001% Tween 20 (Beyotime), CD146 and DCBLD2 were detected by WB.
2.19 Sample processing for proteomic analysis
Sample processing for the TurboID approach was conducted following the modified method proposed by the Alice Y Ting's Laboratory [14]. Briefly, cells were resuspended in serum medium supplemented with 200 μmol/L biotin for 20 min at 37°C with 5% CO2. After proximity labeling, cells were washed four times with cold PBS and lysed in RIPA lysis buffer (50 mmol/L Tris pH 7.5, 150 mmol/L NaCl, 0.1% SDS, 0.5% sodium deoxycholate, 1% Triton X-100, 1× protease inhibitor cocktail, and 1 mmol/L PMSF). The lysates were then sonicated at 30 W for 30 s and centrifuged at 12,000 × g for 15 min at 4°C. The supernatants were filtrated through a 3K MWCO column and then transferred to streptavidin magnetic beads (Thermo Fisher) prewashed with RIPA lysis buffer (Beyotime) and incubated overnight at 4°C with rotation.
For the Affinity Purification Mass Spectrometry (AP-MS) approach, cells were fixed on plates in 0.5% formaldehyde for 15 min at room temperature with rocking and quenched with 125 mmol/L glycine for 5 min. The plates were washed three times with cold PBS and finally lysed in cold RIPA lysis buffer (50 mmol/L Tris pH 7.5, 150 mmol/L NaCl, 0.5% sodium deoxycholate, 1% NP-40, 1× protease inhibitor cocktail, and 1 mmol/L PMSF) in 15 min. The lysates were then centrifuged at 12,000 × g for 15 min at 4°C. Supernatants were transferred to prewashed EZview Red anti-flagM2 agarose beads (Millipore) and incubated overnight at 4°C with rotation.
After incubation, the beads were washed three times with lysis buffer and eluted in elution buffer (1× SDS-PAGE sample loading buffer, 20 mmol/L DTT, 4 mmol/L biotin). A portion of the eluted samples was analyzed for protein enrichment by WB.
For mass spectrometry, samples were run on an SDS-PAGE gel and visualized using the Sliver Staining Kit (Thermo Fisher). Samples were split, minced, and collected in a tube for in-gel digestion as previously described [15]. Peptides extracted from the gels were desalted on C18 Stage Tips and lyophilized in a vacuum concentrator for mass spectrometry. All samples were measured using the nanoLC-Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher).
2.20 Proteomic data analysis
Raw files were analyzed by Proteome Discoverer (version 2.2.0.388, Thermo Fischer) using the integrated peptide search engine SEQUEST HT (Thermo Fisher). Tandem Mass Spectrometry (MS/MS) spectra were searched against a human UniProt database (Homo sapiens, last modified 10/01/2017). Filtering of spectra was done with Percolator, delta CN less than 0.1, FDR was set to 1%, peptide filtering parameters selected peptide confidence was high, and FDR at protein level was similarly set to 1%.
2.21 Statistical analysis
Statistical analyses, which included two-tailed Student's t-test, Tukey's post-hoc test, chi-squared test, Kaplan-Meier (KM) curves/log-rank test, Spearman order correlations, and receiver operator curve (ROC) analysis, were conducted using Graphpad Prism 9 (Graphpad Software Inc., San Diego, CA, USA). Multivariate Cox proportional hazards analysis was performed using SPSS 24 (IBM, Armonk, NY, USA). Data were presented as mean ± standard error of the mean (SEM). Statistical significance is indicated as P < 0.05.
3 RESULTS
3.1 CD146 was up-regulated in malignant PTs and was an independent prognostic marker for PT patients
First, to ascertain the cellular components in the PT microenvironment, scRNA-seq was performed on a variety of PT samples using 10× genomic sequencing. These samples included 7 benign PT samples (53,190 cells), 3 borderline PT samples (18,174 cells), and 7 malignant PT samples (38,001 cells). Details of patient characteristics are included in Supplementary Table S1. Following standardized processing, 33,682 high-quality cells were obtained after setting parameters for cell filtering. After gene expression normalization, PCA and UMAP were applied for dimensionality reduction and clustering. This led to the tentative classification of the cells into 7 distinct subpopulations: HSCs, natural killer (NK) cells and T cells, B cells, myeloid cells, endothelial cells, epithelial cells, and fibroblasts (Figure 1A, Supplementary Figure S1A). The distribution of cell types across different grades of PTs indicated a significantly larger subpopulation of fibroblasts compared to epithelial cells, particularly in malignancies (Figure 1A, Supplementary Figure S1A). Notably, fibroblasts predominated in all sample types, accounting for nearly 50% of both benign and malignant tumors. This was in line with earlier findings of fibroblasts being malignant components of PTs [6]. Subsequent reclassification of all fibroblasts led to the identification of 13 major subpopulations, with clusters 2, 3, 4, 6, 7, 8, and 10 predominating in malignant PTs compared to benign and borderline PTs (Figure 1B). Common fibroblast markers, such as actin alpha 2 (ACTA2), collagen type I (COL1A1), collagen type II (COL1A2), S100 calcium binding protein A4 (S100A4), fibroblast activation protein alpha (FAP), and the mesenchymal cell marker Vimentin (VIM) were expressed in all fibroblast subpopulations, confirming their fibroblast identity (Figure 1C). Interestingly, ACTA2 (encoding α-SMA) [6], S100A4 [16], and VIM [17] were found to be upregulated in fibroblasts that predominate in malignant PTs. It was found that these malignant markers were predominantly overexpressed in cluster 8, but relatively low or moderate expression in other subclusters (Figure 1C). Given earlier findings that α-SMA is a critical predictor of PT recurrence [6], this group of cells was of particular interest. Subsequent identification of the highly expressed genes in cluster 8 revealed signature genes, such as MCAM (encoding CD146), Caveolin 1 (CAV1), regulator of G protein Signaling 5 (RGS5), C-C motif chemokine ligand 2 (CCL2), and C-C motif chemokine ligand 8 (CCL8) (Figure 1D). Of note, the expression of CD146, known as a membrane protein, displayed dramatic increase in expression in borderline and malignant PTs compared to fibroadenoma and benign PTs. This was validated by examining the expression of CD146 in paraffin-embedded samples including 4 fibroadenoma, 146 benign, 90 borderline, and 62 malignant PTs by IHC (Figure 1E, Supplementary Figure S1B). It suggested that CD146 could be a potential therapeutic target against malignant PTs. To further confirm the expression of CD146 in PT cells, IF staining for CD146 and α-SMA was performed and showed the high expression of CD146 in α-SMA+ fibroblasts in PT tissues (Figure 1F), which was consistent with the result of scRNA-seq. Of note, IF staining of CD146 in PT tissue showed that CD146 mainly located at cellular membrane (Figure 1F). Furthermore, the expression of CD146 was upregulated in malignant PT tissues compared to benign ones by WB (Supplementary Figure S1C) and by qRT-PCR (Supplementary Figure S1D). Examination of the level of CD146 in benign and malignant PT cell lines (benign, SYSH-BPT-01; malignant, SYSH-MPT-01, SYSH-MPT-02, SYSH-MPT-03, and SYSH-MPT-04) by qRT-PCR demonstrated that CD146 was significantly increased in SYSH-MPT-02, -03, and -04 cells compared to the benign PT cell line (Supplementary Figure S1E). Cumulatively, the above results demonstrated that CD146 was associated with the malignant progression of PTs.
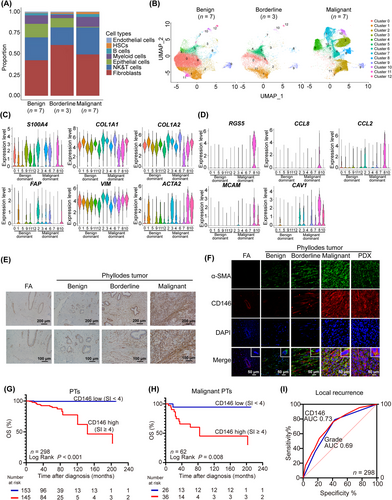
CD146 is up-regulated in malignant PTs and is an independent prognostic marker for PT patients.(A) scRNA-seq on 7 benign PT samples, 3 borderline PT samples, and 7 malignant PT samples were performed. Bar plots showed the proportion cell types in different subtypes of benign, borderline and malignant PTs.(B) UMAP plots showed the total fibroblasts color coded for 13 clusters in different subtypes of benign (n = 7), borderline (n = 3) and malignant PTs (n = 7).(C) Violin plots showed the expression level of selected fibroblasts marker genes in distinct fibroblast subclusters.(D) Violin plots showed the expression level of marker genes for the α-SMA+ fibroblasts.(E) IHC staining showed the level of CD146 in paraffin-embedded FA (n = 4), benign PTs (n = 146), borderline PTs (n = 90), and malignant PTs (n = 62) tissues. Scale bars: 100 μm and 200 μm.(F) IF staining for CD146 and α-SMA in paraffin-embedded FA, PT and PDX tissues. Scale bars: 50 μm.(G) Kaplan-Meier OS curve of PT patients with low level (CD146 SI < 4) and high level (CD146 SI ≥ 4) of CD146 staining in tumor tissues.(H) Kaplan-Meier OS curve of malignant PT patients with low level (CD146 SI < 4) and high level (CD146 SI ≥ 4) of CD146 staining in tumor tissues.(I) ROC curve was performed to the estimate the power of CD146 and histologic grade for predicting the local recurrence of PTs.Abbreviations: PT, phyllodes tumors; scRNA-seq, single-cell RNA sequencing; α-SMA, alpha-smooth muscle actin; IHC, immunochemistry; SI, staining index; ROC, receiver operator curve; HSCs, hematopoietic stem cells; NK, natural killer; UMAP, uniform manifold approximation and projection; S100A4, S100 calcium binding protein A4; COL1A1, collagen type I; COL1A2, collagen type II; FAP, fibroblast activation protein alpha; VIM, vimentin; ACTA2, actin alpha 2; RGS5, regulator of G protein signaling 5; CCL8, C-C motif chemokine ligand 8; CCL2, C-C motif chemokine ligand 2; MCAM, melanoma cell adhesion molecule; CAV1, caveolin 1; FA, fibroadenoma; DAPI, 4’,6-diamidino-2’-phenylindole; AUC, area under curve; IF, Immunofluorescence.
The clinical significance of CD146 in PT patients was also evaluated. The 298 PT patients from Sun Yat-sen Memorial Hospital were followed up for 2-211 months (median, 50 months). During follow-up, 67 cases developed local recurrence (14, 29, and 24 cases in the benign, borderline, and malignant groups, respectively). In addition, 12 cases progressed to metastasis (1 and 11 cases in the borderline and malignant groups, respectively).
Further investigation was conducted to establish the relationship between CD146 and the clinicopathologic status of PTs. Immunostaining of CD146 and Ki67 revealed a positive correlation between the levels of CD146 and Ki67 in PT tissues (Supplementary Figure S1F-G). Further examination of the association of CD146 expression with the clinicopathologic status of PTs are shown in Table 1. It indicated that the level of CD146 in PT tissues increased with higher tumor grade (P < 0.001), mitotic activity (P < 0.001), and stromal overgrowth (P = 0.001), but was not related to age and tumor size (Table 1). In addition, the level of CD146 was increased in PTs with local recurrence (P < 0.001) and metastasis (P = 0.002) (Table 1). KM curve showed that PT patients with low CD146 expression had longer OS (Figure 1G) as well as LRFS (Supplementary Figure S1H) than those with high expression (P < 0.05). Notably, a similar trend in OS (Figure 1H) and LRFS (Supplementary Figure S1I) was observed in the malignant group as well as in the benign and borderline group (Supplementary Figure S1J-K, P <0.05). To ascertain whether CD146 could serve as a predictor for local recurrence of PTs, ROC curve analysis was performed, demonstrating that CD146 could indeed be a suitable predictor (P <0.001; AUC = 0.73; Figure 1I). Notably, CD146 was a better predictor of local recurrence than histologic grade (AUC = 0.69, Figure 1I). Furthermore, multivariate Cox proportional hazards analysis revealed that the level of CD146 in PT tissues, stromal overgrowth in PT tissues, and tumor grade were independent prognostic predictors for OS and LRFS (Table 2). Taken together, these results suggested that CD146 was a more appropriate prognostic marker for PTs than histologic grade.
| CD146 expression, n (%) | ||||
|---|---|---|---|---|
| Clinical characteristic | Total, n (%) |
SI < 4 (n = 153) |
SI ≥ 4 (n = 145) |
P value |
| Age, years | 0.063 | |||
| <40 | 136 (45.6) | 78 (51.0) | 58 (40.0) | |
| ≥40 | 162 (54.4) | 75 (49.0) | 87 (60.0) | |
| Tumor grade | <0.001 | |||
| Benign | 146 (49.0) | 99 (64.7) | 47 (32.4) | |
| Borderline | 90 (30.2) | 28 (18.3) | 62 (42.8) | |
| Malignant | 62 (20.8) | 26 (17.0) | 36 (24.8) | |
| Tumor size, cm | 0.066 | |||
| <5 | 216 (72.5) | 118 (77.1) | 98 (67.6) | |
| ≥5 | 82 (27.5) | 35 (22.9) | 47 (32.4) | |
| Local recurrence | <0.001 | |||
| No | 231 (77.5) | 137 (89.5) | 94 (64.8) | |
| Yes | 67 (22.5) | 16 (10.5) | 51 (35.2) | |
| Metastasis | 0.002 | |||
| No | 286 (96.0) | 152 (99.3) | 134 (92.4) | |
| Yes | 12 (4.0) | 1 (0.7) | 11 (7.6) | |
| Mitoses | <0.001 | |||
| <5/HPF | 168 (56.4) | 104 (68.0) | 64 (44.1) | |
| 5-10/HPF | 85 (28.5) | 33 (21.6) | 52 (35.9) | |
| >10/HPF | 45 (15.1) | 16 (10.4) | 29 (20.0) | |
| Stromal overgrowth | 0.001 | |||
| Absence | 192 (64.4) | 112 (73.2) | 80 (55.2) | |
| Present | 106 (35.6) | 41 (26.8) | 65 (44.8) | |
- Abbreviations: SI, staining index; HPF, high power field.
| OS | LRFS | |||||
|---|---|---|---|---|---|---|
| Variable | HR | 95% CI | P value | HR | 95% CI | P value |
| CD146 | 19.434 | 2.204-171.336 | 0.008 | 2.660 | 1.451-4.876 | 0.002 |
| Age | 0.766 | 0.264-2.222 | 0.624 | 0.801 | 0.484-1.325 | 0.387 |
| Tumor size | 2.020 | 0.585-6.976 | 0.266 | 1.176 | 0.682-2.027 | 0.559 |
| Mitosis | 0.803 | 0.367-1.756 | 0.582 | 0.709 | 0.429-1.170 | 0.179 |
| Stromal overgrowth | 4.004 | 1.126-14.231 | 0.032 | 3.468 | 1.947-6.176 | <0.001 |
| Tumor grade | 7.625 | 2.326-25.003 | 0.001 | 2.067 | 1.263-3.382 | 0.004 |
- Abbreviations: OS, overall survival; LRFS, local recurrence-free survival; HR, hazard ratio; CI, confidence interval.
3.2 CD146 accelerated malignant progression of PT cells
The role of CD146 in the malignant progression of PT cells was further examined. Malignant PT cell lines SYSH-MPT-02 and -03, which express CD146 at relatively high levels, and the benign PT cell line SYSH-BPT-01, which expresses CD146 at relatively lower levels, were utilized. Initially, the expression of CD146 was knocked down in the malignant PT cell line SYSH-MPT-02, and this knockdown was successfully confirmed via qRT-PCR and WB (Figure 2A-B). According to the MTS assay, the knockdown of CD146 dramatically inhibited the proliferation of SYSH-MPT-02 cells (Figure 2C). Similarly, the colony formation assay indicated that the clonogenic capacity of SYSH-MPT-02 cells was impaired following the knockdown of CD146 (Figure 2D-E). In addition, the migratory and invasive abilities of SYSH-MPT-02 cells were suppressed by the knockdown of CD146 (Figure 2F-G). Consistent with these findings, levels of the cell cycle regulator cyclin D1 as well as invasion marker Vimentin were decreased in SYSH-MPT-02 cells treated with CD146 siRNAs (Figure 2H). Given the characteristic ability of PT cells for collagen contraction, the effect of CD146 on this ability was also assessed. It was observed that in SYSH-MPT-02 cells, knockdown of CD146 significantly reduced the ability of contract collagen (Figure 2I-J). Likewise, the knockdown of CD146 significantly suppressed the viability (Supplementary Figure S2A), migration and invasion (Supplementary Figure S2B-C) of SYSH-MPT-03 cells. Consistently, overexpression of CD146 (Supplementary Figure S2D-E) enhanced the vitality (Supplementary Figure S2F-H), migration, and invasion (Supplementary Figure S2I-J) of the benign SYSH-BPT-01 cell line, as well as increasing the levels of Vimentin and cyclin D1 (Supplementary Figure S2K). Furthermore, overexpression of CD146 also heightened the ability of collagen to contract in the benign cell line (Supplementary Figure S2L-M). These findings collectively suggested that CD146 accelerates the malignant progression of PT cells.
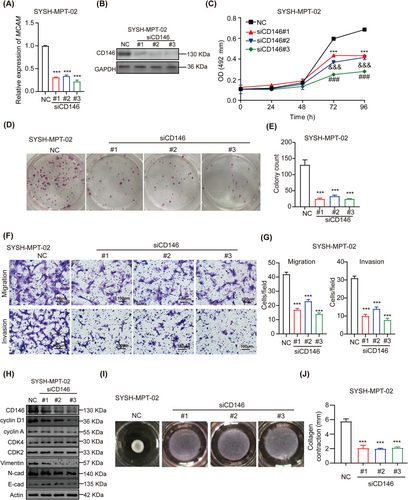
CD146 accelerates malignant progression of PT cells.(A) Efficiency of si-CD146 transfection on malignant SYSH-MPT-02 was determined by qRT-PCR. Data were presented as a histogram representative of mRNA expression normalized to GAPDH.(B) Efficiency of si-CD146 transfection on malignant SYSH-MPT-02 was determined by WB.(C) MTS assay was conducted to evaluate the proliferation of malignant SYSH-MPT-02 cells transfected with NC, siCD146#1, siCD146#2, and siCD146#3. Data were presented in a growth curve.(D) Colony formation assay was conducted to evaluate the proliferation of malignant SYSH-MPT-02 cells transfected with NC, siCD146#1, siCD146#2, and siCD146#3.(E) Bar chart represented the colony number of malignant SYSH-MPT-02 cells transfected with NC, siCD146#1, siCD146#2, and siCD146#3.(F) Photographs of migration and invasion assay of malignant SYSH-MPT-02cells transfected with NC, siCD146#1, siCD146#2, and siCD146#3. Scale bars: 100 μm.(G) Bar chart represented the number of migrant cells and invasive cells under 200× field of malignant SYSH-MPT-02 cells respectively transfected with NC, siCD146#1, siCD146#2, and siCD146#3.(H) The protein level of CD146, cyclin D1, cyclin A, CDK4, CDK2, Vimentin, N-cad, E-cad, and actin in malignant SYSH-MPT-02 cells respectively transfected with NC, siCD146#1, siCD146#2, and siCD146#3 were detected by WB.(I) Images of collagen contraction assay of malignant SYSH-MPT-02cells transfected with NC, siCD146#1, siCD146#2, and siCD146#3.(J) Bar chart represented degree of contraction of malignant SYSH-MPT-02cells transfected with NC, siCD146#1, siCD146#2, and siCD146#3.Data were shown as mean ± SEM. ***, &&&, ### P < 0.001.Abbreviations: PT, phyllodes tumors; qRT-PCR, quantitative real-time-polymerase chain reaction; mRNA, message RNA; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MTS, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2h-tetrazolium; NC, negative control; CDK2, cyclin-dependent kinase 2; CDK4, cyclin-dependent kinase 4; N-cad, N-cadherin; E-cad, E-cadherin; OD, optical density; WB, western blotting.
3.3 CD146 accelerated malignant progression of PTs through activating the PI3K-AKT signaling pathway
In order to explore the mechanism by which CD146 modulates the progression of PT, a GSEA of scRNA-seq from PT patients was conducted. This revealed that mRNAs differentially expressed in CD146 high and low fibroblasts were enriched in pathways such as tumor necrosis factor-alpha (TNFα) signaling pathway via nuclear factor kappa-B (NFκB), epithelial-mesenchymal transition, Interferon-gamma response, complement, Kirstenrat sarcoma viral oncogene homolog (Kras), and PI3K-AKT-mammalian target of rapamycin (mTOR) pathways (Figure 3A). Notably, GSEA showed that the PI3K-AKT-mTOR pathway was activated in CD146-high fibroblasts (Figure 3B).
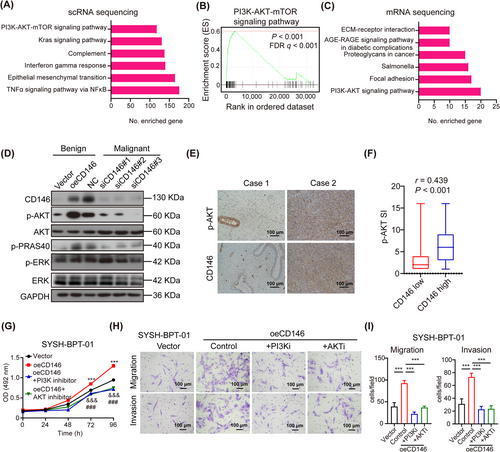
CD146 accelerates malignant progression of PTs through activating PI3K-AKT signaling pathway.(A) GSEA pathway enrichment analysis of the differential gene between CD146 high and low fibroblasts from scRNA-seq of PT tissue.(B) GSEA results of the PI3K-AKT-mTOR signaling pathway which was enriched between CD146 high and low fibroblasts from scRNA-seq of PT tissue.(C) KEGG pathway enrichment analysis of the differential mRNA which were upregulated in SYSH-BPT-01 cells overexpressing CD146 and down-regulated in SYSH-MPT-02 cells silencing CD146.(D) Protein level of CD146, AKT, phospho-AKT, phospho-PRAS 40, ERK, phospho-ERK, and GAPDH in SYSH-BPT-01 cells overexpressing vector and CD146, and in SYSH-MPT-02 cells transfected with NC, siCD146#1, siCD146#2, and siCD146#3 was detected by WB.(E) Immunostaining of CD146 and phospho-AKT in human PT tissues. Scale bars: 100 μm.(F) Association of level of CD146 and phospho-AKT in human PT tissues (n = 153 for CD146-low tissues; n = 145 for CD146-high tissues) was analyzed by spearman's rank correlation coefficient analysis.(G) MTS assay was performed to evaluate the proliferation of SYSH-BPT-01cells overexpressing vector, overexpressing CD146, overexpressing CD146 in addition to PI3K inhibitor and overexpressing CD146 in addition to AKT inhibitor.(H) Photographs of migration and invasion assay of SYSH-BPT-01cells overexpressing vector, overexpressing CD146, overexpressing CD146 in addition to PI3K inhibitor and overexpressing in addition to AKT inhibitor. Scale bars: 100 μm.(I) Bar chart represented the number of migrant cells and invasive cells under 200× field of each group.Data were shown as mean ± SEM. ***, &&&, ### P < 0.001.Abbreviations: PT, phyllodes tumors; PI3K, phosphoinositide 3-kinase; AKT, protein kinase B; GSEA, Gene Set Enrichment Analysis; scRNA-seq, single-cell RNA sequencing; mTOR, mammalian target of rapamycin; KEGG, Kyoto Encyclopedia of Genes and Genomes; PRAS 40, the proline-rich AKT substrate of 40KDa; Erk, extracellular signal-regulated kinase; MTS, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2h-tetrazolium; NC, negative control; OE, overexpress; OD, optical density; AKTi, AKT-inhibitor; PI3Ki, PI3K-inhibitor; WB, western blot.
In further investigation, we conducted experiments by using CD146-knockdown (siCD146) SYSH-MPT-02 cell line and CD146-overexpressing (oeCD146) SYSH-BPT-01 cell line. Transcriptome sequencing was then performed to compare the transcript profiles between 4 groups (benign, control vs. oeCD146; malignant, control vs. siCD146) (Supplementary Figure S3A). The criteria for screening of differentially expressed mRNA were FC >1.41 in the benign PT cell line overexpressing CD146 and FC <0.71 in the malignant PT cell line silencing expression of CD146. Importantly, 48 mRNAs were upregulated in benign PT cell line overexpressing CD146 while downregulated in malignant PT cell line silencing expression of CD146 (Supplementary Figure S3B). According to the KEGG pathway enrichment analysis, the differentially expressed mRNAs were enriched in pathways such as PI3K-AKT signaling, focal adhesion, Salmonella, proteogylcans in cancer, advanced glycation end products-AGEs receptor (AGE-RAGE) signaling pathway, and extracellular matrix (ECM)-receptor interaction pathway (Figure 3C). Interestingly, the PI3K-AKT signaling pathway was investigated in both scRNA-seq in PT tissues (Figure 3A) and mRNA sequencing in cell lines (Figure 3C). Previous reports suggested that activation of the AKT pathway may be associated with malignant transformation and recurrence of PT [18, 19]. To confirm these sequencing results, the pathway was detected by WB, revealing that the PI3K-AKT pathway was activated in malignant PT cells compared to benign PT cells (Figure 3D). Furthermore, the levels of phospho-AKT and phospho-PRAS40 were increased in SYSH-BPT-01 cells overexpressing CD146 and decreased in SYSH-MPT-02 cells with silenced expression of CD146 (Figure 3D). IHC Staining for CD146 and phospho-AKT in human PT tissues revealed a positive correlation between CD146 and phospho-AKT levels (Figure 3E-F).
To verify whether CD146 promotes the progression of PTs by activating the PI3K-AKT signaling pathway, we overexpressed CD146 in SYSH-BPT-01 cells with or without the addition of a PI3K inhibitor or AKT inhibitor. It was observed that overexpression of CD146 promoted proliferation, migration, invasion, and collagen contraction of SYSH-BPT-01 cells, while application of PI3K inhibitor or AKT inhibitor was able to suppress the enhanced proliferation (Figure 3G, Supplementary Figure S3C-D), migration and invasion (Figure 3H-I), and collagen contraction (Supplementary Figure S3E-F) of SYSH-BPT-01 cells by CD146. These findings collectively suggest that CD146 accelerates the malignant progression of PTs by activating the PI3K-AKT signaling pathway.
3.4 Application of anti-CD146 antibody suppressed malignant progression and PI3K/AKT signaling pathway of PT cells In vitro and in vivo
Given the observed promotion of malignant progression in PT cells via the activation of the PI3K-AKT signaling pathway by CD146, the focus was shifted towards exploring a potential therapeutic strategy against malignant PTs by targeting CD146. Previous studies have indicated that the CD146-blocking antibody, AA98, exhibited significant inhibitory activity against tumor progression in various cancer types [20-23]. Accordingly, the anti-CD146 antibody AA98 [24] was tested In vitro for its therapeutic efficacy against malignant PTs. The MTS assay and colony formation assay demonstrated that the anti-CD146 antibody AA98 effectively suppressed the proliferation of SYSH-MPT-02 cells (Figure 4A, Supplementary Figure S4A-B). Notably, a malignant PT organoid system was established in which primary malignant PT cells were able to develop into an organ-like structure that mimicked the internal environment of the tumor. The introduction of AA98 was observed to inhibit the growth of the malignant PT organoid (Figure 4B, Supplementary Figure S4C). Moreover, application of AA98 also inhibited the migration, invasion (Supplementary Figure S4D-E) and collagen contraction (Supplementary Figure S4F-G) of SYSH-MPT-02. The role of anti-CD146 antibody in the activation of PI3K-AKT signaling pathway was confirmed by a WB assay, demonstrating that the application of AA98 downregulated the level of phospho-AKT in SYSH-MPT-02 cells (Figure 4C). These findings collectively suggested that anti-CD146 suppressed malignant progression and the PI3K-AKT signaling pathway of PT cells In vitro.
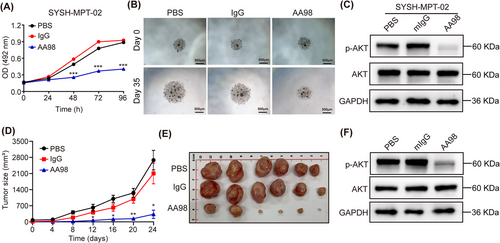
Application of anti-CD146 antibody suppresses malignant progression and PI3K-AKT signaling pathway of PT cells In vitro and in vivo.(A) MTS assay was conducted to evaluate the proliferation of SYSH-MPT-02 cells treated with PBS, IgG, and AA98. Data were presented in a growth curve.(B) Malignant PT organoid was treated with PBS, 150 μg/mL IgG, or 150 μg/mL AA98 and the diameter of organoid was measured. Scale bars: 500 μm.(C) The protein level of AKT, phospho-AKT and GAPDH in SYSH-MPT-02cells treated with PBS, 150 μg/mL IgG, or 150 μg/mL AA98 was detected by WB.(D) The growth curve represented the tumor volume of PDX mice treated with PBS, IgG, or AA98 (n = 6 per group).(E) Representative pictures of malignant PT PDX from mice treated with PBS, IgG, or AA98, respectively.(F) The protein level of AKT, phospho-AKT, and GAPDH in PDX from mice treated with PBS, IgG, or AA98 was measured by WB.Data were shown as mean ± SEM. * P < 0.05; ** P < 0.01; *** P < 0.001.Abbreviations: PT, phyllodes tumors; PI3K, phosphoinositide 3-kinase; AKT, protein kinase B; MTS, 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2h-tetrazolium; PBS, phosphate buffer saline; IgG, immunoglobin G; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; PDX, patient-derived xenograft; OD, optical density; WB, western blot.
The therapeutic efficacy of anti-CD146 antibody against malignant PT was further tested in vivo. A PDX model of malignant PT was established for the application of the anti-CD146 antibody AA98. Donor information is provided in Supplementary Table S4. AA98 was found to substantially inhibit the growth of the PT PDX (Figure 4D-E, Supplementary Figure S4H). It was also observed that AA98 decreased the level of phospho-AKT in PDX tissues as indicated by immunostaining (Figure 4F). Additionally, IHC revealed that the staining index of Ki67 and phospho-AKT was lower in PT cells from PDX tissues treated with AA98 compared to those treated with PBS or IgG (Supplementary Figure S4I-J). Given that CD146 has been reported to be expressed in endothelial cells and associated with the growth of vascular endothelium [20], we also tested the density of vascular endothelium (CD31+ vessel) in malignant PT PDX tissues through IHC, revealing that AA98 also decreased the density of CD31+ vascular endothelium in malignant PT PDX tissues (Supplementary Figure S4I-J). In line with this, scRNA-seq of patient PT tissues revealed that CD146 expression was higher in fibroblasts and endothelial cells than in other cell types (Supplementary Figure S4K), suggesting that CD146 was a suitable and safe therapeutic target. These results collectively demonstrated that targeting CD146 could inhibit the malignant progression and PI3K-AKT signaling pathway of PTs in vivo.
3.5 Identified DCBLD2 as an interaction protein to CD146
While CD146 has previously been associated with the activation of PI3K-AKT pathways in a variety of tumors and endothelial cells, the specifics remain elusive [25]. This study aimed to define CD146 interacting partners to better elucidate its role in malignant PTs. TurboID, a recent proximity labeling method capable of detecting weak or transient protein-protein interactions [14], was incorporated and optimized with the traditional epitope tag-based AP-MS pipeline to identify functional CD146-protein interactions (Figure 5A). Two constructs, CD146-TurboID and MEM-TurboID, carrying CD146 cDNA or a transmembrane peptide as a control respectively, were each tagged with optimized BirA* and Flag epitope. These constructs were transfected into SYSH-MPT-02 cell line. WB analysis indicated that CD146-TurboID was expressed at similar levels as wild-type CD146 and was able to activate PI3K-AKT pathways (Supplementary Figure S5A-B). To identify CD146 interaction proteins, CD146-TurboID and MEM-TurboID cells were subjected to biotin-added cultures, followed by streptavidin pull-down or followed by flag affinity antibody pull-down and mass spectrometry analysis. The TurboID assay identified 316 CD146 proximal proteins and AP-MS identified 227 potential interacting proteins, including known CD146 interactors such as AP2B1 [26], ARHGDIA [21], and other interactors (ROBO1, GJA1, STX4, RAC1, VDAC3, and MARCKS) identified in other high-throughput experiments [27-30] as well as novel interactors (Figure 5B, Supplementary Table S5). Analysis of the biological processes involving CD146 interactors revealed associations with cell adhesion, migration, and growth factor response, processes intimately linked to the malignant phenotype of tumors (Supplementary Figure S5C).
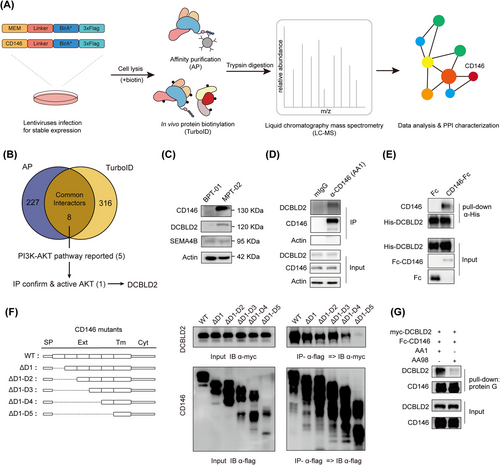
Identified DCBLD2 as an interaction protein to CD146.(A) The workflow for identification of protein complexes and interactions of CD146 in PT cells. Vectors were designed containing transmembrane peptides or CD146 cDNA and C-terminal tagging with BirA* and flag. The expression vector can then be transfected into malignant PT cells to establish the transgenic stably expressing isogenic cell lines. For the AP-MS and TurboID analysis approaches, the cell line is separated into 2 cultures, TurboID cells receiving addition of 50 μmol/L biotin in their culture medium. In the following protein extraction process, optimized lysis and affinity purification conditions for both analysis approaches were used. The interacting proteins were then analyzed by quantitative mass spectrometry and high confidence proteins were inferred via stringent statistical filtering.(B) Venn diagram showing the interactors identified in AP-MS/TurboID assays and the strategy to identify CD146 downstream protein in phyllode tumor.(C) WB showed the changes in the expression level of CD146, DCBLD2, and SEMA4B in benign/malignant cell lines.(D) Immunoprecipitation assay showed the interaction between CD146 and DCBLD2 from SYSH-MPT-02. The immunoprecipitants were precipitated using anti-CD146 mAb AA1 and mIgG as control.(E) Pull-down assay revealed the direct interaction between CD146 and DCBLD2. Purified His-tagged DCBLD2 and Fc-CD146 proteins were incubated in PBS. CD146 and DCBLD2 were pulled down using Protein G PLUS Agarose.(F) Co-IP assays for association of CD146 mutants with DCBLD2 in HEK293T cells showed CD146 D4-D5 domain is necessary for binding.(G) Pull-down assay revealed that anti-CD146 AA98 antibody blocked the CD146-DCBLD2 interaction.Abbreviations: CD146, cluster of differentiation 146; DCBLD2, discoidin, CUB And LCCL domain containing 2; Co-IP, co-immunoprecipitation; AP-MS, affinity purification mass spectrometry; HCIP, high-confidence interaction proteins; PPI, protein-protein interaction networks; PI3K, phosphatidylinositol-3-kinase; AKT, protein kinase B; IP, immunoprecipitation; IB, immunoblotting; SP, signal peptide; Ext, extracellular domain; Tm, transmembrane domain; Cyt, Intracellular domain.
Next, a focus was placed on the interactors co-identified by AP-MS and TurboID (Figure 5B). Among these overlapping candidates, 8 proteins were narrowed down to 5 (CD44 [31], DCBLD2 [32], CD166 [33], EGFR [34], STK24 [35]) with high potential involvement in the PI3K-AKT pathway. The potential interactions between these candidate proteins and CD146 were then examined (Supplementary Figure S5D). Notably, DCBLD2 and CD166 induced significant AKT phosphorylation (Supplementary Figure S5E). Nevertheless, knockdown of CD166 expression in malignant phyllode tumor cell line SYSH-MPT-02 had minimal effect on AKT signaling and affected CD146 expression (Supplementary Figure S5F), which aligns with prior reports positioning CD166 upstream of CD146 in signaling transduction [36]. Consequently, the role of DCBLD2 in PTs was further investigated. DCBLD2 is a type I transmembrane protein implicated in the malignant phenotype of cancer [37] and possesses a CUB domain identified as a potential binding site with CD146 [38]. The expression profile of DCBLD2 across various types of human cancer was assessed using TIMER and CCLE datasets, revealing a positive correlation with CD146 (Supplementary Figure S5G). Further, CD146-DCBLD2 proximity was confirmed by TurboID WB, which detected strong DCBLD2 proximity to the MEM-TurboID control (Supplementary Figure S5H). DCBLD2 protein expression level was upregulated in SYSH-MPT-02 malignant PT cells compared to SYSH-BPT-01 benign PT cells (Figure 5C). IP of PT endogenous CD146 detected DCBLD2 in the immunoprecipitants (Figure 5D), a finding replicated in HEK293T cells (Supplementary Figure S5I). Pull-down assay also revealed direct interaction between DCBLD2 and CD146 (Figure 5E). Both CD146 and DCBLD2 are located on the cell membrane, and their co-localization was further validated by IF (Supplementary Figure S5J). Additionally, the CD146 domain essential for DCBLD2-CD146 interaction using flag-tagged CD146 truncated variants was mapped (Figure 5F). Co-IP assays showed that their interactions could be affected by the 5th Ig-like region, which is also important for AKT activation (Supplementary Figure S5K). Previously developed anti-CD146 antibodies [24, 39], including AA98, which recognizes the 4th to 5th Ig domain of CD146 and disrupts protein dimerization, and AA1, which recognizes the 1st to 2nd domain of CD146 and does not affect protein dimerization, were used. A pull-down assay revealed that AA98, but not AA1, blocked the interaction between DCBLD2 and CD146 (Figure 5G). Consistently, overexpression of truncated variants of CD146, which promote its dimerization as reported [23] in malignant cells, increased AKT phosphorylation (Supplementary Figure S5L). These findings suggest a direct interaction between DCBLD2 and CD146.
3.6 CD146 promoted PI3K-AKT pathway by stabilizing DCBLD2
To investigate whether the interaction between CD146 and DCBLD2 plays a vital role in malignant PTs, their potential to affect the AKT pathway was verified. WB analysis revealed that reducing CD146 expression through siRNA in malignant tumor cells led to decreased DCBLD2 protein levels and downstream PI3K-AKT pathway activity (Figure 6A). Correspondingly, this reduction in CD146 did not impact the transcript levels of DCBLD2 and other proteins in the same family (Supplementary Figure S6A-B). Conversely, lowering DCBLD2 expression in PT cell lines resulted in decreased AKT phosphorylation levels, while CD146 expression remained unaffected (Figure 6B). To further elucidate the relationship between CD146 and DCBLD2, CD146 was knocked out (KO) in malignant tumor cells using CRISPR/Cas9. The WB analysis revealed similar decreases in DCBLD2 protein levels and AKT phosphorylation (Supplementary Figure S6C). An attempt was made to rescue the DCBLD2 expression in CD146 KO cells by transfection, which triggered the activation of AKT signaling, suggesting that CD146 can activate the PI3K-AKT pathway through DCBLD2 (Figure 6C). Furthermore, the re-expression of CD146 in CD146 KO cells also increased DCBLD2 protein level and AKT phosphorylation but had no effect on DCBLD2 and other same family member transcript levels (Supplementary Figure S6A and D). These findings indicate that CD146 indeed regulates AKT phosphorylation via DCBLD2.
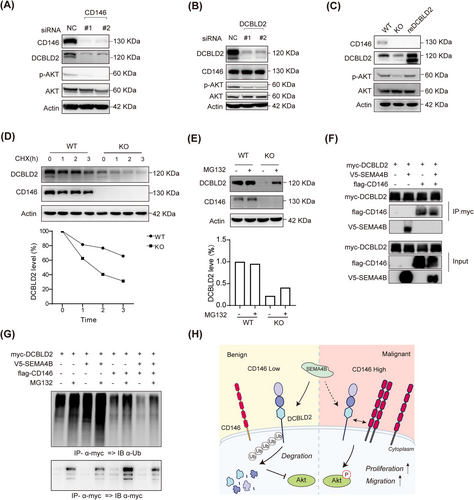
CD146 promotes PI3K-AKT pathway by stabilizing DCBLD2.(A-B) IB showed the changes in the expression level of DCBLD2 and AKT pathway proteins in CD146-knock down (A) or DCBLD2-knock down (B) in SYSH-MPT-02 cells.(C) IB showed the changes in the expression level of DCBLD2 and AKT pathway in DCBLD2 rescued KO cell lines.(D) Analysis of DCBLD2 stability with CHX chase assay in control and CD146-KO SYSH-MPT-02 cells. Immunoblot images (upper panel) and quantitative analysis (lower panel) of DCBLD2 in the upper image.(E) Immunoblots with the indicated antibodies on lysates from control and CD146-KO SYSH-MPT-02 treated with MG132. The images and their quantifications are presented in the upper and lower panels, respectively.(F) Co-IP assays showed the interactions of CD146, DCBLD2, and SEMA4B in HEK293T cells. Proteins were precipitated by indicated anti-tag mAb and examined by immunoblot.(G) Immunoblots with the ubiquitination antibodies on lysates from transfected HEK293T cell lines treated with MG132.(H) Graphical illustration of the working model.Abbreviations: DCBLD2, discoidin, CUB and LCCL domain containing 2; SEMA4B, semaphorin 4B; Co-IP, co-immunoprecipitation; NC, negative control; WT, wild type; KO, knock out; CHX, Cycloheximide, AKT, protein kinase B IP, immunoprecipitation; IB, immunoblotting; Ub, ubiquitination.
Since CD146 appeared to influence DCBLD2 protein levels rather than its mRNA, it is plausible that CD146 might stabilize the DCBLD2 protein against degradation. Previous studies have reported that DCBLD2 interacts with SEMA4B, resulting in increased ubiquitylation of membrane localized DCBLD2 and subsequent degradation [40, 41]. In NSCLC, SEMA4B also inhibits tumor cell migration through the AKT pathway [42]. Thus, the role of CD146 in mediating DCBLD2 stability was explored. It was observed that the DCBLD2 protein had a significantly shorter lifespan in CD146 KO cells than in controls (Figure 6D). Furthermore, the inhibition of the proteasome by MG132 could restore DCBLD2 protein levels that were reduced by CD146-KO, suggesting that CD146 may be involved in inhibiting DCBLD2 degradation through the ubiquitin-proteasome degradation pathway (Figure 6E). To further examine whether CD146 influences SEMA4B levels, qRT-PCR and WB were performed, revealing that disrupting CD146 did not affect SEMA4B expression in SYSH-MPT-02 cells (Supplementary Figure S6A-C). Consistently, there was no significant difference in SEMA4B levels between benign and malignant tumor cells (Figure 5C, Supplementary Figure S6E). Co-IP experiments in HEK293T were then conducted to determine whether CD146 could affect the DCBLD2-SEMA4B interaction. The results showed that CD146 could inhibit the interaction between SEMA4B and DCBLD2 (Figure 6F). The ubiquitination status of DCBLD2 in HEK293T was further investigated, revealing that ubiquitination of DCBLD2, while readily detected in cells co-transfected with empty vector and SEMA4B, was significantly reduce after co-transfection with CD146 (Figure 6G). This reduction may be due to CD146's interaction with DCBLD2, which occupies the CUB domain of DCBLD2, which is necessary for SEMA4B binding. Finally, an analysis of clinical specimens confirmed a positive correlation between CD146 and DCBLD2 levels in PT tissues (Supplementary Figure S6F-G). These results collectively suggested that CD146 acts as a “shield” to protect the DCBLD2 protein by suppressing its proteasome-mediated degradation, in turn activating the PI3K-AKT signaling pathway (Figure 6H).
4 DISCUSSION
This research employed a unique approach, utilizing droplet-based single-cell transcriptomics strategy to characterize both benign and malignant PT tumors. The findings revealed a distinct fibroblast subtype marked by elevated expressions of α-SMA and CD146, which serve as key indicator of malignant PTs. Furthermore, it is a study to document a significant rise in CD146 levels during the malignant progression from benign to malignant PTs, illustrating a significant association with adverse clinical outcome. Additionally, it was discovered that CD146 aggravated PT cell proliferation and invasion by stabilizing DCBLD2, subsequently activating the PI3K-AKT pathway. This process, however, could be effectively mitigated by the introduction of an anti-CD146 antibody.
Given the relatively low incidence of malignant PTs, the exploration into their cellular components has been limited. Previous studies have identified a substantial presence of fibroblast-like cells in phyllode tumors, specifically myofibroblast-like cells (also referred to as tumor fibroblast-like cells), which have been found to correlated with the malignant degree of phyllode tumors [43, 44]. This study not only corroborated the presence of α-SMA+ fibroblasts in PTs, but also revealed the expression of CD146 in these cells. In contrast to a-SMA, CD146 expression was found to be more specific in malignant phyllodes and upregulated in line with tumor malignancy. This pivotal discovery complements the existing research on the components involved in the malignant transformation of PTs. Currently, the classification of PTs is largely based on pathological estimates and histological grading, which suffer from a lack of consensus and objectivity due to the morphologic continuity from benign to malignant PTs [45]. However, a significant challenge in this approach is the absence of universally agreed upon “cut-off” value to determine histologic grading. This lack of consensus and objectivity, compounded by the morphologic continuity from benign to malignant PTs, results in low sensitivity and specificity in the prognosis predictions for PT patients [46]. As a result, the prognostic value of pathological grade is considerably constrained. Date from this study, however, revealed that patients expressing low levels of CD146 had longer OS compared to those with high expression. It has been noted that approximately 10%-46% of benign or borderline PTs may relapse, and a significant portion of these cases may progress to malignant PTs or even metastasize following multiple recurrences. By identifying these recurrence-prone tumors and implementing early interventions. For example, re-excision with wide margins or targeted therapy for CD146 and the likelihood of recurrence and malignant progression could potentially be diminished. Local recurrence rates were found to be 9.59% (14 of 146), 32.22% (29 of 90), and 38.71% (24 of 62) for benign, borderline, and malignant PT patients, respectively. Importantly, the local recurrence rate for patients with low CD146 expression was 10.46% (16 of 153), whereas it stood at 35.17% (51 of 145) for patients with high CD146 expression. In addition, ROC curve analysis showed that CD146 (AUC = 0.73) was a superior predictor of recurrence compared to histologic grade (AUC = 0.69). Cox proportional hazard analysis further confirmed that CD146, unlike histologic grade, is an independent prognostic factor for OS and LRFS in PT patients. overall, these results demonstrate the pivotal role of CD146 as a novel biomarker in predicting the prognosis of PT.
For decades, wide margin surgical excision has been the primary treatment strategy for patients diagnosed with breast PT [47]. However, the effectiveness of adjuvant therapy for PT remains uncertain. There are limited randomized trials of investigating adjuvant chemotherapy. Only a handful have reported that postoperative chemotherapy has minimal effect on LRFS of malignant PT patients [47, 48]. The therapeutic role of radiotherapy in malignant PT patients is also controversial. Though adjuvant radiotherapy has been reported to reduce the local recurrence rate, it does not appear to confer survival benefit [5, 49]. Notably, no targeted agents against malignant PTs currently exist. Therefore, the identification and development of novel therapeutic targets against PTs hold significant clinical value. Over the years, CD146 has been verified to simulate proliferation, angiogenesis, and metastasis of various malignant tumors, including melanoma, prostate cancer, and epithelial ovarian cancer [7]. Prior research from our team showed that CD146 overexpression in breast cancer cells suppressed the epithelial phenotype, induced a mesenchymal phenotype, and increased migratory and invasive behavior along with cancer stem cell (CSC)-like properties [22]. Nevertheless, the role of CD146 in stroma-derived tumors is scarcely reported. The current research data indicates that CD146 dramatically exacerbates the proliferation, migration, invasion, and collagen contraction of PT cells. Notably, the therapeutic efficacy of the anti-CD146 monoclonal antibody AA98 against malignant PT was validated in a malignant PT organoid and PDX model. AA98 specifically binds to the CD146 D4-D5 domain, inhibiting dimerization and subsequently inhibiting its downstream signaling [23]. AA98 was found to significantly suppress PT growth In vitro and in vivo. In PDX tissues, a notable decrease in the density of vascular endothelium was observed in the group treated with anti-CD146. This observation is consistent with previous research findings. Prior studies have identified CD146 as a marker of vascular endothelial cells. It has also been found that AA98 can inhibit angiogenesis [19]. Interestingly, in the AA98 application group, the size of 2 PDX tumors (from Donor 1 with lower level of CD146 compared to Donor 2 and 3) was larger than the size of 4 other PDX tumors (from Donor 2 and 3). It implies that the effect of AA98 may rely on the level of CD146 in PT tissues. Based on these observations, it is hypothesized that the anti-tumor effect of AA98 in vivo might be a combined result of anti-tumor cell growth and anti-angiogenesis. This finding could potentially be of significance, as it suggests that CD146 might serve as a novel and promising therapeutic target, potentially having important implications in malignant PTs.
The role of CD146 in accelerating tumor cell proliferation and survival via the PI3K-AKT pathway in melanoma is well established [25]. Interestingly, a case report identified an NRAS mutation alongside PI3K-AKT-mTOR activation in PTs through whole genome sequencing [50]. Further, exome sequencing of 22 PTs and targeted sequencing of 100 breast fibroepithelial tumors revealed canonical activating mutations of PIK3CA exclusively in higher grade PTs. It provides that the PI3K-AKT pathway might contribute to malignant transformation in PTs [51]. Following intervention on CD146 expression, RNA sequencing and pathway enrichment analysis indicated the PI3K-AKT pathway as a predominantly activated pathways in malignant PT. The tumor-promoting influence of CD146 could be counteracted with PI3K or AKT inhibitors. This is consistent with earlier findings that propose the CCL18/NF-kB/miR-21 axis between tumor-associated macrophages and PT cells as a potential inducer of PTs tumorigenesis via activation of the PI3K-AKT pathway [52]. Despite these insights, the process by which the membrane protein CD146 activates the PI3K-AKT pathway remains elusive. Leveraging TurboID integration with a traditional epitope tagging (AP-MS) pipeline, 5 potential interactors (CD44, DCBLD2, CD166, EGFR, and STK24) were identified that may be implicated in CD146-drivern regulation of PTs. While these proteins have not the focus of this paper, they close association with CD146 underscores their potential significance for further exploration.
Both CD166 and CD44, cell surface adhesion receptors frequently co-expressed with CD146 in various cancers or mesenchymal stem/progenitor cells and regulate metastasis [53-56]. In hepatocellular carcinoma, CD166 positive modulates CD146 through the activation of the PI3K-AKT and c-Raf/MEK/ERK signaling pathways [36]. This regulatory mechanism appears to be mirrored in phyllode tumors; however, the non-impact of CD166 knockdown on downstream AKT signaling invites further investigation. Additionally, CD44 activation triggers a significant MMP-associated cleavage of membrane CD146, releasing a soluble isoform [57]. The exact mechanism maintaining the balance between these two forms of CD146 and its significance in tumor progression remain to be uncovered. EGFR-targeted therapies, while crucial for treating metastatic tumors harboring activating mutations or amplification, often encounter resistance [58, 59]. Zhang et al. [60] demonstrated that CD146 mediated the acquisition of a stemness phenotype and enhanced tumor invasion and metastasis following EGFR-TKI resistance in lung cancer cells. This initial association between CD146 and EGFR suggests that further probing into CD146's role in EGFR signaling could potentially enhance the design of new combination therapies and improve patient survival.
In this study, DCBLD2 was discerned to directly interact with CD146. Previously, DCBLD2, a neuropil-like protein, was found to be upregulated in malignant tumors and linked to adverse patient prognosis [37]. Participating in the PI3K-AKT signaling pathway [32], DCBLD2 possesses a recently identified CUB domain with a binding site for CD146 [61]. SEMA4B, a member of the semaphorin protein family, works as a tumorigenesis suppressor [62]. As an inhibitory ligand of DCBLD2, SEMA4B can instigate ubiquitin degradation of DCBLD2 on the membrane, thereby inhibiting tumor migration [62]. Notwithstanding, no reports exist detailing how tumor cells with high DCBLD2 expression evade these inhibitory signals. The results show that CD146 can safeguard DCBLD2 from ubiquitin degradation by obstructing the binding of DCBLD2 and SEMA4B, thereby cooperatively activating the downstream PI3K-AKT signaling pathway. Given the previously reported interaction between CD146 and CUB domain proteins, it can be conjectured that CD146 may function as a “shield”, occupying the CUB site of DCBLD2 and SEMA4B binding, thereby ensuring the stable presence of tumor expressed DCBLD2 on the membrane and fostering cancer progression. In a pan-cancer analysis, a strong co-expression correlation between CD146 and DCBLD2 was observed. It suggests that this mechanism's potential applicability to other tumor types. Additionally, both CD146 and DCBLD2 can interact with RTK receptors such as VEGFR2 [20, 63] and PDGFRβ [64, 65]. This leads to speculation that the activation of PI3K-AKT signaling may still be associated with RTK receptors in tumor cells, a hypothesis that warrants further investigation. However, this study has some limitations: (1) the mechanisms of CD146 upregulation in PTs remain undetected, which might be regulated by transforming growth factor beta (TGF-β) and reactive oxygen species signaling pathways [66, 67]; (2) assessing the therapeutic impact of combining AA98 with chemotherapy is of paramount importance, considering the modest effect chemotherapy has shown on PT patients [68]. Indeed, the data indicates that CD146, by acting as a “shield” to prevent the DCBLD2 protein and degradation, promotes the proliferation and invasion of PT cells and further stimulates the PI3K-AKT signaling pathway.
5 CONCLUSIONS
In summary, this study revealed a critical role of the CD146-DCBLD2/PI3K-AKT axis in the malignant progression of PTs. Enhanced surveillance of CD146 levels and targeted therapeutic strategies against CD146 could markedly augment precision diagnosis and treatment of breast PTs.
DECLARATIONS
AUTHOR CONTRIBUTORS
Yan Nie, Erwei Song, and Xiyun Yan contributed to study conception and design. Yan Nie, Jiewen Chen, Qingji Xu, Xuehui Chen, Xun Li, Mingyan Guo, Rong Lei, and Hongyan Huang contributed to developing methodology. Yan Nie, Dan Liu, and Wende Li contributed to data acquisition (provided animals, acquired and managed patients, provided facilities, etc.). Yan Nie, Jiewen Chen, Qingji Xu, Mingyan Guo, Jianyou Liao, Xun Li contributed to data analysis and interpretation (e.g., statistical analysis, biostatistics, and computational analysis). Phei Er Saw, Yan Nie, Jiewen Chen, Qingji Xu, and Erwei Song contributed to writing, review, and/or revision of the manuscript. Yan Nie, Jiewen Chen, Qingji Xu, Xuehui Chen, and Xun Li contributed to administrative, technical, or material support (e.g., reporting or organizing data and constructing databases). Yan Nie, Erwei Song, and Xiyun Yan contributed to study supervision. All authors read and approved the final manuscript.
ACKNOWLEDGEMENTS
We thank all members of breast cancer center of Sun Yat-sen Memorial Hospital, Sun Yat-sen University for helpful suggestions. We also thank Core Facility for Protein Research, Institute of Biophysics, Chinese Academy of Science for technical support. We appreciate the assistance from the Disease Registry Department of Sun Yat-sen Memorial Hospital, Sun Yat-sen University.
CONFLICT OF INTERESTS STATEMENT
The authors declare that they have no competing interests.
ETHICAL APPROVAL AND CONSENT TO PARTICIPATE
The study protocol was approved and monitored by the Sun Yat-sen Memorial Hospital Institutional Review Board (Approval No. 2021-66). Written informed consent was obtained from all patients prior to trial participation. All operations on animals are in accordance with the requirements of the Sun Yat-sen University Animal Care and Use Committee (Approval No. SYSU-IACUC-2020-B0721).
CONSENT FOR PUBLICATION
Not applicable.
Open Research
DATA AVAILABILITY STATEMENT
The data of transcriptome sequencing was deposited in the NCBI GEO database (accession number: GSE227380; https://www.ncbi.nlm.nih.gov/geo/). The remaining data including scRNA-seq that support the findings of this study are available from the corresponding author upon reasonable request.




